Abstract
Orchestration of the inflammatory response is crucial for clearing pathogens. Although the production of multiple inflammatory cytokines has been thought to be regulated by common mechanisms, recent evidence indicates that the expression of some cytokines is differentially regulated by epigenetic regulatory mechanisms. In this study, we found that IL-6 production is selectively inhibited by the BET bromodomain protein (BRD) inhibitor I-BET151 in RAW264.7 cells stimulated with lipopolysaccharide (LPS), whereas I-BET151 did not alter the production of several other cytokines (TNFα, IL-1β and IL-10) at the concentration of IBET151 used. I-BET151 prevented the binding of CBP to the promoter of IL-6, but I-BET151 did not affect acetylation, phosphorylation, nuclear translocation, or DNA binding of p65-NF-κB. In vivo, I-BET151 treatment in the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis decreased the early clinical symptoms, which are thought to be dependent on cytokine production. Altogether, these data suggest that targeting epigenetic-related proteins, such as BET proteins, may provide a strategy to reduce inflammation and the severity of inflammatory diseases, such as multiple sclerosis.
Keywords: Cytokines, macrophage, I-BET151, NF-κB, lipopolysaccharide
Introduction
The inflammatory response to pathogens involves the orchestrated expression of multiple genes encoding inflammatory and anti-inflammatory proteins, which is critical to eliminate pathogens. Regulation of the timing and amplitude of increased expression of inflammatory genes is vital for an efficient immune defense. Therefore, precise regulation of the nuclear chromatin complexes present on the promoters of each inflammatory gene is critical. Acetylation of histones is considered to be among the most important regulatory triggers for controlling the open and closed state of the chromatin. Recently, a family of acetylation mark “reader” proteins that includes BET bromodomain proteins (BET proteins; e.g., BRD2, BRD3 and BRD4), have been shown to govern the assembly of histone acetylation-dependent chromatin complexes that regulate inflammatory gene expression (Hargreaves et al., 2009; Jang et al., 2005; LeRoy et al., 2008; Yang et al., 2005). Some of the actions of BET proteins have been identified through the use of newly designed small molecules that act as histone mimics, which displace BET proteins from acetylated histones and alter the transcriptional landscape (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Nicodeme et al., 2010; Zuber et al., 2011). These small molecule inhibitors are specific for BRD2, BRD3, BRD4, and BRDT, and generally do not inhibit other epigenetic enzymes (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Nicodeme et al., 2010; Ott et al., 2012; Zuber et al., 2011). Among the most potent and selective BET protein inhibitors is I-BET151, which preferentially inhibits BRD4 (KD=50 nM) (Mirguet et al., 2012; Seal et al., 2012). I-BET151 has been well characterized particularly in models of dysregulated transcription, such as in cancer, and been suggested to control the inflammatory response (Nicodeme et al. 2010).
Pro-inflammatory cytokines, such as interleukin-6 (IL-6) and tumor necrosis factor-α (TNFα), are induced in response to many pathogens, as well as experimentally by lipopolysaccharide (LPS). The induction of these proinflammatory cytokines is required for the initial innate immune response, but the prolonged expression of these cytokines can have a deleterious effect on the host. Therefore, understanding the mechanisms regulating cytokine transcription may lead to novel treatments for chronic inflammation and inflammatory diseases. Most pro-inflammatory cytokines induced by LPS require the activation of NF-κB. NF-κB is activated by phosphorylation, which promotes its translocation to the nucleus, and acetylation of NF-κB has been shown to enhance its transcriptional activity (Smale, 2011). Because BRD4 associates with acetylated NF-κB, we tested the hypothesis that IBET151, which binds in the BRD4-acetyl-lysine-binding pocket, could selectively modulate the expression of some NF-κB dependent pro-inflammatory genes, and that this confers a certain selectivity to the action of NF-κB.
Materials and methods
Reagents
Protein-free E. coli (K235) LPS was prepared as described (Hirschfeld et al., 2000). I-BET-151 (GSK1210151A) was synthesized to >99.5% purity as described (Dawson et al., 2011).
Cell culture
RAW264.7 cells were cultured in DMEM medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin. In order to test if BRD4 inhibition attenuates IL-6 production, the cells were treated with LPS (100 ng/mL), I-BET151 (1–60 µM), or both, in cell culture medium without supplements for 30 min to 24 h before cells were harvested for analyses.
Immunoprecipitation and immunoblotting
For immunoblotting, cells were lysed with a triton lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol) supplemented with protease and phosphatase inhibitors (1 µg/mL leupeptin, aprotinin, pepstatin A, 1 mM orthovanadate, 50 mM NaF, 0.1 µM okadaic acid, 1 mM PMSF). Nuclear and cytosolic fractions were prepared using a nuclear extraction kit (Active Motif). Protein concentrations were evaluated by the Bradford assay. Proteins were resolved with SDS-PAGE and transferred to nitrocellulose membranes, which were immunoblotted with antibodies to phospho-Ser536-p65, acetyl-Lys310-p65, CREB (Cell Signaling Technology), p65 (Rockland), BDR4 (Santa Cruz), or β-actin (Sigma). For immunoprecipitation, cell extracts were incubated with the indicated primary antibody overnight at 4°C with gentle agitation, followed by incubation with protein A or G sepharose beads (Amersham) for 2 h at 4°C. The precipitated immune complexes were washed three times and Laemmli buffer was added to the samples before resolving the proteins on SDS-PAGE gels as described above.
p65 DNA binding
To test if I-BET151 treatment affected the DNA binding activity of p65 NF-κB, nuclei from RAW264.7 cells treated with LPS (100 ng/mL), I-BET151 (1 µM), or both, were isolated with a nuclear extraction kit according to the manufacturer’s instructions (Active Motif). Nuclear proteins (100 µg) from RAW264.7 cells were incubated with a p65 consensus oligonucleotide bound to a 96 well-plate to determine the DNA binding activity of p65 NF-κB (Active Motif) according to the manufacturer’s instructions.
Measurement of cytokines
Mouse IL-6, IL-1β, IL-10 and TNFα in RAW264.7 cell culture supernatants diluted 10-fold were measured with an enzyme-linked immunosorbent assay (ELISA) kit (eBioscience) according to the manufacturer’s instructions.
Viability
Cell viability was assessed by a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide (MTT) assay. A multi-well scanner was used to measure the absorbance at 570–630 nm dual wavelengths.
Chromatin immunoprecipitation (ChIP)
To identify proteins bound to the IL-6 promoter, nuclei from cross-linked RAW264.7 cells were resuspended in Tris-EDTA buffer and sonicated. The soluble chromatin was adjusted into immunoprecipitation assay (RIPA) buffer (0.1% sodium dodecyl sulfate, 1% Triton X-100, 0.1% sodium deoxycholate, 140 mM NaCl) and precleared. Immunoprecipitation was performed with 1 µL of anti-p65 (Rockland), 2 µL of antiacetylated p65 (Cell Signaling), 10 µL of anti-BRD4 (Santa Cruz), 10 µL of anti-CBP (Santa Cruz), or 2 µL of anti-IgG (Cell Signaling) and the immune complexes were absorbed with protein A beads (Millipore) blocked with bovine serum albumin and salmon sperm DNA. Immunoprecipitated DNA was subjected to semiquantitative PCR. The PCR products were resolved in 2.0% agarose gels in 1× Tris/borate/EDTA buffer, and the gels were stained with ethidium bromide. Q-PCR was used to quantify the ChIP results, and all results were normalized by the respective input values. The IgG control was run in parallel to assure no unspecific binding to the promoter of IL-6. The mouse IL-6 promoter was analyzed using primers: IL-6 forward 5'-AAG CAC ACT TTC C CC TTC C-3′, and reverse, 5′-CTA TCG TTC TTG GTG GGC TC-3′.
EAE induction
Mice were injected intraperitoneally (ip) with 3 mg/kg I-BET151 starting 3 days prior to immunization and then daily throughout the experiment. EAE was induced in male C57Bl/6 mice with subcutaneous (s.c.) injection of 50 µg MOG35–55 peptide emulsified in complete Freund’s adjuvant containing 1.5 mg/mL heat-killed Mycobacterium tuberculosis H37Ra (Difco Laboratories). On the day of immunization and 48 h later, mice were injected ip with 50 ng of pertussis toxin in PBS. Mice were examined daily for clinical signs of EAE. Mice were assigned clinical symptom scores as follows: 0, no paralysis; 1, loss of tail tone; 2, flaccid tail; 3, incomplete paralysis of one or two hind legs; 4, complete hind limb paralysis; 5, moribund (animals were humanly euthanized); 6, death. To compare the time course of disease development in different groups of mice, the daily average of the clinical scores was calculated for each group.
Statistical analysis
Data are expressed as means ± SEM. Statistical significance between groups was evaluated by Mann Whitney or ANOVA tests. Differences between groups were considered significant at p < 0.05.
Results
I-BET151 preferentially reduces IL-6 production induced by LPS in RAW267.4 cells
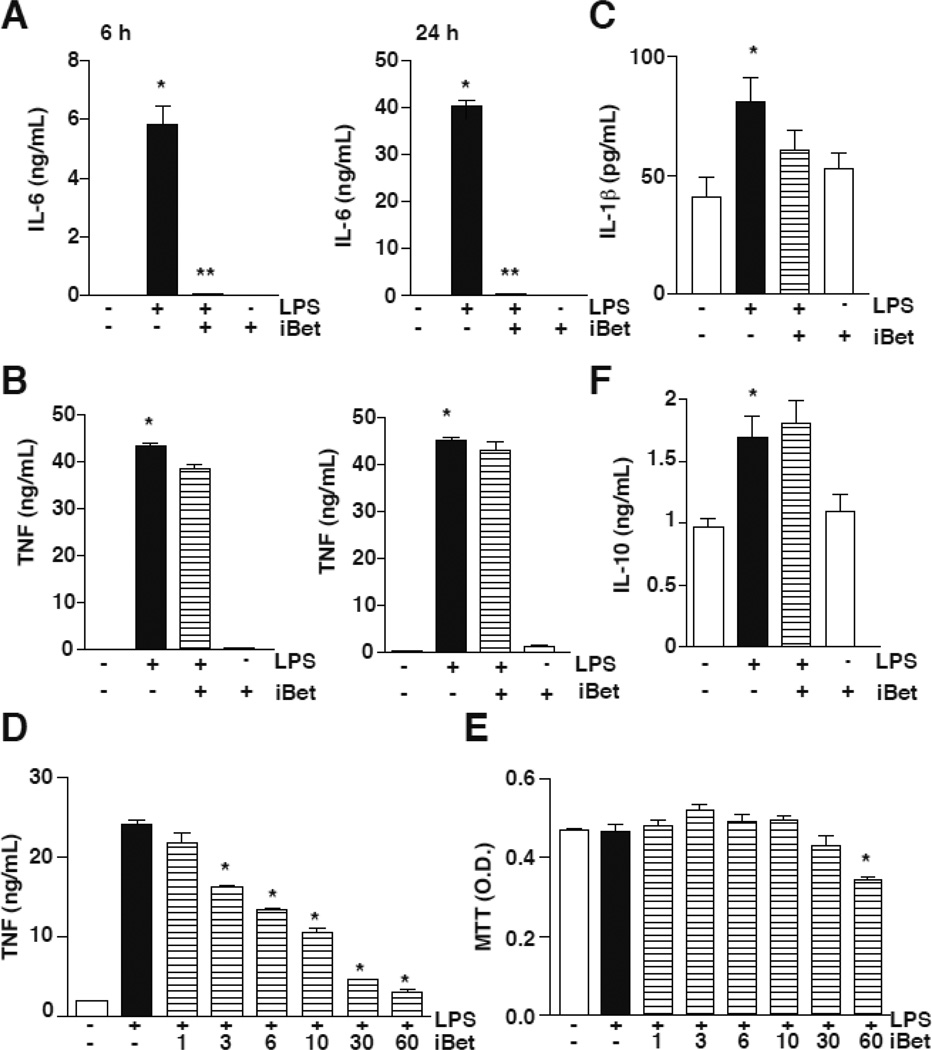
I-BET was reported to downregulate specifically the expression of 151 inflammatory genes while increasing the expression of 5 inflammatory genes in response to LPS in bone marrow-derived macrophages (Nicodeme et al 2010), showing that reader proteins can confer a certain selectivity in regulating gene expression. Consistent with this, examination of pro-inflammatory cytokines in macrophage-like RAW264.7 cells showed that 1 µM I-BET151 preferentially inhibited the production of IL-6 in response to LPS at both 6 and 24 h (Fig. 1A), but had no effect on the production of TNFα at the same times (Fig. 1B), or IL-1β at 24 h (Fig. 1C). In contrast, increased concentrations of I-BET151 decreased TNFα production (Fig 1D) without affecting cell viability except for 60 µM I-BET151 (Fig 1E). Reduced IL-6 production was not the consequence of upregulated production of the anti-inflammatory cytokine IL-10, as the level of IL-10 was unchanged by treatment with I-BET151 (Fig. 1F). Although many pro-inflammatory genes have been shown to be dependent on the activation of the transcription factor NF-κB, I-BET151 appears to have a preferential regulatory effect on the production of the pro-inflammatory IL-6 in response to LPS.
Figure 1. I-BET151 prevents IL-6 production in RAW264.7 cells.
RAW264.7 cells were stimulated with 100 ng/mL LPS in serum-free medium, with or without 1 µM I-BET151, for 6 or 24 h, as indicated. Supernatants were used to measure the production of IL-6 (A), TNF (B), IL-1β (C) and IL-10 (F) by ELISA (n=6; *p < 0.05 LPS treatment compared to untreated samples, **p<0.05 LPS + I-BET compared with samples treated with LPS, Mann Whitney test). RAW264.7 cells were stimulated with 100 ng/mL LPS in serum-free medium, with or without 1, 3, 6, 10, 30, or 60 µM I-BET151, for 6 h. Supernatants were used to measure the production of TNF (D) by ELISA, and cell viability was measured by the MTT assay (F) (n=3; *p < 0.05 LPS treatment compared to I-BET151-treated samples, one-way ANOVA, Bonferroni post-hoc test).
I-BET151 did not affect the activation of NF-κB by LPS
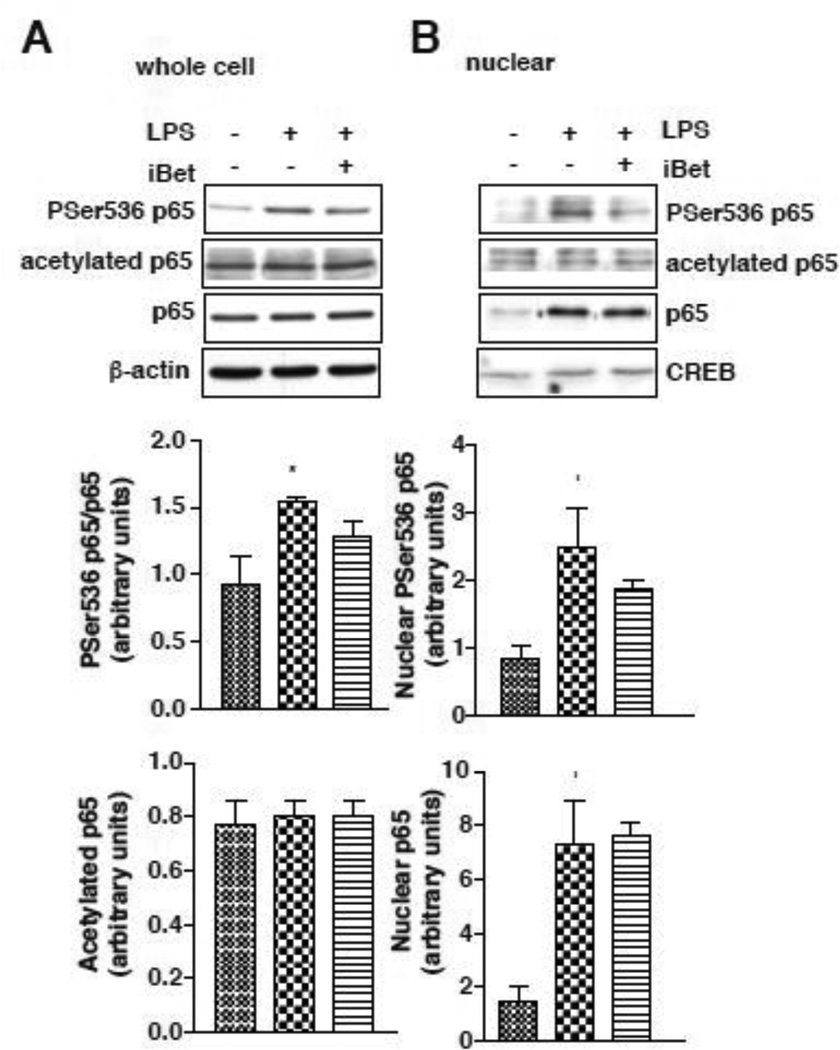
In response to LPS treatment, p65-NF-κB is phosphorylated and translocates to the nucleus to induce the expression of many pro-inflammatory genes. The level of phospho-Ser536-p65 in RAW264.7 cells was increased by LPS treatment, and remained unchanged in the presence of I-BET151 (Fig. 2A). In addition, I-BET151 did not alter either the nuclear translocation of p65, or the nuclear phosphorylation of p65 (Fig. 2B). Acetylation of p65 on Lysine 310, which is critical for the activation of NF-κB in the inflammatory response (Gringhuis et al., 2007; Ishinaga et al., 2007; Ito et al., 2007), was also unaltered in the presence of I-BET151, both in whole cell extracts and in the nuclear fraction (Fig. 2AB). Altogether, these results demonstrate that I-BET151 did not impair the activation and the translocation of p65 to the nucleus, suggesting that the regulation of IL-6 production by I-BET151 occurs at the transcriptional level.
Figure 2. I-BET151 does not affect phosphorylation or acetylation of p65.
RAW264.7 cells were stimulated with 100 ng/mL LPS in serum free medium, with or without 1 µM I-BET151, for 30 min and cell lysates (A) and nuclear extracts (B) were immunoblotted for phospho-Ser536-p65, acetyl-Lys310-p65, and total p65. β-actin (cell lysates) or CREB (nuclear extracts) were used as loading controls. Blots were quantified and graphs represent means ± SEM (n=3, *p<0.05 LPS treatment compared to untreated samples, Mann Whitney test).
I-BET151 reduces the association between BRD4 and acetylated p65
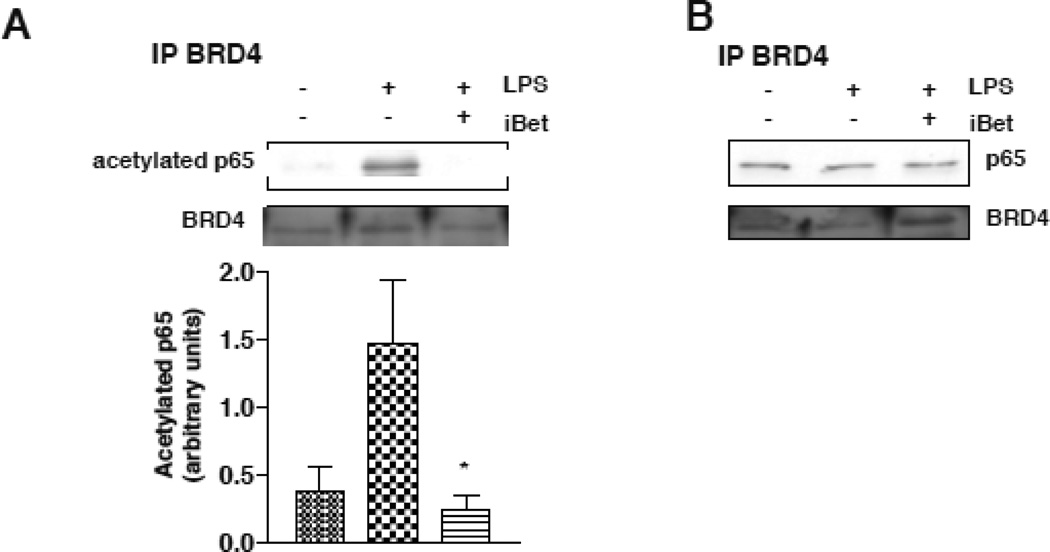
In order to decipher the specific role of I-BET151 in the NF-κB-dependent regulation of IL-6 expression, we also examined if I-BET151 modulates the binding of BRD4 with NF-κB. Acetylation provides specific docking sites for bromodomain proteins, and acetylation of lysines in particular are thought to regulate the functional interactions of proteins with a bromodomain protein (Seet et al., 2006; Yang, 2004). It has been shown that BRD4 associates with acetylated NF-κB, and that BRD4 is recruited to the promoters of a subset of NF-κB target genes (Huang et al., 2009). In RAW264.7 cells, treatment with I-BET151 did not alter the association of BRD4 with p65 after stimulation with LPS (Fig. 3B). However, there was an increased association of BRD4 with acetyl-Lys310-p65 after treatment with LPS, and this was abolished by I-BET151 treatment (Fig. 3A). These results indicate that inhibition of BRD4 by I-BET151 prevents BRD4 association with the transcriptionally active acetylated form of p65.
Figure 3. I-BET151 reduces the association between BRD4 and acetylated p65.
(A) to be consistent with figs 1 and 2 RAW264.7 cells were treated with 100 ng/mL LPS, 1 µM I-BET151, or both, for 30 min. Cell lysates were prepared for immunoprecipitation (IP) of BRD4, and immunoprecipitants were immunoblotted for acetyl-Lys310-p65 and BRD4. Quantified levels of acetyl-Lys310-p65 represent the means ± SEM.; n=3 *p<0.05 compared to LPS treatment (Mann Whitney test). (B) RAW264.7 cells were treated with 100 ng/mL LPS, 1 µM I-BET151, or both, for 30 min. Cell lysates were prepared for immunoprecipitation of BRD4, and immunoprecipitants were immunoblotted for p65 and BRD4.
I-BET151 did not alter the binding of p65 to a DNA consensus sequence but restored acetylated p65 binding to the promoter of IL-6 in response to LPS
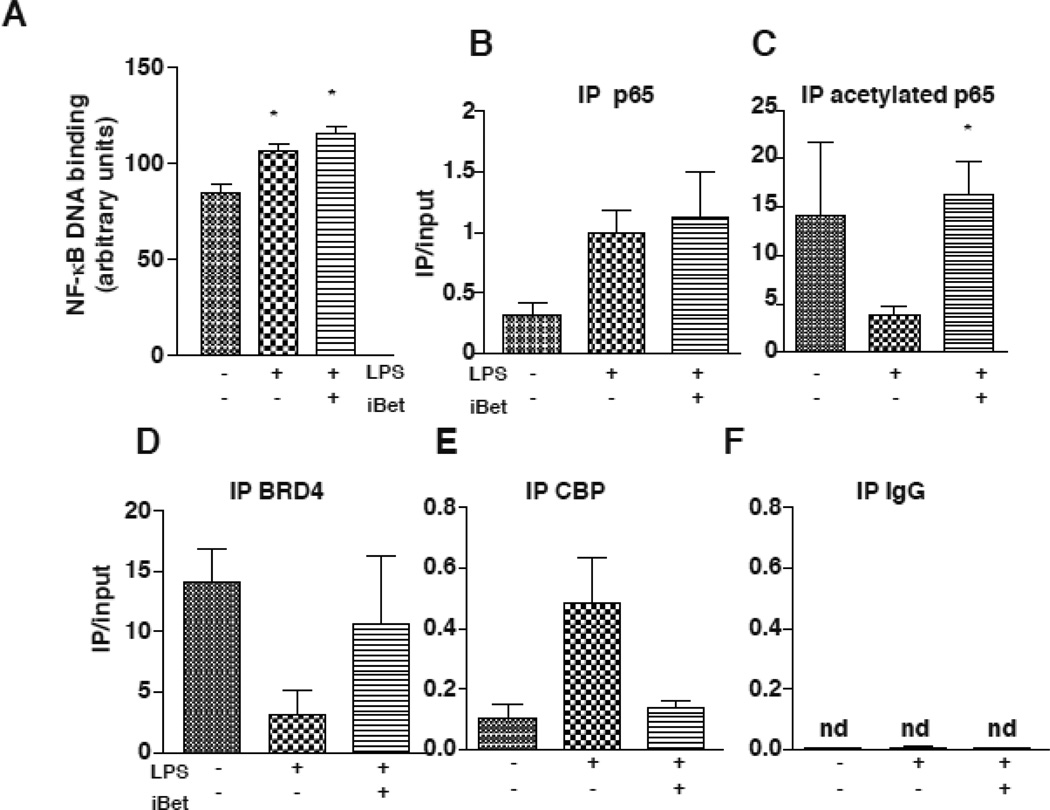
We measured the DNA binding activity of p65 in response to treatment with IBET151 and LPS to determine if impairment of this process may underlie the mechanism of action of I-BET151 in the selective regulation of IL-6 production. The LPS-induced increase in the binding of p65 to an oligonucleotide consensus sequence and the binding of p65 to the promoter of IL-6 were unaltered by I-BET151 treatment after stimulation with LPS (Fig. 4A). LPS reduced acetylated p65 binding to the promoter of IL-6, and this was blocked by I-BET151 treatment (Fig. 4C). LPS also reduced BRD4 binding to the IL-6 promoter, and this was reversed by I-BET151 treatment (Fig. 4D). These results show that I-BET151 lead to the restoration of acetylation of p65 on the promoter of IL-6 and therefore the binding of BRD4 in the IL-6 promoter (Fig. 4D). LPS also promoted CBP recruitment to the IL-6 promoter, and I-BET151 blocked this recruitment (Fig. 4E). Immunoprecipitation with a nonspecific IgG did not pull down any DNA covering the IL-6 promoter region, as we were unable to amplify any DNA by Q-PCR (Fig 4F). Altogether, these results show that I-BET151 blocks the LPS-induced decrease of acetylated p65 in the promoter of IL-6 by promoting BRD4 binding rather than CBP binding to the IL-6 promoter.
Figure 4. I-BET151 selectively reduces the binding of CBP to the promoter of IL-6 in response to LPS.
(A) p65 DNA binding activity was measured in RAW264.7 cells after stimulation with 100 ng/mL LPS, 1 µM I-BET151, or both, for 30 min. Nuclear extracts were incubated with a p65 consensus sequence using TRANSAM technology (n=3, *p<0.05 compared to untreated samples, ANOVA, Bonferroni post test). (B-E) RAW264.7 cells were treated with 100 ng/mL LPS, 1 µM I-BET151, or both, for 30 min. Cell lysates were prepared for chromatin immunoprecipitation. Samples were immunoprecipitated (IP) for p65 (B), acetylated-p65 (C), BRD4 (D), CBP (E) and a nonspecific IgG control (F), and both immunoprecipates and input were amplified by quantitative PCR with primers for the promoter of IL-6. Data represent the mean of the ratio of immunoprecipitate/input ± SEM.; n=3 *p<0.05 compared to LPS treatment (unpaired T test) (nd, non-detectable).
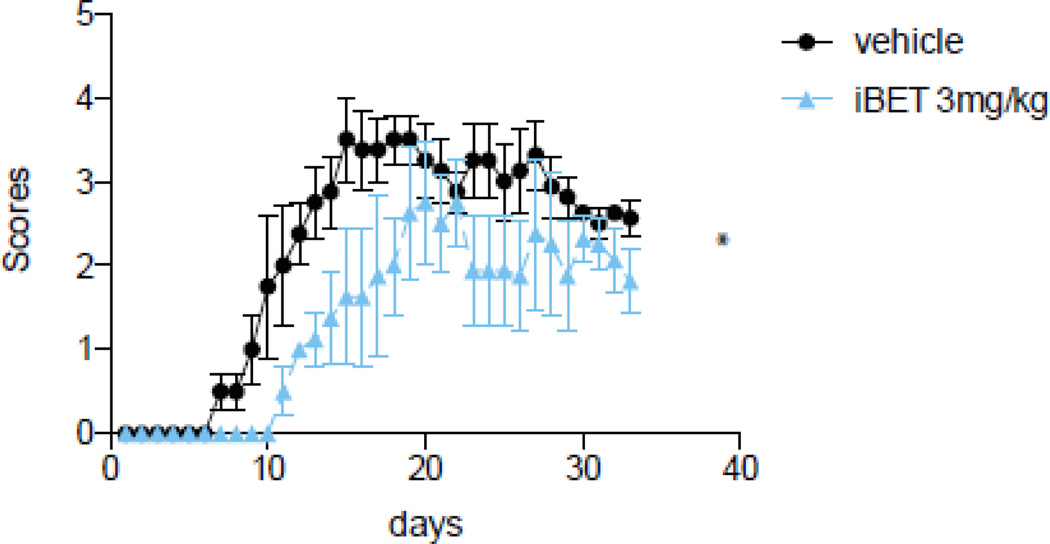
I-BET151 delays onset of EAE in mice
Because I-BET151 appears to be a critical regulator of the proinflammatory cytokine IL-6, we also tested if I-BET151 administration has a beneficial effect in a chronic model of inflammation involving IL-6, experimental autoimmune encephalomyelitis (EAE), which is widely used as a model of multiple sclerosis. Mice were pretreated before immunization with I-BET151 for 3 days, and daily after immunization throughout the development of EAE. Mice treated with I-BET151 had a significant delay in the onset of clinical symptoms and attenuated the maximal severity of EAE (Fig. 5). This result demonstrates that targeting chronic inflammation by I-BET151 is beneficial in the development of EAE.
Figure 5. I-BET151 reduces onset and severity of EAE.
Mice were pretreated with 3 mg/kg I-BET151 for 3 days prior to immunization with MOG35–55 peptide and daily throughout the experiment. Clinical symptoms were scored daily and values shown are Means ± SEM. (n=4–5) *p<0.05, t-test. Data represent 1 out of 2 independent experiments
Discussion
Cytokines are critical mediators of immune responses. The duration and amplitude of their production is tightly regulated, because either excess or insufficient cytokine production can have detrimental consequences in the clearance of pathogens or the health of the host. The expression of cytokines is likely regulated predominantly at the level of transcription factors. In particular, NF-κB is considered the master regulator of the production of multiple inflammatory cytokines, especially in response to LPS stimulation (Smale, 2011). Recently, another level of regulation has been uncovered that involves epigenetic tuning of cytokine production.
Here we report that a bromodomain protein inhibitor (I-BET151) selectively blocks the production of IL-6, without affecting the production of two other pro-inflammatory cytokines (TNFα, IL-1β) or the anti-inflammatory cytokine IL-10, suggesting that bromodomain proteins inhibited by I-BET151 are selectively involved in the control of the production of IL-6. This is partially in agreement with results recently reported by Belkina et al, 2013, who also found a reduction of IL-6 production after treatment with the BRD inhibitor JQ1. However, they also found that JQ1 treatment caused a reduction of TNF production, which we did not find after treatment with 1 µM I-BET151. This discrepancy may be due to the concentration of inhibitors used, and we confirmed that higher concentrations of I-BET151 reduce the production of TNF (Fig 1D). Similarly, the production of IL-1β may be reduced with higher concentrations of I-BET151, consistent with the findings of Nicodeme et al., 2010 where IL-1β was reduced by I-BET151. It is likely that other cytokines in addition to IL-6 may also be regulated by high concentrations of I-BET151. Although BRD proteins are thought to regulate histone acetylation-dependent complexes, it appears that they carry a certain specificity in their mechanism of action since they are able to modulate specific promoters (Prinjha et al., 2012). However, the mode of selectivity of these BRD proteins remains to be determined.
Here we report that Treatment with I-BET151 decreased the binding of CBP, which has been shown to be required for the action of NF-κB in the production of IL-6, as well as other cytokines, suggesting the possibility that the binding of BRD4 to the promoter of IL-6 prevents the binding of CBP. We found that I-BET151 decreases the binding of BRD4 to acetylated NF-κB, suggesting that BRD4 is required for the proper recruitment of transcriptional machinery to acetylated NF-κB. This is consistent with the findings of (Huang et al., 2009) showing that in response to TNF stimulation, BRD4 is recruited to the promoter by acetylated NF-κB. This is also consistent with the finding by Belkina et al., 2013 that BRD4 is tethered to the transcription complex through NF-κB. Other proteins may also be involved in this transcriptional complex, as we hypothesize that BRD4 binding to acetylated NF-κB in the promoter of IL-6 may inhibit recruitment of the transcriptional machinery to promote expression of IL-6 although additional data is required to test this hypothesis. However, it remains to be determined which acetyltransferase is responsible for the acetylation of NF-κB on the promoter of IL-6 in the presence of IBET151. It will also be important to determine which deacetylase reduces acetylation of NF-κB in the promoter of IL-6, because this deacetylase may be also regulated by BRD4 and may compete with BRD4 for binding to the promoter of IL-6. These actions of I-BET151 may be particularly due to its interaction with BRD4, although it also binds other BRD proteins, because as described in Belkina et al, 2013, BRD4 does not bind to the promoter of TNFα, and we found no change in the production of TNF in the presence of I-BET151, suggesting that I-BET151 may preferentially regulate BRD4 to also inhibit IL-6 production. As suggested by Zang et al 2013, signal transduction may also regulate the actions of the BET proteins through kinase cascades.
Although many questions remain about the actions of BRD4 and I-BET151, this study suggests that targeting BRD4 with I-BET151 is an attractive therapeutic strategy (Prinjha et al., 2012) to dampen the production of IL-6 because BET151 nearly eliminates IL-6 production but does not block the production of all NF-κB-dependent cytokines. But further experiments using siRNA will be required to fully clarify the involvement of BRD4 in the action of I-BET151. This potential therapeutic application was further verified by examining the effects of long-term administration of I-BET151 in the mouse EAE model of multiple sclerosis. Administration of I-BET151 in the EAE model delayed onset of the disease, which is thought to be highly dependent on cytokine production by the innate immune system. Furthermore, I-BET151 was well-tolerated by mice over a period of nearly 40 days of treatment. However, I-BET151 administered at this dose was unable to prevent the disease progression, suggesting that other pathogenic mechanisms in EAE were not affected by I-BET151. These could include the production of other cytokines, such as TNF or IL-1β, that may have been unaffected by I-BET151, or the activation of other immune cells. Nevertheless, these results demonstrate a potential therapeutic advantage of using I-BET151 in inflammatory diseases, in addition to LPS-induced endotoxin shock or cancer (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Nicodeme et al., 2010; Zuber et al., 2011), since EAE is a complex disease with an unknown etiology and I-BET151 caused no apparent side effects during the long-term chronic treatment. BET bromodomain protein inhibitors were also shown to inhibit CD4+ T cell cytokines production (Bandukwala et al., 2012), which may also contribute to the EAE effects we observed.
Highlights.
I-BET selectively inhibits IL-6 production in RAW264.7 cells in response to LPS.
I-BET prevented the binding of CBP to the promoter of IL-6, without affect p65.
In vivo, I-BET treatment in EAE decreased the early clinical symptoms.
Acknowledgement
This research was supported by a grant from the NIMH (MH090236).
Abbreviations
- BRD4
bromodomain protein 4
- IL-1β
interleukin 1β
- IL-6
interleukin-6
- IL-10
interleukin 10
- EAE
experimental autoimmune encephalomyelitis
- ELISA
enzyme-linked immunosorbent assay
- LPS
lipopolysaccharide
- NF-κB
nuclear factor kappa B
- TNFα
tumor necrosis factor-α
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AM, Smithers N, Lee K, Witherington J, Tough DF, Prinjha RK, Peters B, Rao A. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A. 2012. 2012;109:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis SI, den Dunnen J, Litjens M, van Het Hof B, van Kooyk Y, Geijtenbeek TB. C-type lectin DC-SIGN modulates Toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappaB. Immunity. 2007;26:605–616. doi: 10.1016/j.immuni.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Molecular and cellular biology. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishinaga H, Jono H, Lim JH, Kweon SM, Xu H, Ha UH, Xu H, Koga T, Yan C, Feng XH, et al. TGF-beta induces p65 acetylation to enhance bacteria-induced NF-kappaB activation. The EMBO journal. 2007;26:1150–1162. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Charron CE, Adcock IM. Impact of protein acetylation in inflammatory lung diseases. Pharmacology & therapeutics. 2007;116:249–265. doi: 10.1016/j.pharmthera.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Molecular cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirguet O, Lamotte Y, Donche F, Toum J, Gellibert F, Bouillot A, Gosmini R, Nguyen VL, Delannee D, Seal J, et al. From ApoA1 upregulation to BET family bromodomain inhibition: discovery of I-BET151. Bioorganic & medicinal chemistry letters. 2012;22:2963–2967. doi: 10.1016/j.bmcl.2012.01.125. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends in pharmacological sciences. 2012;33:146–153. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Seal J, Lamotte Y, Donche F, Bouillot A, Mirguet O, Gellibert F, Nicodeme E, Krysa G, Kirilovsky J, Beinke S, et al. Identification of a novel series of BET family bromodomain inhibitors: binding mode and profile of I-BET151 (GSK1210151A) Bioorganic & medicinal chemistry letters. 2012;22:2968–2972. doi: 10.1016/j.bmcl.2012.02.041. [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nature reviews Molecular cell biology. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Smale ST. Hierarchies of NF-kappaB target-gene regulation. Nature immunology. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zang K, Wang J, Dong M, Sun R, Wang Y, Huang Y, Liu X, Li Y, Wang F, Yu M. Brd2 inhibits adipogenesis via the ERK1/2 signaling pathway in 3T3-L1 adipocytes. PLoS One. 2013;8:e78536. doi: 10.1371/journal.pone.0078536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]