Abstract
A promising class of potential new antibiotics are the antimicrobial peptides or their synthetic mimics. Herein we assess the effect of the type of cationic side chain (i.e., guanidino vs. amino groups) on the membrane perturbing mechanism of antimicrobial α-peptide–β-peptoid chimeras. Two separate Langmuir monolayers composed of 1,2-dipalmitoyl-sn-glycero-3-phosphatidylglycerol (DPPG) and lipopolysaccharide Kdo2-lipid A were applied to model the outer membranes of Gram-positive and Gram-negative bacteria, respectively. We report the results of the measurements using an array of techniques, including high-resolution synchrotron surface X-ray scattering, epifluorescence microscopy, and in vitro antimicrobial activity to study the molecular mechanisms of peptidomimetic interaction with bacterial membranes. We found guanidino group-containing chimeras to exhibit greater disruptive activity on DPPG monolayers than the amino group-containing analogues. However, this effect was not observed for lipopolysaccharide monolayers where the difference was negligible. Furthermore, the addition of the nitrobenzoxadiazole fluorophore did not reduce the insertion activity of these antimicrobials into both model membrane systems examined, which may be useful for future cellular localization studies.
Keywords: antimicrobial peptidomimetics; peptide–peptoid chimeras; guanidinium cation; bacterial membrane; phosphatidylglycerol, X-Ray scattering
1. Introduction
Antimicrobial peptides (AMPs) are ubiquitous in Nature; present in virtually all organisms they serve as endogenous antibiotics through the innate immune response.[1, 2] Members of this class of compounds have been studied extensively due to their potential as promising alternative antibiotics to treat disease caused by the growing number of resistant pathogenic microbes.[1-4] It is generally believed that AMPs exert their direct killing of invading pathogens by selectively interacting with the negatively charged bacterial surfaces over the globally neutral (zwitterionic) eukaryotic cell membranes. The mechanism by which the membranes are permeated is not completely understood, and several models have been proposed based on studies conducted with various peptidic structures.[1] Moreover, recent studies have shown that some of these chemotypes are endowed with additional intracellular modes of action such as interference with cell wall biosynthesis or immunomodulatory effects.[5-9] These findings complicate the understanding of this class of compounds even further and have called for the use of a perhaps more appropriate class designation, host-defense peptides (HDPs).[3]
Despite their diversity in amino acid sequence, lipophilicity and secondary structure,[10] most HDPs share common features includig positive net charge and generally amphipathic nature, separating hydrophilic and hydrophobic residues to the opposite faces of the molecule.[11-13] Typically, positive net charge of naturally occurring peptides is contributed by the guanidino groups of the arginine (Arg)[14, 15] and/or amino groups of the lysine (Lys) residues.[16-18] Both Arg and Lys side chains are generally thought to promote the initial long range electrostatic attractive forces that guide antimicrobials towards the negatively charged bacterial membranes.[19] However, guanidino groups have higher acid dissociation constant (pKa) due to efficient resonance stabilization of the charged protonated state together with efficient solvation in water, which makes them stronger bases and, thus, better suited for stable electrostatic interactions with the negatively charged phosphodiester and phosphomonoester groups of phospholipids.[20-24] Examples of naturally occurring AMPs containing arginine rather than lysine residues include several members of the cathelicidin family, such as indolicidin and tritrpticin.[25, 26] Also, in peptides having high content of both arginine and lysine residues such as the defensins, these residues are not randomly distributed within their sequence and their ordering implies a significance greater than just a net positive charge.[27] Muhle and Tam[28] found that Arg-to-Lys substitution in a cyclic disulfide-stabilized peptide decreased activity against Gram-negative bacteria. Nakase et al. demonstrated improved membrane permeability of antimicrobial peptide (RLA) with lysine substituted by arginine.[29] Other studies have shown that for lactoferricin B and bactenecin 5, which have no hemolytic activity, the replacement of arginine for lysine reduced antibacterial activity.[30] So, the incorporation of guanidino groups into the peptide side chains may have its appeal in drug design.[31-33]
However, there are concerns related to the use of α-peptides in a clinical setting due to their high cost of manufacturing[34] and inherent susceptibility to proteases,[35] which has led to numerous studies aimed at mimicry of peptides using non-natural compounds. Thus, a variety of classes such as β-peptides,[36-38] oligoureas,[39] arylamides,[40, 41] N-substituted oligoglycines (peptoids),[42-44] cyclic D,L-α-peptides,[45-47] hybrid peptidomimetics,[33, 48-50] and polymers[51-53] have been designed to mimic the function of AMPs.
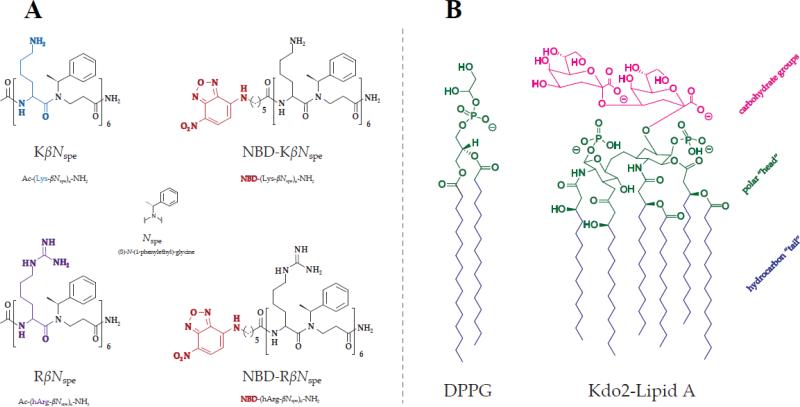
α-peptide–β-peptoid chimeras represent a distinct class of peptidomimetics with backbone composed of alternating peptide and β-peptoid residues.[33, 50, 54-56] In the present study we elucidate the role of the cation type on the antimicrobial properties of this type of synthetic AMP mimics using two α-peptide–β-peptoid chimeras (KβNspe and RβNspe), which differ from each other solely in the identity of cationic functionality [amine (lysine) vs. guanidino group (homoarginine)]. In addition, because fluorophore-labeled analogues of AMPs, which retain antimicrobial activity, constitute powerful tools for studying mechanisms of action and cellular localization, we also prepared and evaluated nitrobenzoxadiazole (NBD)-labeled oligomers NBD-KβNspe and NBD- RβNspe (Fig. 1A).
Figure 1.
Molecular structures of the tested chimeras (A) and lipids used for modeling bacterial cell membranes (B).
Regardless of whether the primary mode of action is of a membrane-disrupting nature or entails perturbation of intracellular targets, the initial interaction between antimicrobial and bacteria involves the cell surface. A fundamental understanding of these lipid–antimicrobial interactions is therefore important for the future design of improved antibiotics for potential clinical use. Since cell membranes have a complex structure and are currently not applicable for highly sensitive surface X-ray scattering methods, the model systems are generally employed to undertake detailed mechanistic studies of membrane-associated processes.[57-61] Previously, the membrane-destabilizing effects of the α-peptide–β-peptoid chimeras have only been investigated in model liposomes prepared from phosphatidylcholine (PC), a phospholipid found predominantly in eukaryotic cells.[55] However, PC-containing systems do not adequately represent bacterial envelope, and furthermore, these compounds have not been investigated using sensitive X-ray methods before.
In order to model the outer surface of Gram-positive and Gram-negative bacteria we have employed insertion assay experiments on two separate Langmuir monolayers composed of 1,2-dipalmitoyl-sn-glycero-3-phosphatidylglycerol (DPPG) and truncated lipopolysaccharide (LPS) Kdo2-Lipid A, respectively (Fig. 1B). The reason behind this choice of lipids is that Kdo-2-lipid A constitutes the hydrophobic core of outer LPS envelope in most Gram-negative bacteria, while PGs are predominat anionic phospholipid species within cytoplasmic membranes of both Gram-negative and Gram-positive strains. This approach has been successfully used in conjunction with liquid surface X-ray scattering to study bacterial membrane lysis by human antimicrobial peptide LL-37,[60] protegrin-1[57, 62] gramicidin[63] and SMAP-29[61] antimicrobial peptides as well as by peptide mimics.[44, 59, 64, 65]
2. Experimental Section
2.1 Monolayer construction
Both DPPG and Kdo2-Lipid A were purchased from Avanti Polar Lipids (Alabaster, AL) and were used without further purification. To form the monolayer systems both DPPG and Kdo2-Lipid A were first dissolved in chloroform–methanol (65:25) at a concentration of 0.2 mg/mL. Using a microliter syringe (Hamilton) the solutions were then spread on the surface of a Dulbecco's phosphate buffered saline (DPBS) (Invitrogen, Carlsbad, Ca) void of calcium and magnesium ions contained in a single barrier Langmuir trough. Over 15 minutes the organic solvents evaporated to form a self-assembled monolayer. The monolayer was then compressed to a biologically relevant packing density of 30 mN × m–1, which was monitored by a Wilhelmy plate. This surface pressure and a temperature of 22 ± 0.5 °C were maintained throughout the experiment. As a result, if changes in the surface pressure occur, the barrier will have to move in order to maintain the set surface pressure. Such change in barrier position then allows for change in area/lipid molecule or area/LPS molecule ΔA to be calculated. Once the monolayer was compressed the chamber containing the Langmuir trough was sealed and purged with helium to lower the oxygen levels in the chamber, which minimizes background X-ray scattering during the X-ray experiments.[58, 66]
2.2 X-ray reflectivity (XR)
XR gives the information about electron density gradient along the plane perpendicular to the surface of monolayer as well as about the film thickness. [67-69] A slab-model, also known as a box model, was used to analyze XR data. This model is based on the Parratt recursive method[70] that describes the interface as a stack of slabs with distinct electron densities (ρ), and thicknesses (l).[71-75] The final fit was achieved by minimizing the χ2-square value while ensuring that parameters obtained were physically relevant. The software used to fit experimental XR data was RFit2000.[76-78] In addition to model-dependent fits, model-independent fits where the electron density profile of the film versus the vertical position along film perpendicular was generated using the software StochFit.[79]
2.3 Grazing-incidence X-ray diffraction (GIXD)
Grazing incidence X-ray diffraction measurements were performed to monitor the effect of compound insertion on the molecular packing of the lipid monolayers.[80] Lipid films spread at the air-water interface may be described by a large number of two-dimensional crystalline domains of ordered hydrocarbon chains randomly oriented around the surface normal.[81] In a GIXD experiment, the momentum transfer has a horizontal and vertical component, Qxy and Qz.[82] The Qxy positions of the observed Bragg peaks yield the repeat distances, dhk = 2π/qhk for the 2D lattice, from which specific parameters (a, b, γ) of the crystal system can be extracted. From the full-width half-maximum (FWHM) values of Bragg peaks, the in-plane coherence length, Lxy was calculated using the Scherrer formula, Lxy = 0.9 × 2π/FWHM. The intensity distribution along the Bragg rod was measured at Bragg peak positions to evaluate the tilt of acyl chains.
All X-ray measurements were done at sector 9-ID at the Advanced Photon Source (APS) of Argonne National Labs (Chicago IL) with an X-ray wavelength of 0.9202 Å. After XR and GIXD were performed on a given monolayer system α-peptide–β-peptoid hybrids were introduced into the system using a bent needle syringe (Hamilton). The needle is placed underneath the barrier and the compounds were injected underneath the monolayer to mimic the approach of the compound from the extracellular fluid to the outer leaflet of the membrane. After injection both XR and GIXD measurements were taken for comparison.
2.4 Real-time epifluorescence microscopy (EFM) imaging
Morphological changes of DPPG films were studied on a microscopic level before and after the introduction of α-peptide–β-peptoid chimeras according to protocols previously described.[83, 84] Briefly, the Langmuir trough setup and procedures used in the formation of the lipid monolayers were essentially the same, except that a 0.1 mol % of lipid-linked Texas Red dye [TR-DHPE (Molecular Probes, Eugene, OR)] was premixed with stock DPPG solution. A heated glass-plate was placed over the trough to reduce contamination and evaporation of the subphase during the experiment.
2.5 Chemical synthesis
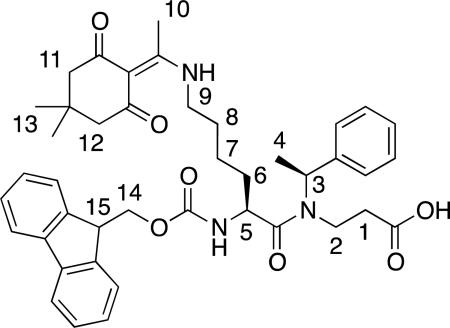
Fmoc-Lys(Dde)-βNspe-OH (8)
Fmoc-Lys-βNspe-OH (7) (1.61 g, 2.96 mmol) and iPr2NEt (1.4 mL, 8.0 mmol) were dissolved in DMF (30 mL), and added acetyl dimedone (913 mg, 5.0 mmol). After stirring for 18 h, the solvent was evaporated in vacuo and the crude product redissolved in EtOAc (100 mL). The solution was washed with 1 M HCl (aq) (2 × 100 mL) and water (2 × 100 mL), dried (Na2SO4), filtered, and evaporated in vacuo to give 1.22 g (82%) of the desired product as a white solid. 1H NMR (300 MHz, CD3OD) δ 1.48 (m, 2H, H-7), 1.66/1.56* (2 × d, 3H, J = 7.0 Hz, H-4), 1.68–1.82 (broad m, 4H, H-6, H-8), 2.17/2.51 (3 × m, 2H, H-1), 2.27*/2.28/2.51*/2.52 (4 × m, 6H, H-11, H-12) 3.19/3.38 (2 × m, 2H, H-2), 3.48 (m, 5H, H-9, H-10), 4.17 (m, 1H, H-15), 4.27–4.43 (broad m, 2H, H-14), 4.52*/4.81 (2 × m, 2H, H-5) 5.42/5.81* (2 × q, 1H, J = 7.0 Hz, H-3), 7.23–7.41 (broad m, 9H, Ph, Fmoc ArH), 7.66 (m, 2H, Fmoc ArH) 7.79 (d, 2H, J = 7.5 Hz, Fmoc Ar). [α]589.2: –46° (c = 1.0, 293 K, CHCl3). UPLC-MS gradient A, tR = 2.20 min (>95), MS: (m/z) [M + H]+ calcd. for C32H38N3O5+ : 708.9, found: 708.6. HRMS: (m/z) [M + H]+ calcd. for C32H38N3O5+ : 708.3643, found: 708.3649 (ΔM = 0.8 ppm).
Solid-phase synthesis of 9
Fmoc-protected Rink amide resin (590 mg, 0.25 mmol) was treated with piperidine–DMF (1:4, 5 mL, 2 × 20 min), and washed with DMF, MeOH, and CH2Cl2 (3 × 5 mL). Oligomerization was performed with a mixture of Fmoc-Lys(Dde)-βNspe-OH (8) (750 mg, 1.1 mmol, 4.5 equiv), HBTU (417 mg, 1.1 mmol, 4.5 equiv), and iPr2NEt (0.38 mL, 2.2 mmol, 9 equiv) in DMF (5 mL), which were preincubated for 10 min before being added to the Rink amide resin and shaken for 18 h. After each coupling the resin was washed with MeOH, DMF and CH2Cl2 (3 × 5 mL). Fmoc deprotection was achieved with piperidine–DMF (1:4, 5 mL, 2 × 20 min) followed by DBU–piperidine–DMF (2:2:96, 5 mL, 2 × 20 min), after each deprotection step the resin was washed using the same procedure as above. This three-step coupling/deprotection sequence was performed 6 times to give the resin-bound oligomer.
Ac-(Lys-βNspe)6-NH2 (KβNspe)
The terminal amino groups of (9) (100 mg, 0.024 mmol) were capped with a mixture of Ac2O–iPr2NEt–DMF (1:2:3, 2 mL, 2 h) and the resin was washed with DMF, MeOH, and CH2Cl2 (3 × 2 mL). The side chains were deprotected using 2% hydrazine in DMF (2 × 2 mL, 45 min). The crude product was cleaved from the support with 50% TFA–CH2Cl2 (2 mL, 2 × 1 h). The TFA was co-evaporated with toluene (3 × 30 mL), toluene–CH2Cl2 (3 × 30 mL), and CH2Cl2 (3 × 3 mL). The residue was purified by preparative RP-HPLC (gradient C) and fractions were lyophilized to give KβNspe as white fluffy material [12.3 mg, 15% (90% per step)]. HPLC gradient D, tR = 10.47 (>95%). HRMS: m/z [M+3H]3+ calcd for C104H158N19O13 3+ : 627.07567, found: 627.07553 (ΔM: 0.22 ppm).[49]
Ac-(hArg-βNspe)6-NH2 (RβNspe)
The terminal amino group of (9) (75 mg, 0.024 mmol) was capped with a mixture of Ac2O–iPr2NEt–DMF (1:2:3, 2 mL, 2 h) and the resin was washed with DMF, MeOH, and CH2Cl2 (3 × 2 mL). The side chains were deprotected using 2% hydrazine in DMF (2 × 2 mL, 2 × 45 min), and washed as above. Boc-protected guanidino groups were introduced by addition of a mixture of N,N’-bis(tert-butoxycarbonyl)-1H-pyrazole-1-carboxamidine (11) (285 mg, 0.92 mmol) and iPr2NEt (0.32 mL, 1.84 mmol) in DMF for 18 h, followed by the above washing procedure. The crude guanidinium-containing product was simultaneously deprotected and cleaved from the support with TFA–CH2Cl2 (1:1, 2 mL, 2 × 1 h). The TFA was co-evaporated with toluene (3 × 30 mL), toluene–CH2Cl2 (3 × 30 mL) and CH2Cl2 (3 × 30 mL). The residue was purified by preparative RP-HPLC (gradient C) and fractions were lyophilized to give RβNspe as white fluffy material [12.9 mg, 20% (90% per step)]. HPLC gradient D, tR = 10.39 (>95%). HRMS: m/z [M+3H]3+ calcd for C110H170N31O13 3+ : 711.1193, found: 711.1190 (ΔM: 0.35 ppm).[49]
NBD-(Lys-βNspe)6-NH2 (NBD-KβNspe)
A Rink amide resin-bound oligomer with Boc protected lysine side chains (150 mg, 0.039 mmol) was prepared as described for 9 using the Fmoc-Lys(Boc)-βNspe-OH[84] building block. The N-terminal was then functionalized with a mixture of N-NBD-6-aminohexanoic acid (73 mg, 0.25 mmol), iPr2NEt (87 μL, 0.5 mmol), and PyBOP (156 mg, 0.3 mmol) in DMF (2 mL). After shaking for 18 h, the resin was washed with DMF, MeOH, and CH2Cl2 (3 × 2 mL), and the compound was cleaved from the support using TFA–CH2Cl2 (1:1, 2 mL, 2 × 1 h). Trifluoroacetic acid was co-evaporated with toluene (3 × 30 mL), toluene–CH2Cl2 (3 × 30 mL), CH2Cl2 (3 × 3 mL), and the residue was purified by preparative RP-HPLC (gradient C). Lyophilization of the fractions containing the title compound furnished a yellow fluffy material [12.5 mg, 15% (88% per step)]. HPLC gradient D, tR = 10.29 (>95%). HRMS: m/z [M+3H]3+ calcd. for C114H168N23O16 3+ : 705.4352, found: 705.4361 (ΔM: 1.3 ppm), and m/z [M+4H]4+ calcd. for C114H169N23O16 4+ : 529.3252, found: 529.3261 (ΔM: 1.7 ppm).
NBD-(hArg-βNspe)6-NH2 (NBD-RβNspe)
Crude NBD-(Lys-βNspe)6-NH2 (NBD-KβNspe) (30 mg, 0.014 mmol) and iPr2NEt (29 μL, 0.16 mmol) were dissolved in DMF (2 mL), followed by addition of N,N’-bis(tert-butoxycarbonyl)-1H-pyrazole-1-carboxamidine (40 mg, 0.13 mmol). After stirring for 3 h, the solvent was evaporated in vacuo, and excess reagent removed by vacuum silica gel chromatography [2 × 6 cm, CH2Cl2–MeOH 0.5% gradient 0→10% (containing 1% concentrated aqueous NH3)]. The product was then deprotected with TFA–CH2Cl2 (1:1, 2 mL, 2 × 1 h) and TFA was removed by co-evaporation with toluene (3 × 30 mL), toluene–CH2Cl2 (3 × 30 mL), and CH2Cl2 (3 × 3 mL). The compound was purified by preparative RP-HPLC (gradient C) to give NBD-RβNspe as a yellow fluffy material (5 mg, 15%). HPLC gradient D, tR = 11.05 (>95%). HRMS: m/z [M+4H]4+ calcd. for C120H181N35O16 4+ : 592.5609, found: 592.5603 (ΔM: 1 ppm).
Details of synthetic procedures, charaterization data, as well as 1H and 13C NMR spectra for all new compounds are presented in Supporting Information.
2.6 Bacterial strains and culture conditions
Activity experiments (Minimum Inhibitory Concentration and Minimum Bactericidal Concentration) were carried out with eight bacterial species representing common laboratory strains and clinical strains derived from both food-borne and nosocomial infections. The strains also represented Gram-positive and Gram-negative species. Stock cultures were stored at –80 °C in 4 % (w/v) glycerol, 0.5% (w/v) glucose, 2% (w/v) skimmed milk powder, and 3 % (w/v) tryptone soy powder. All experiments were carried out with bacteria incubated for one night (approximately 18 hours) at 37 °C. Experiments were performed in cation-adjusted Mueller Hinton II broth [MHB (Becton Dickinson 212322)] adjusted to pH 7.4. MHB was supplemented with 1.25% defibrinated horseblood (Statens Seruminstitut REF23699) to ensure growth of B. cereus and S. pyogenes. Brain Heart Infusion (CM1135)with 1.5% agar (VWR 20768.292) as gelling agent was used throughout for colony plating.
2.7 Antimicrobial activity assay
MIC and MBC were determined using the micro-dilution method according to guidelines of the Clinical and Laboratory Standards Institute (CLSI). Two-fold serial dilutions of the peptidomimetic hybrids were prepared from 1024 μg/mL stock solutions in Milli-Q water to give a final range of 512–0.5 μg/mL in the wells. Colonies grown on BHI agar for approximately 18 hours were suspended in 0.9% saline to give a turbidity of 0.13 at OD546 (approximately 1 × 108 CFU/mL), and then diluted in MHB pH 7.4 to a final concentration of approx. 5 × 105 CFU/mL in each well. Polypropylene plates (Nunc 442587) were used to minimize peptide binding, and the incubation time was 18–20 hours at 37 °C. MIC values, i.e., the lowest concentration of the peptide analogue at which no visible growth was observed, were determined in duplicate. Platings were done from all wells where no visible growth was observed and the lowest concentration of peptide analogue at which no growth occurred on BHI plates were denoted the MBC, the Minimal Bactericidal Concentration Activity is expressed in μg/mL.
3. Results
3.1 Synthesis
The syntheses of chimeras were achieved by preparation of dimeric building blocks followed by oligomerization on solid support using variations of previously described methods.[50, 85, 86] In order to enable an on-resin functionalization of the lysine ε-amino groups we installed an orthogonal 1-(4,4-dimethyl-2,6-dioxacyclohexylidene)ethyl (Dde) group[87, 88] on the lysine side chain functionality to give building block 8 (Scheme 1).
For the standard Fmoc solid-phase peptide synthesis (SPPS) oligomerization, a Chem-Matrix® resin was chosen due to its excellent swelling properties in a variety of solvents. After six rounds of coupling/deprotection (9), the N-terminal was acetylated and the Dde group was removed to give 10, which upon cleavage afforded KβNspe (Scheme 2). Functionalization of the free amines in 10 by guanidinylation,[89] followed by simultaneous deprotection and cleavage furnished RβNspe. Unfortunately, introduction of the fluorophore proved incompatible with our new protecting group strategy, most likely due to sensitivity towards hydrazine during the Dde deprotection step. For the syntheses of labeled analogues NBD-KβNspe and NBD-KβNspe, a different strategy involving guanidinylation in solution was therefore adopted as shown in Supplementary Scheme S1 and Scheme S2.
3.2 Antimicrobial activities
MIC and MBC assays clearly demonstrated greater bacteriostatic and bactericidal properties of RβNspe against all eight examined strains. The favourable effect of guanidinium cation was highly pronounced for Gram-positive bacteria (fourfold difference in MICs and MBCs) and moderate for Gram-negative ones. The toxicity of KβNspe and RβNspe against human erythrocytes was previously determined using hemolytic assay and considered to be negligible.[33]
In our recent studies, fluorescein-labeled versions of α-peptide–β-peptoid chimeras have been prepared to investigate their potential as cell-penetrating peptides,[54, 55] and subsequent antimicrobial testing of these showed a severe decrease in potency when introducing the fluorescein label.[33] Herein, we therefore tested the NBD-labeled chimeras for their antimicrobial activity against a selection of pathogens to determine if the novel fluorescent-labeled antimicrobial could inhibit bacterial growth. Interestingly, MIC and MBC values of NBD-tagged amino-containing chimera were found to be lower against all Gram-positive and Gram-negative strains tested (except V. vulnificus). However for guanidino-containing antimicrobials the trend is opposite. Since the effect of NBD-fluorophore can be either attenuating or enhancing but within acceptable range of the parent compounds, we decided to include fluorescently tagged chimeras in the model study along with KβNspe and RβNspe.
3.3 Epifluorescence microscopy
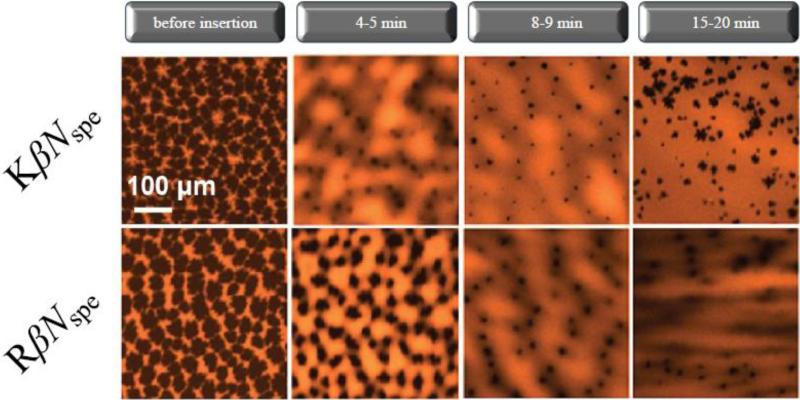
EFM images of the DPPG monolayer at 30 mN × m–1 displays an array of branched dark domains of condensed phase ~25–50 μm in diameter separated from each other by brightly colored “fluid” (disordered) areas. Fig. 2 shows the dynamics of surface morphology changes in lipid film after injection of KβNspe and RβNspe into the subphase. Both compounds caused a decrease in the size of condensed-phase domains starting from 4th min and followed by their complete elimination with transition of the majority of the film to a liquid-disordered phase after 15–20 min. Structurally ordered regions in this case might be either fully destroyed or reduced in size to become smaller than the microscope resolution (<1 μm). This points out to a crystallinity-disruptive behavior of the studied α-peptide–β-peptoid chimeras against DPPG monolayers regardless of the identity of the cations they contain, at least on micrometer scale.
Figure 2. Epifluorescence images of DPPG monolayers after injection of NspeK (A) and NspeR (B) at concentrations corresponding to 20% of their MIC values observed against S. aureus respectively.
Lipid-linked Texas Red-DHPE fluorescence probe (1 mol %) was added to the phospholipid solutions for EFM experiments. Because of steric hindrance, the dye is located in the liquid-disordered phase, rendering it bright whereas the liquid-ordered phase remains dark.
3.4 Specular X-ray reflectivity
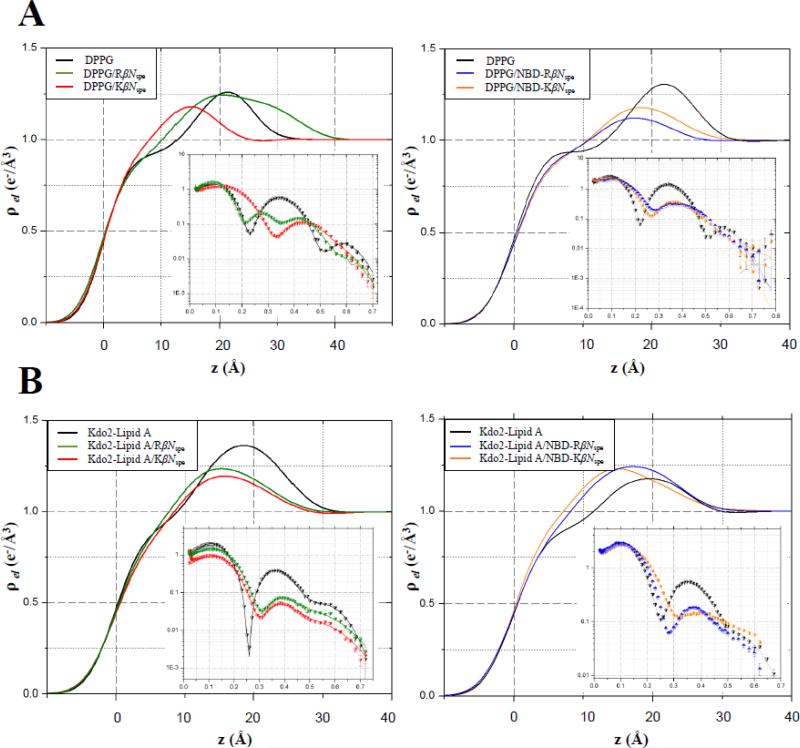
Fig. 3 shows electron density profiles along the surface normal extracted from reflectivity data by model-independent stochastic fitting. The graphs are combined in such a way as to allow visual comparison of amino- and guanidino-containing chimeras. For the lipid monomolecular films, the electron density is zero at the air-water interface, then rises sharply through the hydrocarbon tail region, and comes to a plateau reaching its maximum values for the head groups (at a distance of ~20–25 Å from the air side of the film) before slightly decaying to the subphase electron density. In addition, model-dependent analyses were performed on XR data. Pure DPPG monolayers were modeled as two slabs, with the first slab corresponding to the phospholipid acyl chains, and the second reperesenting the lipid head groups. XR analysis yielded the thickness of the slab related to acyl chains to be 16.5 Å with an electron density of 0.312 e−/Å3. The thickness of the slab used to model the head groups was found to be 8.3 Å with an electron density of 0.477 e−/Å3. Two-slab model-dependent fitting of Kdo-2 Lipid A data yielded 12.0 Å long upper hydrocarbon chain region with electron density of 0.31 e−/Å3. The second slab corresponding to the complex of head moieties and the outer layer of carbohydrate 3-deoxy-D-mannooctulosonic acid known as Kdo has the thickness of 12.8 Å and an electron density of 0.485 e−/Å3. Insertion of antimicrobials into the membrane mimic results in extra electrons per lipid molecule in each slab and is calculated using formula (1).
| (eq. 1) |
Figure 3. Electron density profiles and corresponding Fresnel-divided reflectivity curves against the scattering vector (q) in the z direction (qz) for DPPG (A) and Kdo2-Lipid A (B) monolayers at 30 mN × m–1.
Electron density profiles were normalized to the electron density of the subphase buffer. On the XR graphs, the scatter plots are experimental values and solid lines are the best fits of the models to the experimental data. Fresnel reflectivity is the reflectivity from ideal smooth surface.
Here, l slab and ρ slab are thickness and electron density of the slab, respectively; Alipid + ΔAlipid is the area per lipid molecule upon insertion and Ninitial e−slab minus the number of electrons in the slab in the original untreated monolayer.
Preliminary information about the mode of antimicrobial interaction with membane mimics can be obtained directly from the electron density profiles (Fig.3A). KβNspe and RβNspe displayed a drastic difference in their mode of action against DPPG monolayers. Following injection of KβNspe the first minimum of reflectivity curve shifted from qz ≈ 0.24 Å−1 to a higher qz value with the peak of electron density moved towards the air–water interface. This indicates a decrease in thickness of the film as a result of its insertion. However, RβNspe instead of thinning DPPG monolayer, led to appearance of two minima on the reflectivity profile at qz ≈ 0.21 Å−1 and 0.35 Å−1 and a notable bump of the electron density curve within the range of 20–40 Å away from air-water interface. This might be due to an additional layer of distinct electron density higher than the electron density of subphase present underneath the head group region. These data are corroborated by the model-dependent analysis and are summarized in Supplementary Table S1. Injection of KβNspe into DPPG resulted in an experimental XR curve, which was again best fit with two layers. However, an additional box was required to fit XR data upon introduction of RβNspe. The lower increase in area per lipid molecule observed upon insertion of RβNspe as compared with KβNspe could possibly be explained by partial dimerization or aggregation of guanidino-containing chimera on the outer surface of lipid monolayer. According to the number of extra electrons contributed by incorporated antimicrobials, both KβNspe and RβNspe readily insert into the polar moieties of DPPG and Kdo-2 Lipid A resulting in reduced electron density of bottom slab, but they are both unable to penetrate deeply into overlying hydrophobic core of lipid monolayers. The more substantial decrease in electron density of the DPPG head group region, along with three times more additional electrons present upon introduction of RβNspe points to a higher Gram-positive membrane disruptive potential of guanidino-containing compound versus its amino-containing counterpart. The same trend was observed for the NBD-tagged chimeras. Here compound NBD-RβNspe permeated the entire depth of DPPG film including hydrophobic acyl chains, whereas NBD-KβNspe was found only in the hydrophilic outer shell of the lipid monolayer. Furthermore, the introduction of NBD-RβNspe led to a greater contribution of additional electrons in toto, as well as to a four-fold larger increase in area per single DPPG molecule (ΔAlipid) indicating a favorable effect of arginine residues on the antimicrobial insertion.
In contrast to DPPG, the reflectivity curves of Kdo-2 Lipid A monolayer after introduction of KβNspe and RβNspe look nearly identical (Fig. 3B). For model-dependent analysis two boxes were sufficient to fit experimental XR data and revealed very similar mechanism of action utilized by guanidino- and amino-containing chimeras against Gram-negative bacteria LPS. This consistency in mode of action between KβNspe and RβNspe, as well as between NBD-KβNspe and NBD-RβNspe was confirmed by similar changes in thickness of respective slabs within Kdo-2 Lipid A monolayer and by similar number of contributed extra electrons. The area increase per lipid molecule in both pairs of compounds was also about the same.
Additionally, the effect of NBD-fluorophore was investigated by comparing KβNspe and RβNspe to their NBD-tagged fluorescent analogues NBD-KβNspe and NBD-RβNspe respectively. According to the results of XR analysis, functionalization of α-peptide–β-peptoid chimeras by NBD does not reduce their capability to interact with model bacterial membranes. Moreover, fluorophore-carrying chimeras have provided even greater contribution of additional electrons to the lipid head-groups. This implies a higher number of chimeras to be inserted into the lipid films.
3.5 Grazing-incidence X-ray diffraction
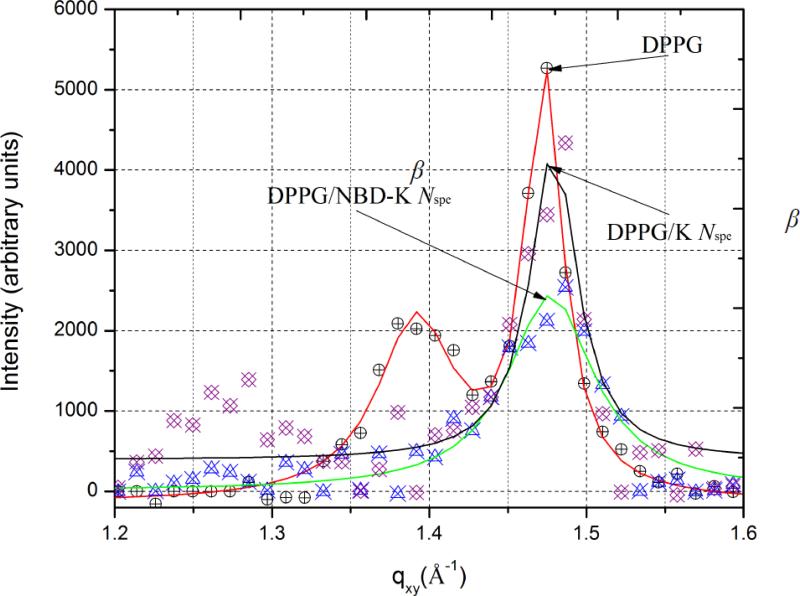
GIXD data for DPPG monolayer before and after injection of antimicrobials are presented in Fig. 4. The corresponding values of unit cell dimensions, d-spacings and sizes of crystallized domains are presented in Table 2. At the surface pressure of 30 mN × m−1 pure DPPG yields two distinct Bragg peaks at Qxy = 1.39 Å−1 and Qxy = 1.47 Å−1 corresponding to d-spacings of 4.51 and 4.26 Å, respectively. This indicates the presence of ordered structure with the centered rectangular packing (a ≠ b, γ = 90°) having unit cell dimensions a = 5.32 Å and b = 8.54 Å and an area of 45.5 Å2 per single DPPG molecule. For Kdo2-Lipid A, on the other hand, no Bragg peak was observed. This means that there were no diffractable 2D crystalline regions within the monolayer, which does not allow a detailed analysis of the surface morphology. According to GIXD data, RβNspe and NBD-RβNspe fully destroy the lateral crystallinity of DPPG monolayers evidenced by complete disappearance of Bragg peaks. Conversely, both KβNspe and NBD-KβNspe, instead of disordering, caused structural rearrangement of the crystal lattice from a centered rectangular crystal packing to a hexagonal (a = b, γ = 120o) resulting in appearance of a single Bragg peak. The coherence length was also reduced, which might explain the disappearance of ordered regions upon introduction of KβNspe observed by EFM. Furthermore, NBD-KβNspe was shown to decrease the size of crystallized domains as well as the order of their crystallinity to a greater extent than its non-labeled counterpart, even though the main parameters of crystal lattice didn’t change much. This supports the hypothesis that labelling of antimicrobials with NBD may enhance their disruptive potential against phosphatidylglycerol-containing membranes without drastically changing the primary mechanism of action.
Figure 4.
Bragg peaks plot of scattering vector Qxy as a function of intensity.
Table 2.
Structural Parameters of Crystal Monolayer Lattice
| Sample | Peak position (Å-1) | D-spacing (Å) | Unit cell parameters | Lxya (Å) | Area unit cell (Å2) |
|---|---|---|---|---|---|
| DPPG | Qxy1=1.39, Qxy2=1.47 | d11=4.51, d02=4.26b | a = 5.32 Å b = 8.54Å γ = 90° θ=27° | L11=93 Lo2=l 96 | 44.51 |
| DPPG/KβNspe | 1.48 | 4.25 | a = 6.93 Å γ = 120° θ = 0° | 156 | 41.62 |
| DPPG/NBD-KβNspe | 1.48 | 4.25 | a = 6.93 Åγ = 120°. θ=0° | 41.69 | |
| DPPG/RβNSpe and DPPG/NBD-RβNSpe displayed no visible GIXD peaks | |||||
Table Coherence length (Lxy) = the average distance in the direction of the reciprocal lattice vector Qxy over which the domain is ordered.
“11” and “02” are used to denote (hk) vectors in reciprocal space.
4. Discussion
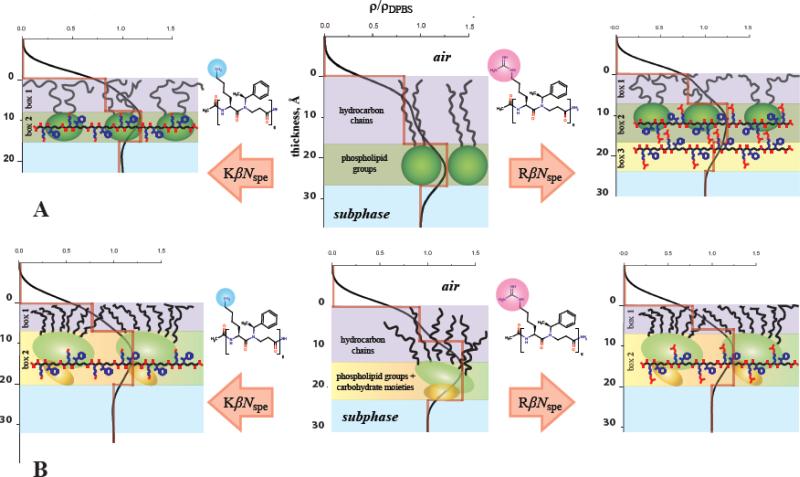
Overall, our data provide solid mechanistic evidence of higher membrane activity against Gram-positive strains displayed by guanidino-containing α-peptide–β-peptoid chimeras as compared to their amino-substituted counterparts. Guanidino groups were shown to considerably improve the capability of antimicrobial peptidomimetics to compromise the integrity of DPPG monolayers mimicking the external leaflet of Gram-positive bacteria cell membranes. These XR data are in excellent agreement with the previously published results.[33, 56] Same trend however was not observed for lipopolysaccharide (Kdo-2 Lipid A) monolayers, which model the outer membrane surface of Gram-negative species. The higher antimicrobial activity of guanidino-containing chimeras in vitro in this case might be due to that fact that Gram-negative bacteria have both outer and cytoplasmic membrane. When passing through the outer (LPS-rich) shell both amino- and guanidino-containing chimeras follow similar self-promoted uptake mechanism, by which bivalent cations are displaced causing a destabilization of the LPS core and the entry into the periplasmic space. However, in order to kill bacteria, it might be not enough just to permeate/damage its outer membrane; the cytoplasmic membrane needs to be affected as well. Because of the high content of phosphatidylglycerol lipid species within the inner membrane guanidino-containing chimeras have better capability to disrupt it that leads to their stronger bactericidal properties. A full explanation of this finding, however, would require extensive experiments beyond the scope of this work. A schematic cartoon illustrating the proposed mechanism of membrane interaction utilized by tested antimicrobials is represented in Fig. 5.
Figure 5. Cartoon schematic of possible interactions of KβNspeK and RβNspeR with (A) DPPG and (B) Kdo-2 Lipid A monolayers at 30 mN × m–1.
Chimeras carrying amino groups are solely located in the polar head-moieties of DPPG accompanied with considerable thinning of the entire monolayer, whereas their guanidino-substituted analogues form an extra layer on the surface of lipid film resulting in more compact distribution of inserted molecules within the model membrane. Unlike DPPG, the insertion mechanisms of KβNspe and RβNspe into Kdo-2 Lipid A model look nearly identical.
As both types of cation are fully protonated at physiological pH we hypothesize that the ability of the guanidino group to form a more stable bidentate electrostatic interaction with negatively charged phosphodiester moieties affects the DPPG lipids to a greater extent than the more structurally rigid Kdo-2 Lipid A. These findings thus provide fundamental insights that should be useful in the future design of optimized synthetic peptidomimetics with selective antibiotic effects.
Finally, addition of the NBD fluorophore did not significantly reduce the insertion activity of the tested chimeras into model membranes that also correlate with their retained antimicrobial potency in vitro, especially for NBD-KβNspe. Moreover, the NBD-labeled chimeras demonstrated even greater ability to destroy both DPPG and Kdo-2 Lipid A monolayers, than their non-tagged analogues. It is assumed that this is a result of increased lipophilicity of modified molecules due to incorporation of the hydrophobic benzofurazan ring of NBD. The resulting amphiphilic properties may reduce the energy penalties associated with penetration of antimicrobials into hydrophobic core and, thus, favor the disruption of membrane structure. The use of fluorescently tagged AMP mimics might facilitate future cellular localization studies aimed at the elucidation of the mechanism of action of oligomeric AMPs in general.
Supplementary Material
Highlights.
Guanidinium cations promote disruption of bacterial cytoplasmic membranes
The effect of guanidine against Gram-negative outer membrane is negligible
Addition of NBD fluorophore does not reduce membrane activity of antimicrobials
Table 1.
Antimicrobial and Hemolytic Activities of α-Peptide-β-Peptoid Chimeras
| Activity | Target strain | MIC,MBC AND HC10, measurements for selected pathogens(μgxmL-1)a |
|||
|---|---|---|---|---|---|
| KβNspe | NBD-KβNspe | RβNspe | NBD-RβNspe | ||
| MIC | |||||
| Gram-negative | E colib | 64 | 64 | S | 64 |
| E. colic | 128 | 32 | 16 | 64 | |
| K. pneumoniaed | 256 | 128 | 32 | 128 | |
| V. vulnificuse | 16 | 16-32 | 8 | 64 | |
| Gram-positive | S. aureusf | 256 | 64 | 32 | 128 |
| S. epidawidisg | 16-64 | 8-16 | 4 | 16 | |
| S. pyogenesh | 16-32 | 8-16 | 4-8 | 16-32 | |
| B cereusi | 64-128 | 15-32 | 4-8 | 16 | |
| MBC | |||||
| Gram-negative | E. colib | 64 | 64 | 8 | 64 |
| E. colic | 128 | 64 | 16 | 64 | |
| K. pneumoniaed | 256 | 128 | 64 | 128 | |
| V vulnificuse | 32 | 64 | 8 | 64 | |
| Gram-positive | S. aureusf | >256 | 128 | 32 | 128 |
| S. epidermidisg | 32-64 | 8-16 | 4-8 | 16-32 | |
| S. pyogenesh | 16-32 | 8-16 | 4-8 | 16-32 | |
| B cereusi | 128 | 16-32 | 4-8 | 16 | |
| HC10 hRBCs[33] | >500 | ND | >500 | ND | |
MIC = Minimum Inhibitory Concentration: lowest concentration without visible growth; MBC = Minimum Bactericidal Concentration: lowest concentration where cell growth could not be detected by plating. The values are based on two individual experiments conducted in duplicate. HC10=concentration that causes 10% hemolysis, hRBCs = human red blood cells, ND = not determined.
Escherichia coli ATCC 25922.
Escherichia coli AAS-EC-009 [Extended Spectrum Beta-Lactamase (ESBL)- producing clinical sample isolated from a Danish patient in 2007].
K. pneumoniae = Klebsiella pneumoniae ATCC 13883.
V vulnificus = Vibrio vulnificus cmcP6 (clinical isolate provided by Joon Haeng Rhee).[90]
S. aureus = Staphylococcus aureus 8325-4.
S.epidermidis = Staphylocuccus epidermidis RP62A.
S. pyogenes = Streptococcus pyogenes GAS-1 [clinical isolate kindly provided by the Statens Serum Institute, Copenhagen, Denmark].
B. cereus =Bacillus cereus ATCC 11778.
Acknowledgements
We thank Dr. Andrey Ivankin for assistance with synchrotron X-ray scattering experiments, Ms. Anne Hector and Dr. Charlotte H. Gotfredsen for assistance with NMR spectroscopy, and Ms. Tina Gustafsson for technical assistance with UPLC-MS and HRMS. The National Center for Antimicrobials & Infection Control, Statens Serum Institut, Denmark is acknowledged for providing the Danish clinical sample of ESBL-producing E. coli and the S. pyogenes strain.
This work was supported by funds from NIH (R01 AI073892, D.G.), DARPA (W911NF-09-1-378, D.G.), the Danish Research Council for Technology and Production (09-065902 and 09-066098), the Danish Independent Research Council | Natural Sciences (Steno Grant no. 10-080907, C.A.O.), and the Technical University of Denmark. Use of the APS was supported by DOE under contract no. W-31-109-Eng-38. C.A.O. is a Lundbeck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature Biotechnology. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 3.Yeung ATY, Gellatly SL, Hancock REW. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JL. Antimicrobial peptides stage a comeback. Nature Biotechnology. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- 5.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 6.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Ravents DS, Neve S, Ravn B, Bonvin AMJJ, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. Plectasin, a Fungal Defensin, Targets the Bacterial Cell Wall Precursor Lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren S, Wittrup A, Cheng F, Jonsson M, Eklund E, Busch S, Belting M. The human antimicrobial peptide LL-37 transfers extracellular DNA plasmid to the nuclear compartment of mammalian cells via lipid rafts and proteoglycan-dependent endocytosis. Journal of Biological Chemistry. 2004;279:17951–17956. doi: 10.1074/jbc.M311440200. [DOI] [PubMed] [Google Scholar]
- 8.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 9.Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, Wang AK, Lee K, Doria S, Hamill P, Yu JJ, Li YX, Donini O, Guarna MM, Finlay BB, North JR, Hancock REW. An anti-infective peptide that selectively modulates the innate immune response. Nature Biotechnology. 2007;25:465–472. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- 10.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 11.Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 12.Hancock RE. Host defence (cationic) peptides: what is their future clinical potential? Drugs. 1999;57:469–473. doi: 10.2165/00003495-199957040-00002. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers. 2008;90:369–383. doi: 10.1002/bip.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg DA, Hurst MA, Fujii CA, Kung AHC, Ho JF, Cheng FC, Loury DJ, Fiddes JC. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Ch. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu MH, Hancock REW. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. Journal of Biological Chemistry. 1999;274:29–35. doi: 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- 16.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge Y, MacDonald DL, Holroyd KJ, Thornsberry C, Wexler H, Zasloff M. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob Agents Chemother. 1999;43:782–788. doi: 10.1128/aac.43.4.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savoia D, Guerrini R, Marzola E, Salvadori S. Synthesis and antimicrobial activity of dermaseptin S1 analogues. Bioorganic & Medicinal Chemistry. 2008;16:8205–8209. doi: 10.1016/j.bmc.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 20.Su YC, Doherty T, Waring AJ, Puchala P, Hong M. Roles of Arginine and Lysine Residues in the Translocation of a Cell-Penetrating Peptide from C-13, P-31, and F-19 Solid-State NMR. Biochemistry. 2009;48:4587–4595. doi: 10.1021/bi900080d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong M, Su YC. Structure and dynamics of cationic membrane peptides and proteins: Insights from solid-state NMR. Protein Sci. 2011;20:641–655. doi: 10.1002/pro.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gale PA. Special issue: Anion coordination chemistry II - Preface. Coordin Chem Rev. 2006;250:2917–2917. [Google Scholar]
- 23.Mavri J, Vogel HJ. Ion pair formation of phosphorylated amino acids and lysine and arginine side chains: a theoretical study. Proteins. 1996;24:495–501. doi: 10.1002/(SICI)1097-0134(199604)24:4<495::AID-PROT8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Woods AS, Ferre S. Amazing stability of the arginine-phosphate electrostatic interaction. Journal of proteome research. 2005;4:1397–1402. doi: 10.1021/pr050077s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen LT, de Boer L, Zaat SA, Vogel HJ. Investigating the cationic side chains of the antimicrobial peptide tritrpticin: hydrogen bonding properties govern its membrane-disruptive activities. Biochimica et biophysica acta. 2011;1808:2297–2303. doi: 10.1016/j.bbamem.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Yang ST, Shin SY, Lee CW, Kim YC, Hahm KS, Kim JI. Selective cytotoxicity following Arg-to-Lys substitution in tritrpticin adopting a unique amphipathic turn structure. FEBS letters. 2003;540:229–233. doi: 10.1016/s0014-5793(03)00266-7. [DOI] [PubMed] [Google Scholar]
- 27.Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. Electropositive Charge in alpha-Defensin Bactericidal Activity: Functional Effects of Lys-for-Arg Substitutions Vary with the Peptide Primary Structure. Infect Immun. 2009;77:5035–5043. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhle SA, Tam JP. Design of Gram-negative selective antimicrobial peptides. Biochemistry. 2001;40:5777–5785. doi: 10.1021/bi0100384. [DOI] [PubMed] [Google Scholar]
- 29.Nakase I, Okumura S, Katayama S, Hirose H, Pujals S, Yamaguchi H, Arakawa S, Shimizu S, Futaki S. Transformation of an antimicrobial peptide into a plasma membrane-permeable, mitochondria-targeted peptide via the substitution of lysine with arginine. Chem Commun (Camb) 2012;48:11097–11099. doi: 10.1039/c2cc35872g. [DOI] [PubMed] [Google Scholar]
- 30.Tokunaga Y, Niidome T, Hatakeyama T, Aoyagi H. Antibacterial activity of bactenecin 5 fragments and their interaction with phospholipid membranes. Journal of peptide science : an official publication of the European Peptide Society. 2001;7:297–304. doi: 10.1002/psc.317. [DOI] [PubMed] [Google Scholar]
- 31.Saczewski F, Balewski L. Biological activities of guanidine compounds. Expert opinion on therapeutic patents. 2009;19:1417–1448. doi: 10.1517/13543770903216675. [DOI] [PubMed] [Google Scholar]
- 32.Findlay B, Szelemej P, Zhanel GG, Schweizer F. Guanidylation and tail effects in cationic antimicrobial lipopeptoids. PloS one. 2012;7:e41141. doi: 10.1371/journal.pone.0041141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen CA, Ziegler HL, Nielsen HM, Frimodt-Moller N, Jaroszewski JW, Franzyk H. Antimicrobial, hemolytic, and cytotoxic activities of beta-peptoid-peptide hybrid oligomers: improved properties compared to natural AMPs. Chembiochem : a European journal of chemical biology. 2010;11:1356–1360. doi: 10.1002/cbic.201000232. [DOI] [PubMed] [Google Scholar]
- 34.Giuliani A, Pirri G, Nicoletto SF. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent Eur J Biol. 2007;2:1–33. [Google Scholar]
- 35.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Proteolytic Studies of Homologous Peptide and N-Substituted Glycine Peptoid Oligomers. Bioorg Med Chem Lett. 1994;4:2657–2662. [Google Scholar]
- 36.Hamuro Y, Schneider JP, DeGrado WF. De Novo design of antibacterial Œ≤-peptides. J. Am. Chem. Soc. 1999;121:12200–12201. [Google Scholar]
- 37.Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Non-haemolytic beta-amino-acid oligomers. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 38.Arvidsson PI, Frackenpohl J, Ryder NS, Liechty B, Petersen F, Zimmermann H, Camenisch GP, Woessner R, Seebach D. On the antimicrobial and hemolytic activities of amphiphilic beta-peptides. Chembiochem. 2001;2:771–773. doi: 10.1002/1439-7633(20011001)2:10<771::aid-cbic771>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Violette A, Fournel S, Lamour K, Chaloin O, Frisch B, Briand JP, Monteil H, Guichard G. Mimicking helical antibacterial peptides with nonpeptidic folding oligomers. Chem Biol. 2006;13:531–538. doi: 10.1016/j.chembiol.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Tew GN, Liu D, Chen B, Doerksen RJ, Kaplan J, Carroll PJ, Klein ML, DeGrado WF. De novo design of biomimetic antimicrobial polymers. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodson B, Ehrhardt A, Ng S, Nuss J, Johnson K, Giedlin M, Yamamoto R, Moos WH, Krebber A, Ladner M, Giacona MB, Vitt C, Winter J. Characterization of novel antimicrobial peptoids. Antimicrob Agents Chemother. 1999;43:1429–1434. doi: 10.1128/aac.43.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patch JA, Barron AE. Helical peptoid mimics of magainin-2 amide. J Am Chem Soc. 2003;125:12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 44.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Lopez S, Kim HS, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA, Wilcoxen KM, Ghadiri MR. Antibacterial agents based on the cyclic D,L-alpha-peptide architecture. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 46.Dartois V, Sanchez-Quesada J, Cabezas E, Chi E, Dubbelde C, Dunn C, Granja J, Gritzen C, Weinberger D, Ghadiri MR, Parr TR., Jr. Systemic antibacterial activity of novel synthetic cyclic peptides. Antimicrob Agents Chemother. 2005;49:3302–3310. doi: 10.1128/AAC.49.8.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motiei L, Rahimipour S, Thayer DA, Wong CH, Ghadiri MR. Antibacterial cyclic D,L-alpha-glycopeptides. Chem Commun (Camb) 2009:3693–3695. doi: 10.1039/b902455g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radzishevsky IS, Rotem S, Bourdetsky D, Navon-Venezia S, Carmeli Y, Mor A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat Biotechnol. 2007;25:657–659. doi: 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- 49.Zaknoon F, Sarig H, Rotem S, Livne L, Ivankin A, Gidalevitz D, Mor A. Antibacterial Properties and Mode of Action of a Short Acyl-Lysyl Oligomer. Antimicrob Agents Ch. 2009;53:3422–3429. doi: 10.1128/AAC.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olsen CA, Bonke G, Vedel L, Adsersen A, Witt M, Franzyk H, Jaroszewski JW. Alpha-peptide/beta-peptoid chimeras. Org Lett. 2007;9:1549–1552. doi: 10.1021/ol070316c. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda K, DeGrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 52.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. Mimicry of antimicrobial host-defense peptides by random copolymers. J Am Chem Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 53.Nederberg F, Zhang Y, Tan JPK, Xu KJ, Wang HY, Yang C, Gao SJ, Guo XD, Fukushima K, Li LJ, Hedrick JL, Yang YY. Biodegradable nanostructures with selective lysis of microbial membranes. Nat Chem. 2011;3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 54.Vedel L, Bonke G, Foged C, Ziegler H, Franzyk H, Jaroszewski JW, Olsen CA. Antiplasmodial and prehemolytic activities of alpha-peptide-beta-peptoid chimeras. ChemBioChem. 2007;8:1781–1784. doi: 10.1002/cbic.200700385. [DOI] [PubMed] [Google Scholar]
- 55.Foged C, Franzyk H, Bahrami S, Frokjaer S, Jaroszewski JW, Nielsen HM, Olsen CA. Cellular uptake and membrane-destabilizing properties of alpha -peptide/beta -peptoid chimeras: lessons for the design of new cell-penetrating peptides, Biochimica et Biophysica Acta. Biomembranes. 2008;1778:2487–2495. doi: 10.1016/j.bbamem.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 56.Hein-Kristensen L, Knapp KM, Franzyk H, Gram L. Bacterial membrane activity of alpha-peptide/beta-peptoid chimeras: influence of amino acid composition and chain length on the activity against different bacterial strains. BMC Microbiol. 2011;11:144. doi: 10.1186/1471-2180-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gidalevitz D, Ishitsuka Y, Muresan AS, Konovalov O, Waring AJ, Lehrer RI, Lee KY. Interaction of antimicrobial peptide protegrin with biomembranes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6302–6307. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivankin A, Apellaniz B, Gidalevitz D, Nieva JL. Mechanism of membrane perturbation by the HIV-1 gp41 membrane-proximal external region and its modulation by cholesterol. Biochimica et biophysica acta. 2012;1818:2521–2528. doi: 10.1016/j.bbamem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivankin A, Livne L, Mor A, Caputo GA, Degrado WF, Meron M, Lin B, Gidalevitz D. Role of the conformational rigidity in the design of biomimetic antimicrobial compounds. Angew Chem Int Ed Engl. 2010;49:8462–8465. doi: 10.1002/anie.201003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neville F, Cahuzac M, Konovalov O, Ishitsuka Y, Lee KY, Kuzmenko I, Kale GM, Gidalevitz D. Lipid headgroup discrimination by antimicrobial peptide LL-37: insight into mechanism of action. Biophysical journal. 2006;90:1275–1287. doi: 10.1529/biophysj.105.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neville F, Ivankin A, Konovalov O, Gidalevitz D. A comparative study on the interactions of SMAP-29 with lipid monolayers. Biochim Biophys Acta. 2010;1798:851–860. doi: 10.1016/j.bbamem.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neville F, Ishitsuka Y, Hodges CS, Konovalov O, Waring AJ, Lehrer R, Lee KY, Gidalevitz D. Protegrin interaction with lipid monolayers: Grazing incidence X-ray diffraction and X-ray reflectivity study. Soft matter. 2008;4:1665–1674. doi: 10.1039/b718295c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitehouse C, Gidalevitz D, Cahuzac M, Koeppe RE, Ii, Nelson A. Interaction of gramicidin derivatives with phospholipid monolayers. Langmuir : the ACS journal of surfaces and colloids. 2004;20:9291–9298. doi: 10.1021/la048797l. [DOI] [PubMed] [Google Scholar]
- 64.Sarig H, Livne L, Held-Kuznetsov V, Zaknoon F, Ivankin A, Gidalevitz D, Mor A. A miniature mimic of host defense peptides with systemic antibacterial efficacy. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:1904–1913. doi: 10.1096/fj.09-149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaknoon F, Sarig H, Rotem S, Livne L, Ivankin A, Gidalevitz D, Mor A. Antibacterial properties and mode of action of a short acyl-lysyl oligomer. Antimicrobial agents and chemotherapy. 2009;53:3422–3429. doi: 10.1128/AAC.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivankin A, Kuzmenko I, Gidalevitz D. Cholesterol-phospholipid interactions: new insights from surface x-ray scattering data. Physical review letters. 2010;104:108101. doi: 10.1103/PhysRevLett.104.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Losche M. Surface-sensitive X-ray and neutron scattering characterization of planar lipid model membranes and lipid/peptide interactions. Curr Top Membr. 2002;52:117–+. [Google Scholar]
- 68.Schalke M, Losche M. Structural models of lipid surface monolayers from X-ray and neutron reflectivity measurements. Adv Colloid Interface Sci. 2000;88:243–274. doi: 10.1016/s0001-8686(00)00047-6. [DOI] [PubMed] [Google Scholar]
- 69.Jensen TR, Balashev K, Bjornholm T, Kjaer K. Novel methods for studying lipids and lipases and their mutual interaction at interfaces. Part II. Surface sensitive synchrotron X-ray scattering. Biochimie. 2001;83:399–408. doi: 10.1016/s0300-9084(01)01265-2. [DOI] [PubMed] [Google Scholar]
- 70.Parratt LG. Surface studies of solids by total reflection of x-rays. Physical Review. 1954;95:359. [Google Scholar]
- 71.Kjaer K. Some Simple Ideas on X-Ray Reflection and Grazing-Incidence Diffraction from Thin Surfactant Films. Physica B. 1994;198:100–109. [Google Scholar]
- 72.Kjaer K, Alsnielsen J, Helm CA, Tippmankrayer P, Mohwald H. Synchrotron X-Ray-Diffraction and Reflection Studies of Arachidic Acid Monolayers at the Air-Water-Interface. J Phys Chem-Us. 1989;93:3200–3206. [Google Scholar]
- 73.Pedersen JS, Hamley IW. Analysis of Neutron and X-Ray Reflectivity Data by Constrained Least-Squares Methods. Physica B. 1994;198:16–23. [Google Scholar]
- 74.Mohwald H. Phospholipid and Phospholipid-Protein Monolayers at the Air/Water Interface. Annu Rev Phys Chem. 1990;41:441–476. doi: 10.1146/annurev.pc.41.100190.002301. [DOI] [PubMed] [Google Scholar]
- 75.Thoma M, Schwendler M, Baltes H, Helm CA, Pfohl T, Riegler H, Mohwald H. Ellipsometry and X-ray reflectivity studies on monolayers of phosphatidylethanolamine and phosphatidylcholine in contact with n-dodecane, n-hexadecane, and bicyclohexyl. Langmuir. 1996;12:1722–1728. [Google Scholar]
- 76.Konovalov OV, Feigin LA, Shchedrin BM. Statistical evaluation of the accuracy of structure parameter determination from x-ray and neutron reflectivity data. Kristallografiya+ 1996;41:640–643. [Google Scholar]
- 77.Konovalov OV, Feigin LA, Shchedrin BM. Allowance for apparatus distortions in modeling the structure of Langmuir-Blodgett films from reflectivity data. Kristallografiya+ 1996;41:629–634. [Google Scholar]
- 78.Samoilenko II, Konovalov OV, Feigin LA, Shchedrin BM, Yanusova LG. Processing of experimental reflectivity data within the REFLAN software package. Crystallogr Rep+ 1999;44:310–318. [Google Scholar]
- 79.Danauskas SM, Li D, Meron M, Lin B, Lee KYC. Stochastic fitting of specular X-ray reflectivity data using StochFit. Journal of Applied Crystallography. 2008;41:1187–1193. [Google Scholar]
- 80.Jensen TR, Kjaer K. Structural properties and interactions of thin films at the air-liquid interface explored by synchrotron X-ray scattering. Studies in Interface Science. 2001;11:205–254. [Google Scholar]
- 81.Als-Nielsen J, Jacquemain D, Kjaer K, Leveiller F, Lahav M, Leiserowitz L. Principles and Applications of Grazing-Incidence X-Ray and Neutron-Scattering from Ordered Molecular Monolayers at the Air-Water-Interface. Phys Rep. 1994;246:252–313. [Google Scholar]
- 82.Jacquemain D, Wolf SG, Leveiller F, Deutsch M, Kjaer K, Als - Nielsen J, Lahav M, Leiserowitz L. Two - Dimensional Crystallography of Amphiphilic Molecules at the Air–Water Interface. Angewandte Chemie International Edition in English. 1992;31:130–152. [Google Scholar]
- 83.Sarig H, Livne L, Held-Kuznetsov V, Zaknoon F, Ivankin A, Gidalevitz D, Mor A. A miniature mimic of host defense peptides with systemic antibacterial efficacy. Faseb J. 2010;24:1904–1913. doi: 10.1096/fj.09-149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brockman H. Lipid monolayers: why use half a membrane to characterize protein-membrane interactions? Curr Opin Struc Biol. 1999;9:438–443. doi: 10.1016/S0959-440X(99)80061-X. [DOI] [PubMed] [Google Scholar]
- 85.Bonke G, Vedel L, Witt M, Jaroszewski JW, Olsen CA, Franzyk H. Dimeric building blocks for solid-phase synthesis of alpha-peptide-beta-peptoid chimeras. Synthesis-Stuttgart. 2008:2381–2390. [Google Scholar]
- 86.Laursen JS, Engel-Andreasen J, Fristrup P, Harris P, Olsen CA. Cis-Trans Amide Bond Rotamers in beta-Peptoids and Peptoids: Evaluation of Stereoelectronic Effects in Backbone and Side Chains. Journal of the American Chemical Society. 2013;135:2835–2844. doi: 10.1021/ja312532x. [DOI] [PubMed] [Google Scholar]
- 87.Diaz-Mochon JJ, Bialy L, Bradley M. Full orthogonality between Dde and Fmoc: the direct synthesis of PNA--peptide conjugates. Org Lett. 2004;6:1127–1129. doi: 10.1021/ol049905y. [DOI] [PubMed] [Google Scholar]
- 88.Chan WC, Bycroft BW, Evans DJ, White PD. A Novel 4-Aminobenzyl Ester-Based Carboxy-Protecting Group for Synthesis of Atypical Peptides by Fmoc-Bu(T) Solid-Phase Chemistry. J Chem Soc Chem Comm. 1995:2209–2210. [Google Scholar]
- 89.Bernatowicz MS, Wu YL, Matsueda GR. Urethane Protected Derivatives of 1-Guanylpyrazole for the Mild and Efficient Preparation of Guanidines. Tetrahedron Letters. 1993;34:3389–3392. [Google Scholar]
- 90.Kim YR, Lee SE, Kim CM, Kim SY, Shin EK, Shin DH, Chung SS, Choy HE, Progulske-Fox A, Hillman JD, Handfield M, Rhee JH. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun. 2003;71:5461–5471. doi: 10.1128/IAI.71.10.5461-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.