Abstract
Cardioviruses of the Encephalomyocarditis virus (EMCV) and Theilovirus species, encode small, amino-terminal proteins called Leaders (L). Phosphorylation of the EMCV L (LE) at two distinct sites by CK2 and Syk kinases is important for virus-induced Nup phosphorylation and nucleocytoplasmic trafficking inhibition. Despite similar biological activities, the LE phosphorylation sites are not conserved in the Theiloviruses, Saffold virus (LS, SafV) or Theiler’s murine encephalitis virus (LT, TMEV) sequences even though these proteins also become phosphorylated in cells and cell-free extracts. Site prediction algorithms, combined with panels of site-specific protein mutations now identify analogous, but not homologous phosphorylation sites in the Ser/Thr and Theilo protein domains of LT and LS, respectively. In both cases, recombinant AMP-activated kinase (AMPK) was reactive with the proteins at these sites, and also with LE, modifying the same residue recognized by CK2.
Keywords: Cardiovirus, Leader protein, phosphorylation, AMPK
INTRODUCTION
Encephalomyocarditis virus (e.g. EMCV) and Theiloviruses (e.g. Vilyusik virus, Theiler’s murine encephalitis viruses [TMEV], and Saffold viruses [SafV]) are Cardiovirus species in the Picornaviridae family. All isolates from this genus have small, Leader proteins (L) encoded at the amino terminus of their viral polyproteins. These highly acidic peptides of 67-(EMCV, LE), 71-(SafV, LS) or 76-(TMEV, LT) amino acids (aa), carry out important anti-host functions. For example, the LE and LT proteins have been attributed with pro- or anti-apoptotic functions, depending on the viral strain and cell culture system (1-7). For TMEV (DA), LT expression in a virus context can inhibit stress granule formation in the cytoplasm, thereby reducing the turnover of RNA (8). It is not clear whether all cardioviruses share all these same pathways, or if these particular effects are just observed downstream consequences of the major L function, the common catastrophic inhibition of active cellular nucleo-cytoplasmic trafficking (9-13). The introduction of LE, LT or LS into cells by viral or recombinant means uniformly triggers massive phosphorylation of Phe/Gly-containing nuclear pore proteins (Nups) (11, 14, 15). The modified Nups are refractive to cargo-carrying karyopherins (importins or exportins), halting almost immediately the normal active nuclear-cytoplasmic transport system for proteins and RNAs, to a mere trickle allowed by diffusion-only. This unique cardiovirus phenomenon requires a poorly understood mechanism involving the binding of L to RanGTPase, followed by activation and misdirection of a particular cohort of mitogen-activated protein kinases (MAPKs) (12). For EMCV, the best studied system, MAPKs ERK1/2 and p38, as well as their downstream substrates, MAPKAP-2 (MK2) and RSK, become potently activated in an LE-dependent manner (12). Chemical inhibition of ERK1/2 and p38 pathways abolishes LE-directed Nup phosphorylation and protects the cells (12). Likewise, LE mutations which prevent RanGTPase interactions or unfold the protein are unable to trigger Nup phosphorylation (16).
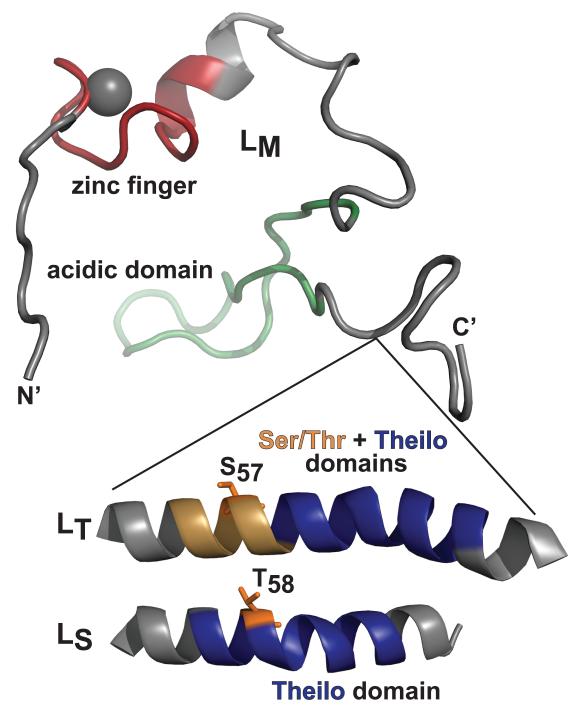
Despite having identity variations that can exceed 65%, all cardiovirus L proteins are homologs, if not presumed analogs. The LM (EMCV, Mengo) structure as solved by NMR (PDB: 2M7Y) shows a flexible coiled-coil configuration distinguished by an unusual, conserved (genus level) CHCC-based zinc finger (aa 10-22) near the amino terminus (Fig 1). A carboxyl-proximal “acidic” domain (aa 37-52) is also shared, conferring a pI of 3.6-3.8 to each protein. A central linker or hinge region (LE aa 35-40) mediates essential interactions with cellular binding partner, RanGTPase, presumably through induced-fit contacts (16). Theilovirus L proteins have shorter amino-termini, but they are longer than LE proteins, because of carboxyl-proximal insertions that add 14- or 19-aa respectively to the LS and LT sequences. The common portion of these insertions, the “Theilo domain” is diagnostic of that species (BeAn aa 60-73). Only the LT display additional, upstream “Ser/Thr-rich” residues (BeAn aa 51-63). The indel domain functions have been partially inferred by genetic mapping with viruses. Point mutations to the LT (DA) Theilo domain reduce viral apoptotic activity, decrease virus persistence in FVB/N mice, reduce Nup98 phosphorylation with consequent decreased inhibition of nucleocytoplasmic trafficking. They also decrease the ability to inhibit IFN-β and RANTES activities, and decrease the ability to inhibit IRF-3 dimerization (14). Such viruses are also unable to inhibit stress granule formation (8). Equivalent mutations to the LT Ser/Thr domain (S51A, S53A, S54A) do not have similar effects (14), although a single amino acid change in the LT of GDVII virus (P57) and DA virus (S57) correlates with differential apoptotic phenotypes (2), and LT intracellular localization differences (17).
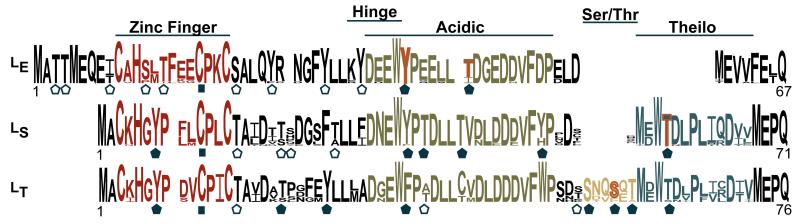
Figure 1.
Cardiovirus Leader sequences. WebLogo depiction summarizes known variation in LE, LS and LT proteins. Ser, Thr, Tyr sites predicted by any online phosphorylation algorithm are highlighted ( ). Sites selected for mutagenesis (
). Sites selected for mutagenesis ( ) are a subset and include also zinc-finger disruption mutations (
) are a subset and include also zinc-finger disruption mutations ( ). Mapped phosphorylation sites from this study (LS, LT) and others (LE) are in orange.
). Mapped phosphorylation sites from this study (LS, LT) and others (LE) are in orange.
The problem with evaluating such results is that even point mutations can have unanticipated long-range effects if they unintentionally disrupt protein structure. Moreover, as has been shown for LE, directed phosphorylation particular to these regions plays a very important role in the observed activity (15). In cells, LE is sequentially phosphorylated by casein kinase 2 (CK2) and spleen tyrosine kinase (Syk) on T47 and Y41, respectively (15). The reactions are not obligatory for RanGTPase interactions, but they are essential for the subsequent trigger of MAPK pathways. LT and LS also become phosphorylated, but at different sites than LE, and in reactions that do not require CK2 (15). Sequence comparisons have suggested S57 (17) and/or T63 (14) as possible sites in LT (DA). We now have experimental validation that LT (BeAn) and LS (SafV-2) and are differentially phosphorylated at S57 (Ser/Thr domain) and T58 (Theilo domain), respectively. Both reactions can be catalyzed by AMP-activated protein kinase (AMPK), an enzyme that can substitute for CK2 in the initial phosphorylation of LE. The identified LS and LT sites have superimposable structural locales in an extended helical motif, modeled for the Ser/Thr and Theilo domain insertions.
MATERIALS AND METHODS
Plasmids and proteins
Bacterial plasmids pLE-GST, pLS-GST and pLT-GST and select point mutations were prepared as described (15) using site-directed mutagenesis via PCR overlap extension methods (18). We thank Dr. Howard Lipton for the generous gift of SafV-2 and TMEV (BeAn) cDNA starting materials. The matched panels linked the LX gene(s) upstream, in-frame with a glutathione S-transferase gene (GST) in a context that allowed direct cDNA transfection, transcription (via a CMV promoter) and expression in eukaryotic cells. LX-GST fusion proteins were expressed in bacteria and purified on Glutathione Sepharose High Performance Column (GE Healthcare Life Sciences) as described (19). Proteins prepared this way can have smaller αGST-reactive bands in addition the full-length fusion, because of alternative bacterial translation start sites in the LX genes (15).
Phosphorylation assays
HeLa or BHK cytosol was prepared via dounce homogenization (20). Phosphorylation reactions (80 μl total, with 30 μl cytosol, 2 μl 10 mM ATP, 2 μg LX-GST, 0.75 μl [γ-32P]-ATP) were in GST binding buffer (50 mM HEPES, 150 mM NaCl, 0.5% NP40, pH 7.4) for 45 min (37°C). Glutathione sepharose 4B beads (10 μl per sample, GE Healthcare Life Sciences) were then added, followed by agitation (4°C, 2h). The beads were collected by centrifugation (500 g), washed with GST buffer (4×) then boiled in SDS gel loading buffer. Protein fractionation was by SDS-PAGE (8 or 10%) with band visualization by phosphorimaging (Typhoon 9200 Variable Mode Imager, GE Healthcare), silver stain, or western analysis.
rAMPK (α1, β1, γ1) was obtained commercially (SignalChem). Specific activity information was from the technical data sheet. Reactions were similar to those described (15). Purified LX-GST in buffer (10 μl, 2 μl 0.5 mM AMP, 1 μl 50 ng/μl AMPK, 2 μg LX-GST, 25 mM MOPS pH 7.2, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT) was reacted with 2 μl AMPK assay cocktail (0.25 mM ATP, 0.167 μCi/μl [γ-32P]-ATP) for 15 min, 30°C. The LX-GST protein was extracted with glutathione beads, washed and then fractionated by SDS-PAGE. Phosphorylation signals were quantified by densitometry using TotalLab Quant (Nonlinear Dynamics Ltd, Newcastle, UK), normalizing to input protein (from Western analysis or silver stain).
Western analyses
Protein bands were electrotransferred to polyvinylidene difluoride membranes and treated with primary and secondary antibodies as described (15). The primary antibodies included: αGST (murine mAb, 71087, Novagen), αCK2 (rabbit pAb, 06-873, Millipore), αtubulin (murine mAb, T4026, Sigma-Aldrich), αP-AMPK 23A3 (rabbit mAb, 2603, Cell Signaling),
RESULTS AND DISCUSSION
Phosphorylation site mapping
A dataset of cardiovirus L proteins (Fig 1) was compiled from GenBank, aligned by ClustalX (21) and is summarized using Weblogo format (22). The LT (BeAn), LS (SafV-2) and LE (EMCV-R) sequences are those from GenBank files, m16020, am922293, and m81861 respectively. Phosphorylation predictions for these proteins used online program suites, including PPSP (set to “high sensitivity” for “all kinases”) (23), NetPhosK1.0 (set to “prediction without filtering” (fast), threshold=0.5) (24), NetPhos2.0 (25), Phosida (26), DiPHOS (specified predict for viruses) (27), Phosphomotif Finder (28) and ScanSite (set to “low stringency”) (29).
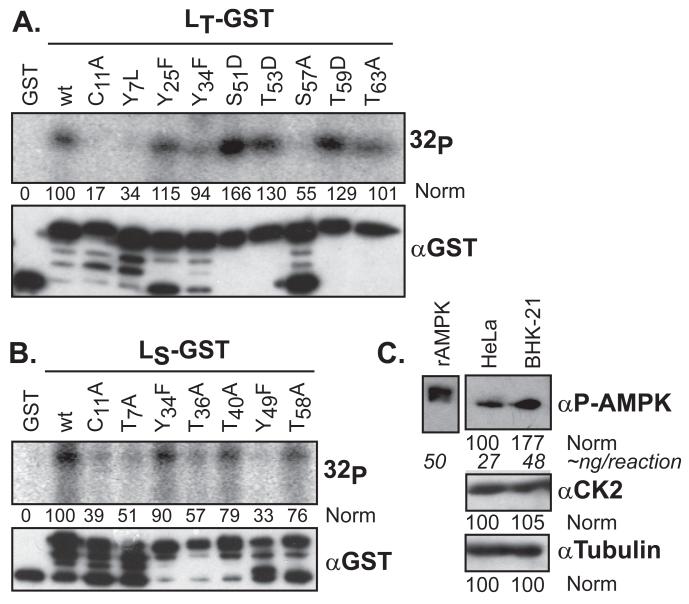
For LE (recombinant or viral), CK2 phosphorylation at T47 is the primary, obligatory event in cells or in vitro, followed by a Syk reaction at Y41 (15). The LT-GST and LS-GST proteins do not react with CK2 and the required site (T47XXD/E) is not conserved in this species. Credible alternative predicted phosphorylation sites, common to each Theilo sequence group included 6 Tyr/Thr residues in LS, and 8 Tyr/Thr/Ser residues in LT (filled symbols in Fig 1). These, and structurally debilitating (C11) mutations in the respective zinc finger domains, were individually constructed into LT- and LS-GST plasmids. Each protein was expressed and purified. When added to HeLa cytosol in the presence of γ-32P-ATP, all of the LT-GST sequences were labeled, except C11A, Y7L and S57A (Fig 2A). Some proteins, especially those with phosphomimetic glutamate mutations (S51D, T53D, T59D), actually registered more strongly than wild-type (wt) when the signals were normalized (densitometry) to the unmodified parental fusion protein band (αGST, top band). When this type of diversity was observed with the LE-GST proteins (15) it was indicative of interdependent phosphorylation events, one of which (at T47) was an obligatory precursor for the other (at Y41).
Figure 2.
LX phosphorylation in cytosol. LT-GST (A) or LS-GST (B) proteins were incubated with HeLa cytosol and [γ-32P]-ATP. After GST extraction and SDS-PAGE, the proteins were detected by autoradiography (32P), or Western analysis (αGST). After densitometry, the signals were normalized to the GST control (“0”) and wt (“100”) samples. The uppermost αGST band is the fusion protein. (C) Equivalent cytosol samples (HeLa, BHK) to A and B, were probed by Western analysis for AMPK, CK2 and tubulin signals, relative to a standardized sample of rAMPK.
The LS-GST proteins (Fig 2B) had more variable responses. The wt protein was only weakly labeled in HeLa extracts (15), and all members of this panel incorporated less label than any reactive LT-GST. The membranes required more than twice the exposure to even visualize the positive bands. Relative to each other, the C11A, Y7L, T36A and Y49F sequences were labeled 2-3 times less efficiently than wt. The T40A and T58A had reduced in label incorporation by about 25%. The most active LS-GST sequences were wt and Y34F.
LX-GST phosphorylation by rAMPK
For LE, disruption of the zinc finger domain at C19, unfolds the protein, as measured by single-proton NMR, and renders it inactive as substrate for reactions with cytosol or recombinant kinases (16). The C11 in LT or LS is the structural analog to LE C19 with presumably similar functions. Indeed, neither Theilovirus protein with this mutation was strongly labelled in Hela cytosol. Given the inability to discriminate between structural and phosphorylation phenotypes at Y7, this location was noted, but not pursued further.
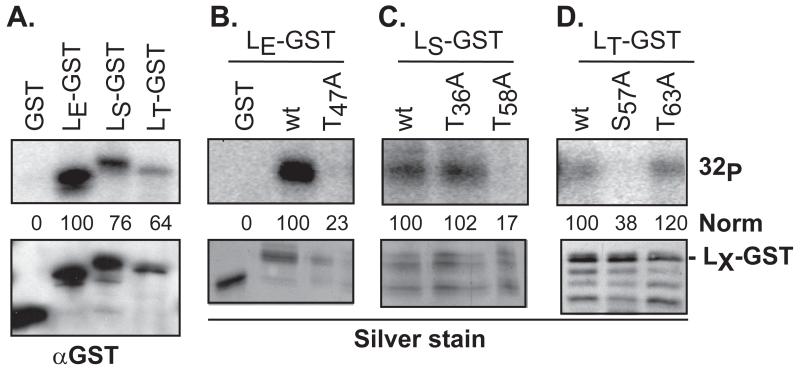
Instead, the S57 site of LT-GST, the only other potent mutation for this protein, was suggested by the algorithms as a potential target for AMP-kinase, a ubiquitous sensor of cellular energy homeostasis, and an enzyme common to all organisms from yeast to mammals. AMPK is involved in the replication cycles of many viruses (for review, see (30)). Recombinant AMPK was active with each of the wt LX-GST proteins but not with GST control (Fig 3A). Surprisingly, LE-GST reacted even more strongly than LT-GST or LS-GST, an unexpected finding since neither of the mapped LE-GST sites (Y41, T47) had been previously assigned to this enzyme (15). Parallel reactions with mutant proteins confirmed AMPK was selecting the same LE site (T47) as the previously mapped CK2 activity (Fig 3B).
Figure 3.
Phosphorylation of LX-GST by rAMPK. Recombinant GST and LX-GST proteins were reacted with rAMPK (50 ng) and [γ-32P]-ATP as in Methods. After pulldown and protein fractionation, gel bands were detected by phosphoimaging (32P) and silver stain, or after transfer to membranes, by Western analyses (αGST). The phosphorylation signals as measured by densitometry were normalized (norm) to the top band (full-length fusion protein) of LE-GST (A, B) LS-GST (C), or LT-GST (D) αGST signal. Other GST-reactive bands in these panels are from inappropriate LX translational start sites during bacterial protein expression.
For LT-GST, mutation at S57, but not T63, also abolished AMPK reactivity (Fig 3D). The LT-dependent apoptotic phenotypes (2), and LT intracellular localization differences (17) previously assigned to the S57 (DA) and P57 (GDVII) sequences, are now logically explained by the phosphorylation status of this residue. We do not yet know if GDVII or other viruses of this subtype use an alternative primary site instead. There was no evidence in cytosol experiments (Hela or BKH, not shown), that BeAn LT-T63 was a viable, reactive site (Fig 2A), and certainly it was unreactive with rAMPK (Fig 3D). With the LS-GST protein, mutation at T58 but not T63 also prevented rAMPK reactions (Fig 3C). The selection of this subset of proteins for testing was based on the site-prediction algorithms, as these were the only LX sites that fit the enzyme profile.
AMPK in cells
With standard reaction conditions, 50 ng of rAMPK put an average of 1 phosphate onto each substrate LE-GST protein (not shown). The cytosol reactions with the same amount of substrate usually required longer exposures, suggesting a lower saturation of label. When tested side-by-side for antibody-band signals, relative to rAMPK, a sample of HeLa cell lysate registered about 27 ng of enzyme, as present in similar reactions (Fig 3). BHK extracts gave a higher concentration, so there is variability in the availability of AMPK in these cell types. This means that native AMPK is probably not a dominant, high concentration enzyme in HeLa cells. The alternative possibility, that LS-GST (and LT-GST, LE-GST) is perhaps a non-ideal substrate is not supported by the incorporation data. The listed specific activity for rAMPK on its preferred acetyl-CoA carboxylase substrate is 737 nmol/min/mg, a value that should label 2 μg of LS-GST (~62 pmol) within 3-5 min, as was observed in the actual reactions.
Structure prediction
The Theilo and Ser/Thr domains of LS-GST and LT-GST contain the respective T58 and S57 mutation-mapped AMPK phosphorylation sites for these proteins (Fig 1). These sites are not homologs because the alignments place the LS S57 within the context of the Theilo domain, paired vertically with LT T63, a non-phosphorylated site. Unique to the Theilovirus proteins, however, the sequences within both inserted domains uniformly show strong, conserved patterns of 3-4 amino acid periodicities (e.g. Ser/Thr, Asp/Gly, Leu/Val). Such patterns typically identify strong α-helical regions. When queried with the Lasergene Protean Suite, every algorithm concurred on this configuration for the short (~4 turn, LS), or longer (~7 turn, LT) insertions. If this model is true, as depicted in Fig 4, the central helical portions, scaled to the determined LM structure, would place both Theilovirus AMPK sites virtually on top of each other, a proximity dictated by the respective helical folds. The Ser/Thr domain (e.g. LT) then would behave physically just like an extended Theilo domain (e.g. LS), with a consequently displaced AMPK site. The LT residue T63, the sequence homolog to LS T58 was not phosphorylated by AMPK. That activity is apparently shifted upstream, by 2 turns of the putative helix, to the structural analog, S57. Possibly, the adjacent proximity to the upstream acidic domain plays a role in this site selection. The LE-GST primary site, recognized by CK2 or AMPK, is just on the other side of this acidic patch, again suggesting an analogous if not homologous function.
Figure 4.
Structure models. An LM configuration as determined by NMR (PDB: 2M7Y) shows the location of Theilovirus Ser/Thr and Theilo domain insertions, modeled here as helices. Within these contexts, the mapped LT and LS AMPK sites occupy analogous locations relative to the start of each helix.
Conclusions
Why do cardiovirus LX use different phosphorylation sites when the end goal of nucleocytoplasmic trafficking inhibition is the same? Perhaps phosphorylation variability could just be fallout of cell-type preference by EMCV, Saffold, and TMEV, i.e. each virus type has adapted to use a kinase abundant in their cell type of choice. Alternatively, the site selection and required kinase(s) could represent distinct regulatory methods. LE from highly virulent EMCV is phosphorylated by the highly expressed and constitutively active CK2 and also at the same site, by AMPK, an enzyme which is less abundant in some cell types. The Theiloviruses, however, are not as robust in their host range, and at least for the sites mapped here, seem to rely on AMPK and not CK2, possibly slowing the viral lifecycle in a way that is helpful for the establishment of persistence. The LT site for AMPK, S57, is a known determinant distinguishing between persistent demyelinating strains of TMEV (DA, BeAn) and neurovirulent strains (GDVII). Recombinant conversion of DA S57 to P57 results in a GDVII phenotype (high titer, large plaques) in BHK-21 cells, while the opposite conversion (GDVII P57 to S57) gives a DA phenotype (low titer, small plaques) (31). Apoptotic phenotypes are also localized to this residue (2). RanGTPase binding is ubiquitous to all cardiovirus L proteins, but the phosphorylation status of the LX apparently does not contribute to this step (16). Therefore, LX phosphorylation must be required for the subsequent ternary (or quaternary?) complexes that actually carry out Nup phosphorylation. Differential partner selection after Ran binding are logical experimental directions predicted by these findings.
ACKNOWLEDGEMENTS
This work was supported by NIH grant AI017331 to ACP. We thank Dr. Adam Swick and Dr. John Yin for BHK-21 cells, Dr. Howard Lipton for the generous gift of SafV-2 and TMEV (BeAn) cDNAs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Arslan SY, Son K-N, Lipton HL. The antiapoptotic protein Mcl-1 controls the type of cell death in Theiler’s virus-infected BHK-21 cells. J Virol. 2012;86:1922–1929. doi: 10.1128/JVI.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stavrou S, Ghadge GD, Roos RP. Apoptotic and antiapoptotic activity of L protein of Theiler’s murine encephalomyelitis virus. J Virol. 2011;85:7177–7185. doi: 10.1128/JVI.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanova L, Lidsky P, Kolesnikova M, Fominykh KV, Gmyl AP, Sheval EV, Hato SV, van Kuppeveld FJ, Agol V. Antiapoptotic activity of the cardiovirus leader protein, a viral “security” protein. J Virol. 2009;83:7273–7284. doi: 10.1128/JVI.00467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuwa T, Taniura N, Saito M, Himeda T, Ohara Y. The opposite effects of two nonstructural proteins of Theiler’s murine encephalomyelitis virus (TMEV) regulates apoptotic cell death in BHK-21 cells. Microb Immun. 2010;54:639–643. doi: 10.1111/j.1348-0421.2010.00260.x. [DOI] [PubMed] [Google Scholar]
- 5.Ghadge GD, Ma L, Sato S, Kim JH, Roos RP. A protein critical for a Theiler’s virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J Virol. 1998;72:8605–8612. doi: 10.1128/jvi.72.11.8605-8612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himeda T, Ohara Y, Asakura K, Kontani Y, Murakami M, Suzuki H, Sawada M. A lentiviral expression system demonstrates that L* protein of Theiler’s murine encephalomyelitis virus (TMEV) is essential for virus growth in a murine macrophage-like cell line. Virus Res. 2005;108:23–28. doi: 10.1016/j.virusres.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Carocci M, Cordonnier N, Huet H, Romev A, Relmy A, Gorna K, Blaise-Boisseau S, Zientara S, Kassimi LB. Encephalomyocarditis Virus 2A Protein Is Required for Viral Pathogenesis and Inhibition of Apoptosis. J Virol. 2011;85:10741–10754. doi: 10.1128/JVI.00394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borghese F, Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 2011;85:9614–9622. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lidsky PL, Hato S, Bardina MV, Aminev AG, Palmenberg AC, Sheval EV, Polyakov VY, van Kuppeveld FJ, Agol V. Nucleo-cytoplasmic traffic disorder induced by cardioviruses. J. Virol. 2006;80:2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardina MV, Lidsky P, Sheval EV, Fominykh KV, van Kuppeveld FJ, Polyakov VY, Agol V. Mengovirus-induced rearrangements of the nuclear pore complex: Hijacking cellular phosphorylation machinery. J Virol. 2009;83:3150–3161. doi: 10.1128/JVI.01456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter FW, Palmenberg AC. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition of cardioviruses. J Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter FW, Brown B, Palmenberg A. Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J Virol. 2010;84:12538–12548. doi: 10.1128/JVI.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhaye S, van Pesch V, Michiels T. The leader protein of Theiler’s virus interferes with nucleocytoplasmic trafficking of cellular proteins. J Virol. 2004;78:4357–4362. doi: 10.1128/JVI.78.8.4357-4362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricour C, Borghese F, Sorgeloos F, Hato SV, van Kuppeveld FJ, Michiels T. Random mutagenesis defines a domain of Theiler’s virus leader protein that is essential for antagonism of nucleocytoplasmic trafficking and cytokine gene expression. J Virol. 2009;83:11223–11232. doi: 10.1128/JVI.00829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basta HA, Bacot-Davis VR, Ciomperlik JJ, Palmenberg AC. Encephalomyocarditis virus leader is phosphorylated by CK2 and Syk as a requirement for subsequent phosphorylation of cellular nucleoporins. J. Virol. 2014;88:2219–2226. doi: 10.1128/JVI.03150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacot-Davis VR, Palmenberg AC. Encephalomyocarditis virus Leader protein hinge domain is responsible for interactions with Ran GTPase. Virology. 2013;443:177–185. doi: 10.1016/j.virol.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniura N, Saito M, Okuwa T, Saito K, Ohara Y. Different subcellular localization of Theiler’s murine encephalomyelitis virus leader proteins of GDVII and DA strains in BHK-21 cells. J Virol. 2009;83:6624–6630. doi: 10.1128/JVI.02385-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho SH, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Cornilescu CC, Porter FW, Zhao Q, Palmenberg AC, Markley JL. NMR structure of the Mengovirus leader protein zinc-finger domain. FEBS Letters. 2008;582:896–900. doi: 10.1016/j.febslet.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Watters K, Palmenberg A. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J Virol. 2011;85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R, Thompson J, Gibson T, Higgins D. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 22.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A Sequence Logo Generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Y, Li A, Wang L, Feng H, Yao X. PPSP: prediction of PK-specific phosphorylation site with Bayesian decision theory. BMC Bioinformatics. 2006;7:163. doi: 10.1186/1471-2105-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 25.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 26.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic acids research. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nuc Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat Biotech. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 29.Obenauer J. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nuc Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mankouri J, Harris M. Viruses and the fuel sensor: the emerging link between AMPK and virus replication. Rev Med Virol. 2011;21:205–212. doi: 10.1002/rmv.687. [DOI] [PubMed] [Google Scholar]
- 31.Takano-Maruyama M, Ohara Y, Asakura K, Okuwa T. Theiler’s murine encephalomyelitis virus leader protein amino acid residue 57 regulates subgroup-specific virus growth on BHK-21 cells. J. Virol. 2006;80:12025–12031. doi: 10.1128/JVI.00693-06. [DOI] [PMC free article] [PubMed] [Google Scholar]