Abstract
Background
Mastocytosis associated with germline KIT activating mutations is exceedingly rare. We report the unique clinicopathologic features of a patient with systemic mastocytosis caused by a de novo germline KIT K509I mutation.
Objectives
To investigate the impact of the germline KIT K509I mutation on human mast cell development and function.
Methods
Primary human mast cells derived from CD34+ peripheral blood progenitors were examined for growth, development, survival and IgE-mediated activation. In addition, a mast cell transduction system which stably expressed the KIT K509I mutation was established.
Results
KIT K509I biopsied mast cells were round, CD25(−) and well differentiated. KIT K509I progenitors, cultured in SCF, demonstrated a ten-fold expansion compared to progenitors from healthy subjects and developed into mature, hypergranular mast cells with enhanced antigen-mediated degranulation. KIT K509I progenitors cultured in the absence of SCF survived, however lacked expansion and developed into hypogranular mast cells. A KIT K509I mast cell transduction system revealed the SCF-independent survival to be reliant on the preferential splicing of KIT at the adjacent exonic junction.
Conclusion
Germline KIT mutations associated with mastocytosis drive a well-differentiated mast cell phenotype, distinct to that of somatic KIT D816V disease, whose oncogenic potential may be influenced by SCF and selective KIT splicing.
Clinical Implications
Mastocytosis associated with reported germline KIT activating mutations, in this case KIT K509I, display a mature, well-differentiated mast cell phenotype distinct to that of somatic KIT D816V disease.
Keywords: KIT, K509I, mastocytosis, germline, mast cells, well-differentiated
INTRODUCTION
Systemic mastocytosis is a myeloproliferative neoplasm characterized by the pathologic expansion and infiltration of mast cells within tissues.(1–3) Affecting mainly adults, the onset is sporadic and often associated with acquired gain-of-function mutations in the receptor tyrosine kinase, KIT. KIT signaling through its ligand, stem cell factor (SCF), influences mast cell proliferation, activation and differentiation. The most common somatic mutation, KIT D816V, is located in the second intracellular tyrosine kinase domain, induces SCF-independent activation and is observed in greater than 90% of adult patients with systemic mastocytosis.(4)
Rarely, mastocytosis may be associated with germline KIT mutations, as underscored by seven reports in the literature.(5–11) The inheritance pattern is generally autosomal dominant and a consequence of non-synonymous point mutations involving either the extracellular, transmembrane or juxtamembrane regions of KIT. These mutations are thought to enhance KIT dimerization and/or impair kinase regulation; while generally maintaining sensitivity to the tyrosine kinase inhibitor, imatinib. An exception is the recent report of a family with cutaneous mastocytosis accompanied by a germline KIT N822I mutation.(10) This mutation is located within the kinase domain and was found to be resistant to imatinib. A germline KIT D816V mutation to date has not been reported.
Cell culture systems to effectively study the primary mast cells of patients with mastocytosis are lacking; mainly due to the limited recovery of neoplastic mast cells from tissues and a lack of significant clonal expansion ex vivo. Therefore, studies to understand the effects of KIT activating mutations in vitro have relied primarily on mast cell lines or transduction experiments, often in non-mast cell lineages. Although much has been learned utilizing these alternative approaches, the capacity to expand and study primary mast cells from patients with mastocytosis is favored.
In this study, we report the unique clinicopathologic features of well-differentiated systemic mastocytosis (WDSM) driven by a de novo germline KIT K509I mutation. WDSM is a rare variant of systemic mastocytois characterized by compact aggregates of mature, round, fully granulated mast cells in the bone marrow; lacking the KIT D816V mutation, as well as the aberrant expression of CD25/CD2 markers.(12–15) The germline nature of this presentation permitted the growth of KIT K509I CD34+ derived human mast cells (HuMCs) from the patient. The HuMCs displayed a mature phenotype with enhanced proliferation, granulation and activation. Moreover, SCF-independent growth and development was determined to be dependent on the preferential splicing of KIT. We propose that the activating potential of germline KIT mutations may retain significant ligand and molecular regulation, thus resulting in a well-differentiated HuMC phenotype.
MATERIALS
Patient
The patient is a white female who, at the age of 6 weeks, was reportedly diagnosed with cutaneous mastocytosis after developing “blisters” on her skin. Throughout childhood, she reported sporadic flushing, pruritus and urticaria (Figure 1A). In addition, she reported recurrent episodes of abdominal discomfort requiring hospitalization on three occasions. By the age of 19, her gastrointestinal symptoms regressed and skin symptoms stabilized to the point of requiring no antihistamines. At 22 years old, she developed significant morning stiffness and arthralgia involving her hands, shoulders and knees; she was subsequently diagnosed with seronegative rheumatoid arthritis.
FIG. 1.
Clinicopathological features of germline KIT K509I well-differentiated systemic mastocytosis. A, Cutaneous presentation as an infant. B, Diffuse cutaneous presentation as an adult with comparison to typical KIT D816V urticarial pigmentosa. C, KIT and CD25 staining of bone marrow mast cells with comparison to typical KIT D816V morphology. D, Diagram of KIT and location of the heterozygous K509I mutation in relationship to GNNK splice site.
At the age of 24, the patient’s mastocytosis-related symptoms flared after moving to Arizona. Symptoms included diarrhea, abdominal pain, musculoskeletal pains, flushing and headaches. Her skin displayed a diffuse pattern of involvement, erythrodermic in nature, accompanied by scattered nodules (subcutaneous benign lipoma) and significant pruritus. This diffuse cutaneous presentation is in contrast to the urticaria pigmentosa classically observed in adult onset KIT D816V systemic mastocytosis (Figure 1B). A bone marrow exam revealed 90% cellularity (almost entirely mast cells) and the aspirate was 75% mast cells with round nuclei and variable granularity. The bone marrow mast cells displayed no evidence of “spindling” or CD25 expression (Figure 1C). A serum total tryptase level was 189 ng/ml. A diagnosis of indolent systemic mastocytosis was established according WHO criteria(2, 3) and the disease was further defined as WDSM. Sanger sequencing of KIT was performed after initial KIT D816V mutation testing was negative. A heterozygous KIT K509I mutation was identified in the cDNA of the bone marrow mononuclear cells (Figure 1D), as well as the gDNA of peripheral blood mononuclear cells, buccal mucosa and hair samples (Supplemental Figure 1). This mutation was not detected in either parent and suggests the KIT K509I mutation was a de novo germline event.
The patient’s significant symptoms, bone marrow mast cell involvement, and known sensitivity of the KIT K509I mutation to imatinib(9) prompted a trial of imatinib 300 mg daily. Although an encouraging decrease in her serum tryptase was noted (57 ng/ml), she temporarily discontinued imatinib secondary to an exacerbation of her headaches. Imatinib was restarted at 100 mg daily and upon increasing to 200 mg daily, her headaches and skin erythema worsened (Figure 2A) requiring imatinib to be held. To control the reactions, prednisone 40 mg daily was started one week prior to the re-initiation of imatinib 100 mg every other day. Prednisone was tapered weekly by 10 mg increments until reaching a stable regimen of predisone 10 mg daily and imatinib 100 mg every other day. At this time, mast cells involved approximately 25% of the bone marrow, 10% of the aspirate and her serum total tryptase level was 37.2 ng/ml. Despite these measures, imatinib was ultimately discontinued by the patient due to intolerance and a desire to possibly conceive.
FIG. 2.
Clinical reaction and response to imatinib. A, Erythematous rash exaggerated by the initiation of imatinib. B, Serum total tryptase levels upon discontinuation and reinitiation of imatinib. C, Tryptase staining of bone marrow biopsy demonstrating mast cell involvement pre and post imatinib.
The patient received symptoms based treatment under guarded observation for approximately 3 years. Following an initial increase, her tryptase level plateaued at approximately 100 ng/ml (Figure 2B). However, her bone marrow mast cell involvement progressively increased to 50%, accompanied by increased daily flushing, pruritus and severe bone pain, limiting her daily activity. Aspirin, Gastrocrom and UVA/UVB phototherapy were added to her antihistamine regimen with minimal relief. At 28 years old, imatinib was restarted at 50 mg daily and was gradually increased over 4 months. She ultimately achieved 100 mg daily which resulted in a normal tryptase level (Figure 2B); clearance of the bone marrow mast cells (Figure 2C), and a modest reduction in symptoms.
All patients and healthy donors provided informed consent on NIH IRB-approved protocols. (NCT00044122, NCT00050193 and NCT00001756)
Mutational analysis
Total RNA, cDNA and gDNA was prepared as described.(16) Overlapping KIT PCR amplification products were purified and directly sequenced by Macrogen USA, Rockville MD. Sequencing data was analyzed using Sequencher (Version 4.5, Softgenetics). Detection of the KIT D816V mutation was assessed by PCR/RFLP as described.(17)
CD34+ derived human mast cell (HuMC) cultures
Leukapheresis and enrichment of peripheral CD34+ cells was performed as described (18) with the exception that a mobilizing agent (G-CSF) was not administered to the patient. The percent of enriched CD34+ cells obtained by leukapheresis was determined using a FITC-conjugated anti-CD34+ antibody (BD Biosciences, San Jose, CA) on a FACSCalibur (BD Biosciences) with CellQuest 3.3 software (BD Biosciences). HuMCs were cultured in StemPro complete media including L-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 mg/ml), IL-3 (30 ng/ml, first week only) and IL-6 (100 ng/ml) in the presence or absence of SCF (100 ng/ml) (Pepro Tech, Rocky Hill, NJ) using an equal starting number of CD34+ progenitor cells. (19) For all HuMC studies, different healthy donors were used as controls for each set of experiments. HMC 1.1, HMC1.2 and LAD2 mast cell lines were cultured as described. (20, 21)
Light and electron microscopy (EM)
Cytospin preparations followed by toluidine blue staining were performed using standard protocols.(18) For EM, HuMCs grown on 13 mm thermanox coverslips (Nunc, Naperville, IL) were fixed overnight at 4° C with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7. Samples were post-fixed 30 min with 0.5% osmium tetroxide/0.8% potassium ferricyanide in 0.1 M sodium cacodylate, 1 h with 1% tannic acid and overnight with 1% uranyl acetate at 4° C. Samples were dehydrated with a graded ethanol series, and embedded in Spurr’s resin. Thin sections were cut with a Leica EM UC6 ultramicrotome (Leica, Vienna, Austria), and stained with 1% uranyl acetate and Reynold’s lead citrate prior to viewing at 120 kV on a Tecnai BT Spirit transmission electron microscope (FEI, Hillsboro, OR). Digital images were acquired with a Hammamatsu XR-100 side mount digital camera system (Advanced Microscopy Techniques, Danvers, MA) and processed using Adobe Photoshop (Adobe Systems Inc., San Jose, CA).
Multiparameter flow cytometry
Bone marrow mast cells and HuMCs were analyzed as described(17) using the following antibodies: CD45-PerCP, CD2-PE, CD25-FITC, CD117-APC, CD69-FITC, CD63-FITC, CD203-PE (BD Biosciences) and/or FcεRI-PE (eBioscience, San Diego, CA) with a FACSCanto II flow cytometer (BD Biosciences).
MTT and apoptosis assays
HuMCs were plated at 2 × 105 cells/ml (MTT assay) or 1×105 cells/ml (apoptosis assay) with different concentrations of SCF for 72 h. The MTT assay (Cell Growth Determination Kit, Sigma, St. Louis, MO) was performed at the final 3 h of incubation according to the manufacturer’s instructions. Apoptosis was evaluated using the cellular Annexin V-FITC Apoptosis Kit (Biovision, Mountain View, CA) according to the manufacturer’s instructions.
HuMC antigen-mediated activation assays
HuMCs were sensitized overnight with biotinylated-human IgE (100 ng/ml) in the absence of SCF and subsequently stimulated with various concentrations of streptavidin (referred to as Ag), in the presence or absence of SCF (100 ng/ml) at 37°C for 30 min.(22) β-hexosaminidase (β-hex) release was determined as described.(23) Prostaglandin D2 (PGD2) release was determined using the PGD2-MOX ELISA kit (Cayman Chemical Company, Ann Arbor, MI) according to the manufacturer’s instructions. Ca2+ flux was measured in sensitized and activated Fura 2-AM (2 μM) (Molecular Probes)-loaded HuMCs as described.(24)
IC2 mast cell transduction system
The IL-3 dependent immature murine mast cell line, IC2, expresses FcεRI, however lacks surface KIT expression and exhibits minimal granule formation.(25) KIT WT and D816V associated with GNNK +/− variants were cloned into the pMX-puro retroviral expression vector (Cell BioLabs, San Diego, CA) as described.(26) KIT K509I was subsequently generated using the QuickChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Transduction of IC2 cells (including empty vector control) was performed as described.(26) For metabolism experiments, transduced IC2 cells were washed free of IL-3 and cultured in the presence or absence of SCF (100 ng/ml) for one week. 2 × 105 cells/ml were subsequently incubated in a 96 well-plate for 24 h and the MTT assay performed as above. To assess proliferation, the transduced IC2 cells maintained in SCF were subsequently seeded at 2 × 105 cells/ml in a 96 well plate in the presence or absence of SCF. Viable cells were counted on day 3 using the Cellometer Auto T4 cell counter (Nexcelom Bioscience, Lawrence, MA).
Immunoblotting
For KIT activation experiments, HuMCs sensitized in the absence of SCF overnight were stimulated with SCF (10 ng/ml) at 37°C for 2 min. Cell lysates were prepared and loaded on to 4 – 12% NuPAGE Bis-Tris gels (Invitrogen) for electrophoretic separation and immunoblotting as described.(27) Anti-human c-Kit mAb (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-human phospho-c-Kit (Tyr703) mAb (Cell signaling technology, Danvers, MA ), anti-human CD226 mAb (Santa Cruz Biotechnology) and anti-β-actin mAb (Sigma) were used for immunoblotting. Immunoreactive proteins were visualized with enhanced chemiluminesence ECL (Perkin Elmer Life Sciences, San Jose, CA).
Statistical analysis
Data are presented as the mean ± standard error of mean (SEM). Comparison between two groups was analyzed by unpaired Student t test. P-values <0.05 were considered statistically significant. Analysis was performed using PRISM software, version 5 (GraphPad Software, San Diego, CA). *p < 0.05, **p < 0.01, ***p < 0.0005, **** p < 0.0001
RESULTS
KIT K509I CD34+ progenitors display enhanced proliferation and develop into hypergranular HuMCs
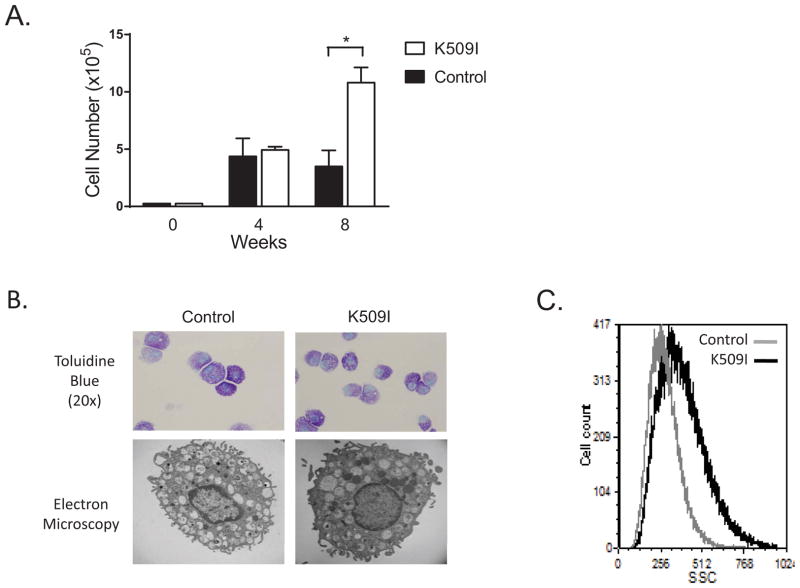
To investigate the impact of the KIT K509I mutation on primary mast cell development, CD34+ progenitors were cultured ex vivo. At 8 weeks, the KIT K509I progenitors cultured in SCF demonstrated a ten-fold expansion compared to progenitors from healthy donors (p < 0.05; Figure 3A). KIT K509I HuMC expansion continued for up to 20 weeks, exceeding normal survival (10–12 weeks). Preservation of the KIT K509I mutation at 8 weeks was confirmed by Sanger sequencing (Supplemental Figure 1). The morphology of the KIT K509I HuMCs was examined by light and electron microscopy (EM). Similar to our bone marrow observations, the KIT K509I HuMCs grown in SCF appeared round and mature as indicated by the well-condensed nuclei, with an apparent increase in granulation by EM (Figure 3B). This hypergranular phenotype was supported by an increase in side scatter when the cells were analyzed by flow cytometry (Figure 3C).
FIG. 3.
KIT K509I CD34+ progenitors display enhanced proliferation and develop into hypergranular HuMCs in the presence of SCF. A, Growth of K509I HuMCs in the presence of SCF. Data are the mean ± SEM of 3 independent experiments performed in duplicate. B, Light microscopy with toluidine blue staining (20x magnification) and electron microscopy. C, Side scatter (SSC) histogram. Representative of 3 experiments.
SCF-depletion impacts KIT K509I HuMC proliferation, development and survival
As HuMCs are dependent on SCF for survival, proliferation, activation and differentiation, we next investigated the oncogenic potential of the KIT K509I HuMCs in the absence of SCF. After 3 days of SCF depletion, a significant decline in the survival of both the control HuMCs (p < 0.0001) and KIT K509I HuMC’s (p < 0.0001) was observed by MTT assay (Figure 4A). However, the KIT K509I HuMCs were less sensitive to SCF depletion and demonstrated a survival advantage over the control HuMCs (85% vs. 26% survival, p < 0.0001). An apoptosis assay, performed in parallel, revealed that SCF withdrawal resulted in minimal apoptosis in the KIT K509I HuMCs (p = 0.055) and a significant induction of apoptosis (p < 0.05) in the control HuMCs (Figure 4B). This observation is further illustrated when the degree of apoptosis in the KIT K509I HuMCs is directly compared to controls (15% vs. 67% apoptosis, p < 0.01). Consistent with these observations, the KIT K509I HuMCs displayed minimal autophosphorylation of KIT in the absence of SCF (Figure 4C).
FIG. 4.
SCF-depletion impacts KIT K509I HuMC proliferation, development and survival. KIT K509I HuMC survival (A) and apoptosis (B) with various SCF concentrations. Data are the mean ± SEM of 3 independent experiments performed in triplicate. C, Immunoblot of KIT phosphorylation pre and post SCF stimulation. Representative of 3 experiments. D, SCF independent KIT K509I HuMC growth. Data are the mean ± SEM of 3 independent experiments performed in duplicate. E, Light microscopy with toluidine blue staining (20x magnification).
With the observation that the KIT K509I HuMCs retained a marginal survival dependence on SCF, we sought to determine if the KIT K509I mutation may support SCF-independent HuMC growth and development. KIT K509I and control CD34+ progenitors were cultured in the absence of SCF for 8 weeks. Only the KIT K509I CD34+ progenitors survived and developed into HuMCs, however these cells displayed minimal expansion by week 8 (Figure 4D). HuMCs accounted for nearly 100% of the culture and appeared mature, although hypogranular (Figure 4E).
KIT K509I transduced IC2 cells are dependent on the KIT GNNK− isoform for SCF independent survival
As a result of alternative mRNA splicing, two major isoforms of KIT are co-expressed in mast cells, characterized by the presence or absence of four amino acids (GNNK) in the juxta-membrane region of the extracellular domain. (26, 28–30) The GNNK− variant is the predominant transcript; as was observed in the patient’s bone marrow mononuclear cells (Figure 1D). The KIT K509I mutation is located adjacent to this GNNK splice region; therefore we hypothesized that the activating potential of the mutation may be influenced by GNNK splicing. We expressed the KIT K509I GNNK− or GNNK+ isoforms in the IL-3 dependent immature mast cell line, IC2, to assess the potential impact on SCF independent growth and survival. As reported, in the presence of SCF, WT KIT metabolism (Figure 5A) and growth (Figure 5C) was enhanced when associated with the GNNK− variant as compared to the GNNK+ variant (p < 0.0005). However, this difference was negated by the presence of either KIT K509I or KIT D816V (Figures 5A, 5C). With SCF absent, WT KIT transduced IC2 cells succumbed, while KIT D816V supported ligand independent metabolism (Figure 5B) and growth (Figure 5D). SCF independent survival of KIT K509I transduced IC2 cells was only observed when associated with the GNNK− isoform (Figure 5B, 5D) and displayed a far weaker oncogenic potential as compared to the KIT D816V mutation. Of note, the GNNK− variant appears to influence KIT D816V ligand independent metabolism (Figure 5B), but not cell proliferation (Figure 5D) as reported.(26)
FIG. 5.
Activating potential of KIT K509I is dependent on the GNNK− isoform. MTT assay of transduced IC2 cells cultured in the (A) presence or (B) absence of SCF. Data are the mean +/− SEM of 3 independent experiments performed in triplicate. Growth and survival of transduced IC2 cells cultured in the (C) presence or (D) absence of SCF. Data are the mean +/− SEM of 2 independent experiments performed in duplicate. Control: empty vector.
KIT K509I HuMCs display enhanced antigen-mediated degranulation, PGD2 release and intracellular calcium flux
Enhanced HuMC degranulation via KIT activation is well recognized.(24, 31) In the presence (Figure 6A) or absence of SCF (Figure 6B), the KIT K509I HuMCs displayed enhanced FcεR1 mediated degranulation with increasing antigen concentrations as compared to controls. No increase in baseline degranulation was observed in the KIT K509I HuMCs in the absence of antigen, irrespective of the presence of SCF. In contrast, SCF alone produced a baseline PGD2 release in both the KIT K509I and control HuMCs (Figure 6C); a reported effect.(32) However, in the absence of SCF, the KIT K509I HuMCs displayed enhanced PGD2 release with increasing antigen concentrations (Figure 6D). Antigen-mediated intracellular calcium flux was also examined in the presence (Figure 6E) or absence of SCF (Figure 6F), with an enhanced signal observed in the KIT K509I HuMCs.
FIG. 6.
KIT K509I HuMCs display enhanced antigen-mediated activation. β-hexosaminidase release (A and B), PGD2 release (C and D) and intracellular Ca2+ flux (E and F) in the presence or absence of SCF. Data are the mean +/− SEM of 3 independent experiments performed in triplicate. Ca2+ flux represents an experiment performed in triplicate. G, FcεR1 surface expression by flow cytometry. Representative of 3 experiments. H, CD226 immunoblotting of whole cell lysates. Representative of 2 experiments.
KIT K509I HuMCs exhibit increased expression of FcεRI and CD226
With enhanced antigen-mediated activation confirmed in the KIT K509I HuMCs, we sought to identify potential contributing features. FACS analysis revealed no difference in CD117, CD25, CD2, CD203c, CD63 or CD69 expression. However, surface expression of the IgE receptor, FcεRI, was markedly increased (Figure 6E). To identify alterations in gene regulation, a limited gene expression array was performed with the KIT K509I HuMCs (not shown). As compared to a control, the gene with the most profound increase in expression (10 fold) was the immunomodulating transmembrane glycoprotein, CD226. CD226 immunoblotting with KIT K509I HuMC lysates confirmed this preliminary finding and minimal CD226 expression was observed in the control HuMCs (Figure 6F). We expanded this analysis and observed moderate CD226 expression in HMC1.1 cells (KIT D816V negative) and a lack of CD226 expression in HMC1.2 cells (KIT D816V positive). LAD2 cells (KIT D816V negative) were found to overexpress CD226, in excess of the positive control, Jurkat cells.
DISCUSSION
In this study, we demonstrate that a germline juxtamembrane KIT K509I activating mutation enhances proliferation and development of mature, round, hypergranular HuMCs which lack the CD2/CD25 activation markers characteristically observed in KIT D816V driven disease. This well-differentiated HuMC phenotype(12, 14, 15) appears common to mastocytosis associated with germline KIT mutations as summarized in Table 1. Interestingly, this shared morphology does not appear entirely dependent on the location of the activating mutation within KIT. Although the mutations tend to cluster in the KIT juxta and transmembrane regions, mutations in the extracellular and tyrosine kinase domains are also reported and suggest other factors may be involved in the tolerance of these oncogenic events.
Table 1.
Mastocytosis associated with germline KIT mutations
| Reference | KIT mutation | Exon | Location | Genotype | Inheritance pattern | Mast cell morphology | CD25 expression | Cutaneous involvement | Systemic progression | Imatinib sensitive |
|---|---|---|---|---|---|---|---|---|---|---|
| Beghini, A. Cancer. 2001;92(3):657–662 | A559V | 10 | JMI | Het | AD | Round, mature | * | Urticaria Pigmentosa | * | * |
| Tang, X. J Med Genet. 2004;41e88 | A533D | 10 | TM | Het | AD | Round, mature | * | Diffuse Cutaneous | yes | * |
| Akin C. Blood. 2004;103(8):3222–3225 | F522C | 10 | TM | Het | De novo | Round, mature, hypergranular | Negative | Urticaria Pigmentosa | yes | yes |
| Hartmann K. Gastroenterology. 2005;129:1042–1046 | Del 419 Asp | 8 | EX | Het | AD | Round, mature∂ | * | Diffuse Cutaneous | yes | * |
| Zhang, LY. Leukemia. Research. 2006;30:373–378 | K509I | 9 | JME | Het | AD | Round, granular | * | Diffuse Cutaneous | yes | yes |
| Wasag B. Exp Hematology. 2011;39:859–865 | N822I | 17 | TK | Het | AD | Round, mature | Negative | Urticaria Pigmentosa | no | no |
| Speight SA. J of Clinical Oncology. 2013; 31:16:e245–e247 | K509I | 9 | JME | Het | De novo | Round, mature | * | Diffuse Cutaneous | yes | no |
Not reported,
Personal communication, JMI= juxtamembrane intracellular, TM=transmembrane, EX=extracellular, JME=juxtamembrane extracellular, TK=tyrosine kinase domain, AD=autosomal dominant
We have herein demonstrated that KIT K509I HuMCs retain a modest dependency on SCF for growth, development and survival. Perhaps germline KIT mutations share a common reliance on SCF and as a result, their oncogenic potential is limited and compatible with a mature HuMC phenotype and long term survival. Consistent with this hypothesis, the KIT K509I HuMCs displayed negligible ligand independent autophosphorylation of KIT and an enhanced response to SCF, as evidenced by increased expansion and activation. It is plausible this hyper-responsiveness coupled with the potential for autocrine SCF production by the KIT K509I HuMCs(33) may account for the survival and protection from apoptosis observed in the “SCF-free” culture conditions. Future studies with the KIT K509I HuMCs, as well as other germline KIT mutations, may define the critical signaling pathways contributing to the enhanced responsiveness to SCF and their association with a well-differentiated phenotype.
Alternative splicing has a recognized role in oncogenesis(34, 35) and the proximity of the K509I mutation to the KIT GNNK splicing region has prompted interest. A normal GNNK− predominance in the patient’s bone marrow mononuclear cells was in agreement with Zhang et al (9), who observed that the K509I mutation did not alter the GNNK splicing profile in a family with mastocytosis. Nonetheless, selective expression of the GNNK isoforms had a significant impact on the KIT K509I oncogenic potential, with SCF-independent survival only achieved when the KIT K509I mutation was associated with the GNNK− isoform in the transduced IC2 mast cells. In contrast, the GNNK isoforms have relatively little influence on the robust oncogenic potential of the KIT D816V mutation.(26, 36) Our observation suggests that the survival, growth and development of the KIT K509I HuMCs in absence of SCF were supported solely by the GNNK− isoform. It is provocative to consider that HuMCs in vivo could regulate the activating potential of the KIT K509I mutation through selective GNNK splicing; however no evidence presently exists to support this conclusion.
In our attempt to identify additional factors contributing to the enhanced degranulation of the KIT K509I HuMCs, we observed increased expression of FcεRI and CD226. Aggregation of surface FcεRI, the high-affinity IgE receptor, is central to antigen mediated HuMC degranulation and it is plausible that upregulation of FcεRI may enhance this activation. CD226 is a transmembrane glycoprotein which mediates cell adhesion, cytokine secretion and cytotoxicity.(37) Moreover, co-engagement of CD226 with FcεRI on mast cells synergistically augments degranulation.(38) Interestingly, CD226 appeared overexpressed in neoplastic mast cell lines lacking the KIT D816V mutation, most profoundly the LAD2 cell line.(23) Taken together, these observations suggest the existence of interplay between CD226, FcεR1 and KIT signaling which enhances mast cell activation.
To our knowledge, this study is the first to evaluate IgE-mediated degranulation and activation of primary mast cells from a patient with mastocytosis. The KIT K509I HuMCs displayed heightened antigen-dependent activation. This data supports the clinical observation that IgE mediated systemic reactions (anaphylaxis) are more prevalent in adult patients with systemic mastocytosis.(39, 40) Indeed, enhanced degranulation was observed at lower antigen concentrations in the KIT K509I HuMCs (most notably in the presence of SCF), however IgE sensitization alone (no antigen) was insufficient to induce basal degranulation in our assays.
In summary, our study contributes to a larger body of evidence suggesting that germline KIT mutations associated with mastocytosis drive a well differentiated mast cell phenotype, markedly different to that of somatic KIT D816V driven disease, whose activating potential may be influenced by SCF and selective KIT splicing.
Supplementary Material
Confirmation of the heterozygous germline KIT K509I mutation in various tissues and cell populations.
Acknowledgments
Funding: This study was supported by the Division of Intramural Research of the NIAID, NIH.
We thank the patient and her family, as well as the LAD and Mayo clinical research staff for their contributions.
Abbreviations
- HuMC
CD34+ derived human mast cell
- SCF
stem cell factor
- EM
electron microscopy
- WDSM
well-differentiated systemic mastocytosis
- PGD2
prostaglandin D2
- NIAID
National Institute of Allergy and Infectious Diseases
- NIH
National Institutes of Health
- IRB
Institutional Review Board
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metcalfe DD. Mast cells and mastocytosis. Blood. 2008;112(4):946–56. doi: 10.1182/blood-2007-11-078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horny H-P, Metcalfe DD, Bennett JM, Bain BJ, Akin C, Escribano L, Valent P. Mastocytosis. In: Swerdlow SH, Campo E, Harris NL, Jaffee ES, Pileri SA, Stein H, Thiele J, Varderman JW, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4. Lyon, France: International Agency for Research on Cancer; 2008. pp. 54–63. [Google Scholar]
- 3.Valent P, Horny HP, Escribano L, Longley BJ, Li CY, Schwartz LB, et al. Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res. 2001;25(7):603–25. doi: 10.1016/s0145-2126(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.Nagata H, Okada T, Worobec AS, Semere T, Metcalfe DD. c-kit mutation in a population of patients with mastocytosis. Int Arch Allergy Immunol. 1997;113(1–3):184–6. doi: 10.1159/000237541. [DOI] [PubMed] [Google Scholar]
- 5.Beghini A, Tibiletti MG, Roversi G, Chiaravalli AM, Serio G, Capella C, et al. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer. 2001;92(3):657–62. doi: 10.1002/1097-0142(20010801)92:3<657::aid-cncr1367>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 6.Tang X, Boxer M, Drummond A, Ogston P, Hodgins M, Burden AD. A germline mutation in KIT in familial diffuse cutaneous mastocytosis. J Med Genet. 2004;41(6):e88. doi: 10.1136/jmg.2003.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akin C, Fumo G, Yavuz AS, Lipsky PE, Neckers L, Metcalfe DD. A novel form of mastocytosis associated with a transmembrane c-kit mutation and response to imatinib. Blood. 2004;103(8):3222–5. doi: 10.1182/blood-2003-11-3816. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann K, Wardelmann E, Ma Y, Merkelbach-Bruse S, Preussner LM, Woolery C, et al. Novel germline mutation of KIT associated with familial gastrointestinal stromal tumors and mastocytosis. Gastroenterology. 2005;129(3):1042–6. doi: 10.1053/j.gastro.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 9.Zhang LY, Smith ML, Schultheis B, Fitzgibbon J, Lister TA, Melo JV, et al. A novel K509I mutation of KIT identified in familial mastocytosis-in vitro and in vivo responsiveness to imatinib therapy. Leuk Res. 2006;30(4):373–8. doi: 10.1016/j.leukres.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Wasag B, Niedoszytko M, Piskorz A, Lange M, Renke J, Jassem E, et al. Novel, activating KIT-N822I mutation in familial cutaneous mastocytosis. Exp Hematol. 2011;39(8):859–65. e2. doi: 10.1016/j.exphem.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Speight RA, Nicolle A, Needham SJ, Verrill MW, Bryon J, Panter S. Rare, germline mutation of KIT with imatinib-resistant multiple GI stromal tumors and mastocytosis. J Clin Oncol. 2013;31(16):e245–7. doi: 10.1200/JCO.2012.42.0133. [DOI] [PubMed] [Google Scholar]
- 12.Akin CEL, Nunez R, et al. Well-differentiated systemic mastocytosis: A new disease variant with mature mast cell phenotype and lack of codon 816 c-kit mutations. Journal of Allergy and Clinical Immunology. 2004;113(S327) [Google Scholar]
- 13.Teodosio C, Garcia-Montero AC, Jara-Acevedo M, Sanchez-Munoz L, Alvarez-Twose I, Nunez R, et al. Mast cells from different molecular and prognostic subtypes of systemic mastocytosis display distinct immunophenotypes. J Allergy Clin Immunol. 2010;125(3):719–26. 26 e1–26 e4. doi: 10.1016/j.jaci.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Munoz L, Alvarez-Twose I, Garcia-Montero AC, Teodosio C, Jara-Acevedo M, Pedreira CE, et al. Evaluation of the WHO criteria for the classification of patients with mastocytosis. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(9):1157–68. doi: 10.1038/modpathol.2011.84. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Twose I, Gonzalez P, Morgado JM, Jara-Acevedo M, Sanchez-Munoz L, Matito A, et al. Complete response after imatinib mesylate therapy in a patient with well-differentiated systemic mastocytosis. J Clin Oncol. 2012;30(12):e126–9. doi: 10.1200/JCO.2011.38.9973. [DOI] [PubMed] [Google Scholar]
- 16.Wilson TM, Maric I, Simakova O, Bai Y, Chan EC, Olivares N, et al. Clonal analysis of NRAS activating mutations in KIT-D816V systemic mastocytosis. Haematologica. 2011;96(3):459–63. doi: 10.3324/haematol.2010.031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maric I, Robyn J, Metcalfe DD, Fay MP, Carter M, Wilson T, et al. KIT D816V-associated systemic mastocytosis with eosinophilia and FIP1L1/PDGFRA-associated chronic eosinophilic leukemia are distinct entities. J Allergy Clin Immunol. 2007;120(3):680–7. doi: 10.1016/j.jaci.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 18.Radinger M, Jensen BM, Kuehn HS, Kirshenbaum A, Gilfillan AM. Generation, isolation, and maintenance of human mast cells and mast cell lines derived from peripheral blood or cord blood. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 7. Chapter 7. 2010. p. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13) Blood. 1999;94(7):2333–42. [PubMed] [Google Scholar]
- 20.Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res. 1988;12(4):345–55. doi: 10.1016/0145-2126(88)90050-1. [DOI] [PubMed] [Google Scholar]
- 21.Kirshenbaum AS, Akin C, Wu Y, Rottem M, Goff JP, Beaven MA, et al. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res. 2003;27(8):677–82. doi: 10.1016/s0145-2126(02)00343-0. [DOI] [PubMed] [Google Scholar]
- 22.Jensen BM, Beaven MA, Iwaki S, Metcalfe DD, Gilfillan AM. Concurrent inhibition of kit- and FcepsilonRI-mediated signaling: coordinated suppression of mast cell activation. J Pharmacol Exp Ther. 2008;324(1):128–38. doi: 10.1124/jpet.107.125237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woolhiser MR, Okayama Y, Gilfillan AM, Metcalfe DD. IgG-dependent activation of human mast cells following up-regulation of FcgammaRI by IFN-gamma. Eur J Immunol. 2001;31(11):3298–307. doi: 10.1002/1521-4141(200111)31:11<3298::aid-immu3298>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 24.Hundley TR, Gilfillan AM, Tkaczyk C, Andrade MV, Metcalfe DD, Beaven MA. Kit and FcepsilonRI mediate unique and convergent signals for release of inflammatory mediators from human mast cells. Blood. 2004;104(8):2410–7. doi: 10.1182/blood-2004-02-0631. [DOI] [PubMed] [Google Scholar]
- 25.Lunderius C, Xiang Z, Nilsson G, Hellman L. Murine mast cell lines as indicators of early events in mast cell and basophil development. Eur J Immunol. 2000;30(12):3396–402. doi: 10.1002/1521-4141(2000012)30:12<3396::AID-IMMU3396>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Chan EC, Bai Y, Bandara G, Simakova O, Brittain E, Scott L, et al. KIT GNNK splice variants: Expression in systemic mastocytosis and influence on the activating potential of the D816V mutation in mast cells. Exp Hematol. 2013 doi: 10.1016/j.exphem.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai Y, Bandara G, Ching Chan E, Maric I, Simakova O, Bandara SN, et al. Targeting the KIT activating switch control pocket: a novel mechanism to inhibit neoplastic mast cell proliferation and mast cell activation. Leukemia. 2013;27(2):278–85. doi: 10.1038/leu.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi S, Kunisada T, Ogawa M, Yamaguchi K, Nishikawa S. Exon skipping by mutation of an authentic splice site of c-kit gene in W/W mouse. Nucleic acids research. 1991;19(6):1267–71. doi: 10.1093/nar/19.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crosier PS, Ricciardi ST, Hall LR, Vitas MR, Clark SC, Crosier KE. Expression of isoforms of the human receptor tyrosine kinase c-kit in leukemic cell lines and acute myeloid leukemia. Blood. 1993;82(4):1151–8. [PubMed] [Google Scholar]
- 30.Piao X, Curtis JE, Minkin S, Minden MD, Bernstein A. Expression of the Kit and KitA receptor isoforms in human acute myelogenous leukemia. Blood. 1994;83(2):476–81. [PubMed] [Google Scholar]
- 31.Jensen BM, Metcalfe DD, Gilfillan AM. Targeting kit activation: a potential therapeutic approach in the treatment of allergic inflammation. Inflamm Allergy Drug Targets. 2007;6(1):57–62. doi: 10.2174/187152807780077255. [DOI] [PubMed] [Google Scholar]
- 32.Lewis A, Wan J, Baothman B, Monk PN, Suvarna SK, Peachell PT. Heterogeneity in the responses of human lung mast cells to stem cell factor. Clin Exp Allergy. 2013;43(1):50–9. doi: 10.1111/cea.12045. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Anderson DF, Bradding P, Coward WR, Baddeley SM, MacLeod JD, et al. Human mast cells express stem cell factor. J Pathol. 1998;186(1):59–66. doi: 10.1002/(SICI)1096-9896(199809)186:1<59::AID-PATH140>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & development. 2010;24(21):2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal S, Gupta R, Davuluri RV. Alternative transcription and alternative splicing in cancer. Pharmacology & therapeutics. 2012;136(3):283–94. doi: 10.1016/j.pharmthera.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen M, Ronnstrand L, Sun J. The c-Kit/D816V mutation eliminates the differences in signal transduction and biological responses between two isoforms of c-Kit. Cell Signal. 2009;21(3):413–8. doi: 10.1016/j.cellsig.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z, Jin B. A novel interface consisting of homologous immunoglobulin superfamily members with multiple functions. Cellular & molecular immunology. 2010;7(1):11–9. doi: 10.1038/cmi.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachelet I, Munitz A, Mankutad D, Levi-Schaffer F. Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J Biol Chem. 2006;281(37):27190–6. doi: 10.1074/jbc.M602359200. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez de Olano D, de la Hoz Caballer B, Nunez Lopez R, Sanchez Munoz L, Cuevas Agustin M, Dieguez MC, et al. Prevalence of allergy and anaphylactic symptoms in 210 adult and pediatric patients with mastocytosis in Spain: a study of the Spanish network on mastocytosis (REMA) Clin Exp Allergy. 2007;37(10):1547–55. doi: 10.1111/j.1365-2222.2007.02804.x. [DOI] [PubMed] [Google Scholar]
- 40.Brockow K, Jofer C, Behrendt H, Ring J. Anaphylaxis in patients with mastocytosis: a study on history, clinical features and risk factors in 120 patients. Allergy. 2008;63(2):226–32. doi: 10.1111/j.1398-9995.2007.01569.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of the heterozygous germline KIT K509I mutation in various tissues and cell populations.