Abstract
The organization of chromatin within the nucleus and the regulation of transcription are tightly linked. Recently, mechanisms underlying this relationship have been uncovered. By defining the organizational hierarchy of the genome, determining changes in chromatin organization associated with changes in cell identity, and describing chromatin organization within the context of linear genomic features (such as chromatin modifications and transcription factor binding) and architectural proteins (including Cohesin, CTCF, and Mediator), a new paradigm in genome biology was established wherein genomes are organized around gene regulatory factors that govern cell identity. As such, chromatin organization plays a central role in establishing and maintaining cell state during development, with gene regulation and genome organization being mutually dependent effectors of cell identity.
Introduction
Gene regulatory processes that govern the establishment and maintenance of cell identity during development occur within the three-dimensional (3D) space of the nucleus. Following the pioneering work of Job Dekker and colleagues in 20021, elucidation of 3D chromosome folding has been greatly spurred by an expanding suite of chromosome conformation capture (3C)-based techniques, including those leveraging the power of high-throughput sequencing2,3 (summarized in Table 1). These methods jointly rely on cross-linking of spatially juxtaposed chromatin, fragmentation of cross-linked chromatin with restriction endonucleases or sonication, ligation of proximal DNA fragments, and amplification of ligation pairs via PCR, with or without sequencing, allowing for the identification of physically interacting chromatin fragments, with more frequently interacting fragments showing a higher prevalence in the resulting PCR-amplified libraries.
Table 1.
Summary of chromosome conformation capture (3C)-based methods
| Method | Acronym | Range | Description |
|---|---|---|---|
| Chromosome conformation capture | 3C | one-to-few | The first step of 3C-based methods is to covalently cross-link spatially adjacent chromatin segments. Restriction endonuclease digestion and ligation of cross-linked chromatin produces chimeric DNA fragments. PCR primer pairs are designed to amplify chimeric DNA fragments consisting of hypothesized interacting regions. As such, this method requires a priori hypotheses about potential interacting chromatin fragments within a population of cells.1 |
| Circular chromosome conformation capture | 4C | one-to-all | Captures the genome-wide interaction profile (“interactome”) of a single locus (“bait” or “viewpoint”). Following 3C library production, a second round of restriction endonuclease digestion and ligation results in circularized, chimeric DNA products. Inverse PCR primers based on the selected bait fragment are designed to amplify intervening interacting sequences, obviating the need to hypothesize interaction regions.57–59 |
| Chromosome conformation capture carbon copy | 5C | many-to-many | An ensemble version of 3C that produces a matrix of interaction frequencies (“contact map”) within specified regions of interes, by tiling high-throughput-sequencing amenable PCR primer pairs across a number of given regions, allowing for the identification of interactions between any two primer pairs.60 |
| Genome-wide chromosome conformation capture | Hi-C | all-to-all | Allows interactions between any two genomic regions to be interrogated simultaneously to produce genome-wide contact maps. Biotinylated nucleotides are incorporated into ligation junctions during 3C library production. Ligated chromatin is then sonicated and isolated with streptavidin beads for identification of interacting fragments via paired-end sequencing.9,61 |
| Tethered genome-wide chromosome conformation capture | TCC | all-to-all | A Hi-C variant wherein proteins are biotinylated in the initial cross-linked complex and tethered to streptavidin-coated beads. Subsequent Hi-C library generation steps can therefore be performed on immobilized chromatin fragments reducing the possibility of spurious ligations between free-floating chromatin fragments.62 |
| Chromatin interaction analysis by paired-end tag sequencing | ChIA-PET | all-to-all interactions of chromatin fragments that are associated with a protein of interest | A Hi-C variant incorporating a chromatin immunoprecipitation (ChIP) step to capture only interactions between chromatin fragments associated with a protein of interest.63 |
The recent explosion of 3C-based genome organization studies, in combination with widespread mapping of linear genomic features (such as transcription factor binding sites, chromatin modifications, and transcription) in cell types of varying developmental stages and across numerous species, has made it clear that genome organization is an important and dynamic contributor to nuclear processes2–8. In particular, the discovery of various cell type-specific and cell type-invariant organizational features of the mammalian genome and their correlation with transcriptional regulators has offered insights into causal relationships between chromatin organization and gene regulation. At the largest scale, these findings include the spatial segmentation of the nucleus into open, transcriptionally permissive and closed, transcriptionally inert compartments9. Developmentally regulated switches of chromatin segments from the open to the closed compartments allow for the sequestration of transcriptionally repressed developmental genes at the nuclear lamina, ensuring their stable silencing10,11. Cell type-specific longrange interactions between distal genomic regions many megabases (Mb) away on the same chromosome (in cis), or on different chromosomes (in trans), have been identified and occur between genomic regions residing in the same compartment (open or closed)12,13. Genomic regions interacting over long distances often exhibit enrichment for common gene regulatory factors, such as chromatin regulators or transcription factors13–16, and appear to occur between megabase-scale self-associating genomic regions termed topologically associating domains (TADs)17–19. Notably, although their long-range interactions can be developmentally regulated, the linear position of TADs have been argued to be largely cell type-invariant and are evolutionarily conserved17,18, and function to restrict the distance over which enhancer-promoter interactions can occur20. Within TADs, however, enhancer-promoter interactions can change in scope, relevance, and dynamics. Finally, recent work has demonstrated that various architectural proteins, including Cohesin, CTCF, and the Mediator complex, are important for the establishment and maintenance of a variety of cell type -specific and -invariant genome organizational features, including enhancer-promoter contacts and long-range inter-TAD chromatin contacts14,21,22, as well as TAD boundaries17,23,24.
In this review, we focus on the latest findings focusing on 3C-based studies conducted in mouse and human cells that have begun to establish causal links between gene regulation and nuclear architecture, and demonstrated the importance of this coupling to mammalian development. We will pay particular attention to the mounting evidence for the role of developmentally regulated linear chromatin features in organizing the genome in 3D. Importantly, these recent findings suggest that chromatin organization contributes to the maintenance and establishment of cell identity in differentiation and reprogramming processes, making the identification of mechanistic links between chromatin organization and the linear genomic features that determine cell type a vitally important task for future work.
The segregated nucleus: Compartmentalization of nuclear function
The mammalian genome is highly organized within the nucleus. Microscopy-based approaches demonstrated that each chromosome resides within a discrete volume of space known as a chromosome territory (CT), with individual CTs exhibiting minimal overlap25–27. More recently, 3C-based methods have demonstrated a further spatial segregation of the genome between transcriptionally permissive, euchromatic regions, and transcriptionally inert regions enriched for features of constitutive heterochromatin and nuclear lamina association, defined as the A and B, or open and closed compartments, respectively9. Chromatin segments residing in specific compartments interact with themselves, and eschew interactions with segments in the alternative compartment4,9. 3C-based approaches have also identified self-associating chromatin domains of approximately 1Mb in size, termed topologically associating domains (TADs), that appear to be very stable across cell types and species, and are composed of complex networks of enhancer-promoter interactions that are restricted by the domains’ boundaries17,18. These TADs appear to be the fundamental modular unit of chromatin organization.
Thus, the genome is structured in a hierarchical manner with promoter-enhancer interactions occurring within TADs, chromosomes being subdivided into many TADs, and co-localization between TADs composed of similarly transcriptionally permissive or inert chromatin, in cis and in trans, leading to the establishment of A and B compartments, and, at the highest level, chromosomes residing in discrete, minimally overlapping CTs. This organizational hierarchy is conserved across mammalian species and Drosophila15,19, which attests to their importance in nuclear biology. Although the necessary and sufficient components of mammalian TAD boundaries are yet to be identified, highly expressed genes are enriched at these boundaries17. Notably, this finding is echoed even in prokaryotes, where the insertion of a highly expressed gene into the Caulobacter crescentus genome was sufficient to demarcate a TAD-analogous “chromosomal interaction domain” despite the absence of a nucleosome-based chromatin structure28.
As described above, TADs look to be the fundamental building blocks of high-order chromosome organization. However, the position of a given TAD within the 3D space of the nucleus with respect to other TADs, or nuclear structures such as the transcriptionally repressive nuclear lamina, can change during development, supporting a role for TAD localization in cell type specification. Mirroring and expanding microscopy-and genomics-based findings that demonstrated a sequestration of lineage-specific loci to the transcriptionally repressive nuclear lamina10,29,30, Lin et al. mapped global chromatin organization during differentiation of pre-pro-B cells to the pro-B stage. Various genes associated with the nuclear lamina in pre-pro-B cells relocate away from the nuclear periphery to the center of the nucleus, switching from the B to the A compartment, concurrent with differentiation to pro-B cells11. Similarly, during the course of mammalian X-chromosome inactivation in early embryonic development, entire TADs on the X-chromosome relocalize to the nuclear lamina18. These reports suggest that TAD sequestration at the nuclear lamina-associated B compartment is an important genome organization-based mechanism for the establishment or maintenance of lineage restricted gene expression during development11,18. The developmentally regulated switch of TADs between the active and inactive compartments is an extreme example of the modular nature of TAD localization. Across cell types, long-range interactions between TADs (inter-TAD interactions), in both cis and trans, also change within the A and B compartments, respectively13,11,12.
Long-distance relationships: Cell type-specific inter-TAD interactions point to a role for gene regulatory factors in higher order genome organization
Several recent 4C-based studies interrogated changes in genome organization upon differentiation of embryonic stem cells (ESCs) and during reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) by expression of the Yamanaka reprogramming factors Oct4, Sox2, Klf4, and cMyc31. These reports revealed a largescale re-organization of long-range, inter-TAD chromatin contacts of pluripotency loci including the Nanog14,15, Dppa2/413,32, Oct413,22, and Sox215 genes during differentiation, and demonstrated that the ESC-specific organization of the genome is re-established upon reprogramming to iPSCs13–15. This pluripotency-specific organization of the mammalian genome suggested a role for pluripotency-associated gene regulatory networks in the organization of long-range chromatin contacts in ESCs and iPSCs. In support of this idea, genomic regions bound by the master pluripotency transcription factors Oct4, Sox2, and Nanog were found to interact with each other over large distances in the ESC nucleus13–15,21,22 (Figure 1).
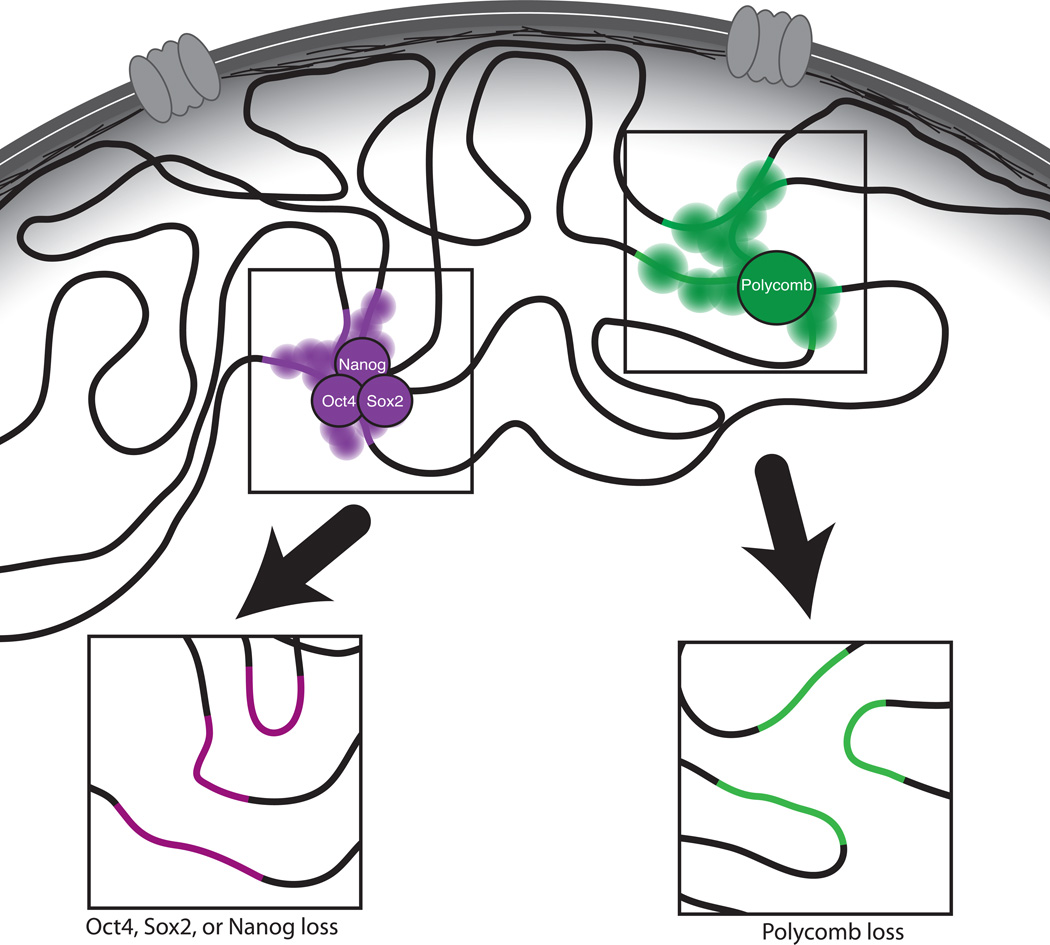
Figure 1. Gene regulatory factors shape inter-TAD chromatin interactions within the ESC nucleus.
Chromatin within the ESC nucleus is compartmentalized based on the preferential co-localization of open, transcriptionally permissive ‘A’ compartment chromatin (white background away from the nuclear periphery) or closed, nuclear lamina-associated ‘B’ compartment chromatin (gray background, nuclear lamina-associated). Within the ‘A’ compartment, genomic regions enriched for binding by pluripotency transcription factors, Mediator, or Cohesin (purple), co-localize, as do regions enriched for Polycomb proteins and the H3K27me3 histone mark (green). Loss off pluripotency transcription factors, Mediator, or Cohesin, or the Polycomb repressive complex 2 (arrows) result in loss of inter-TAD interactions, without disrupting the overall A vs. B compartmental structure of the nucleus.
Similarly, genomic regions enriched for binding by the transcriptionally repressive Polycomb repressive complex 2 (PRC2), which mediates methylation of histone H3 at lysine 27, also co-localize in ESCs, albeit separately from the pluripotency transcription factors13 (Figure 1). Both pluripotency factor and Polycomb-enriched genomic region interactions occur within the context of the A compartment in pluripotent cells13. Specific gene regulatory network-based inter-TAD interactions have also been described within the transcriptionally repressive B compartment in mouse olfactory neurons, within which monogenic olfactory receptor (OR) expression is ensured in part through the formation of OR-exclusive heterochromatic foci formed by aggregation of OR clusters from multiple chromosomes12. Together these results argue for a cell type-specific segregation of genomic compartments based on transcriptionally permissive and inert chromatin, within which specific inter-TAD interactions form between distal regions enriched for similar transcriptional networks (regulators). This in turn begs the question of whether these transcriptional regulators are critical for the formation of these long-range chromatin interactions.
Testing the model wherein particular transcriptional networks drive specific inter-TAD interactions, we found that disruption of the Polycomb/H3K27me3 network by genetic ablation of Eed, a core subunit of PRC2, specifically abolished contacts between genomic regions highly enriched for Polycomb proteins and H3K27me3 in wild-type cells, while not effecting overall chromosome conformation13 (Figure 1). Notably, it was previously shown that the TAD structure within the X chromosome inactivation center is not affected by the Eed knockout18, indicating that different regulatory mechanisms function at different scales of genome organization. The demonstration of Polycomb-dependent chromatin co-localization in mammalian cells echoes findings in Drosophila8,33, suggesting an evolutionarily conserved mechanism of Polycomb-mediated gene silencing and genome organization8.
Supporting a causative relationship between cell type-specific gene regulatory networks and genome organization, loss of Klf422, Nanog14,15, or Oct415 disrupted pluripotency-specific long-range chromatin contacts in pluripotent cells (Figure 1). Furthermore, ectopic recruitment of Nanog to chromatin was sufficient to induce chromatin interactions between the targeted locus and other Nanog-bound regions15. Although these functional studies have made it clear that gene-regulatory factors play causal roles in the establishment and maintenance of chromatin organization, in future studies it will be important to discern between the direct effects of these factors on genome organization and secondary effects due to changes in transcription or chromatin environment upon loss or gain of these factors.
The reprogramming of somatic cells to pluripotency is a useful tool for defining the temporal relationship between the establishment of pluripotency-specific genome organization, pluripotency factor binding, and pluripotency-specific transcription. Analysis of pre-iPSCs, a late reprograming intermediate, showed that pluripotency-specific long-range chromatin interactions are not yet established for pluripotency genes, especially not for those genes that remain inactive and unbound by pluripotency transcription factors in this late intermediate cell type, such as Dppa2 and Zfp4213,32. Another line of experimentation found that pluripotency factor binding at pluripotency genes early during reprogramming is insufficient for induction of gene expression in the absence of intra-chromosomal loops to bring their enhancer and promoters into close proximity34. Interestingly, genomic regions that interact with the Nanog locus in reprogramming intermediates are enriched for the open chromatin mark H3K4me3 and bound by the reprogramming factor Klf4, but, only about half of all genes associated with newly formed 3D-contacts show an increase in expression, either in the intermediate or subsequent fully reprogrammed cells14. Surprisingly, Nanog, itself is not up-regulated in a reprogramming intermediate despite its promoter being looped towards an enhancer already enriched for binding by reprogramming factors at this stage14. Together, these data show that regulatory factor binding and the establishment of distal chromatin interactions correlate with the re-establishment of pluripotency and expression. However, the data also argue that neither binding by key pluripotency factors nor looping alone is always sufficient for the induction of gene expression, indicating the requirement for additional mechanisms for the establishment of the pluripotency transcription program.
The studies introduced thus far suggest a causative relationship between gene regulatory factors and the establishment of 3D chromatin organization, however the requirement of specific inter-TAD chromatin contacts for the induction of gene expression is very difficult to show unequivocally. To this end, Fanucchi and colleagues demonstrated a hierarchy of gene expression among distally located genes35 known to co-localize upon TNF-alpha stimulation16. Among the genes analyzed, SLC6A5 expression is rarely detected without TNFAIP2 and SMAD4A expression, while TNFAIP2 expression is rarely detected without SMAD4A expression, arguing that, for their own expression, genes at the bottom of the hierarchy show a strong reliance on expression of genes above them in the hierarchy35. Remarkably, disruption of the SMAD4A chromatin loop by TALEN-directed double strand DNA break abrogated the expression of both genes lower in the hierarchy, arguing that chromatin loops and co-localization of genes over long distances in cis and in trans are required for gene expression. Similar approaches applied to different interaction scenarios will show how general the requirement for co-localization is for the expression of co-regulated genes.
In summary, the co-localization of distal chromatin fragments bound by members of the same transcriptional network within the 3D space of the nucleus appears to be an important aspect of transcriptional regulation, perhaps due to the resulting increase in the concentration of specific gene regulatory factors at specialized transcription factories36 or Polycomb bodies33. This model also explains how changes in cell identity lead to changes in chromatin organization, as different transcriptional networks bring about the co-localization of different genomic regions during the course of development. How these distal sites find each other and avoid co-localizing with genes regulated by disparate transcription networks within the nuclear volume remains unclear. Another interesting observation is that specific 3D-interactions could be essential for the function of long-noncoding (lnc) RNAs. For instance, we speculated that the interactions observed between Hox clusters could provide the 3D conformation necessary for HOTAIR, a lncRNA transcribed from the HoxC cluster, to find target genes located within the HoxD cluster on a different chromosome, using a mechanism analogous to that employed by another lncRNA, Xist, during X-chromosome inactivation13,37,38.
The logic behind enhancer-promoter-exon looping
Apart from guiding global chromatin organization through the establishment of long-range chromatin contacts, cell type-specific gene regulatory factors also govern short-range enhancer-promoter contacts, forming the foundation for tissue-specific regulation of transcription. Examining promoter interactions in 1% of the genome across three human cell lines (GM12878, K562 and HeLa-S3)39, the ENCODE consortium demonstrated a surprising promiscuity of enhancer-promoter interactions, showing that many promoters in a given cell are contacted by multiple enhancers, and vice-versa, and that gene expression driven from a given promoter positively correlates with the number of enhancers contacting it in a cell population39.
As the primary driver of cell type-specific gene expression, enhancer usage is dynamic during the course of development. Correlation between the chromatin state at enhancers and RNA polymerase II (RNAPII) occupancy at promoters across numerous cell lines allowed for the identification of co-regulated promoters and enhancers20. These enhancer-promoter pairs showed a propensity to cluster linearly in the genome, often falling within TADs, and supporting the model that functional promoter-enhancer interactions are delimited by TAD boundaries20. Genes at TAD boundaries, however, appear to be able to switch their interactions between different TADs. For instance, genes lying at the interface of two TADs within the HoxD cluster switch the set of enhancers with which they interact between sequential TADs, allowing for co-linear gene expression of the HoxD cluster during the course of mouse limb development40. The co-regulation of enhancer chromatin state and RNAPII occupancy, as well as developmentally regulated changes in enhancer usage argues for a role of developmental stage- and cell-specific transcription factors in the orchestration of enhancer promoter contacts.
Within the context of B-cell development, the cell type-specific transcription factors E2A or PU.1, as well as the histone acetyltransferase p300 (indicative of enhancers), are enriched at sites of both intra- and inter-TAD interactions that vary with developmental progression, suggesting that at least some interactions involving enhancer elements can cross TAD boundaries11. In line with these findings, a study by Phillips-Cremins and colleagues showed that Mediator and Cohesin, architectural proteins that are thought to facilitate 3D-chromatin interactions, act together within TAD boundaries to support enhancer-promoter interactions, but are also associated with longer-range interactions21. In the context of stimulus response, enhancers adjacent to 17β-oestradiol-upregulated genes in a human breast cancer cell line exhibited an increase in enhancer–promoter looping upon stimulation, supporting the importance of enhancer-promoter looping in control of gene expression41. Together these findings demonstrate that developmentally and stimulus-driven transcription programs are governed at the level of enhancer-promoter networks within TADs, with rare enhancer-promoter interactions crossing TAD boundaries.
Kieffer-Kwon et al. utilized ChiA-PET to identify 3D-chromatin interactions involving the pre-initiation transcriptional complex at promoters and found differential enhancer utilization across two cell types, not only for tissue-specific genes, but, surprisingly, also for constitutively expressed genes42, implying that highly dynamic enhancer-promoter interactions govern both cell type-specific and cell-type invariant transcriptional programs. A similar approach found that intragenic looping between promoters and exons facilitates alternative splicing in a cell-type-specific manner by bringing promoters and specific exons into close spatial proximity while looping out intronic sequences43. Together, these results suggest that chromatin looping can occur between a variety of genetic elements within a given cell type, linking local genome organization to cis-regulation of both gene expression and alternative splicing.
Remarkably, despite the apparent role for transcription factor-driven enhancer-promoter loops and gene transcription, TNF-α-responsive enhancers are in contact with their target promoters prior to the induction of signaling genome-wide44. This suggests that the 3D chromatin landscape is stable in a given cell type despite signaling activation and that signaling networks act on pre-existing networks of enhancer-promoter contacts. Importantly, this finding also indicates that enhancer-promoter co-localization is insufficient to initiate transcription. A similar case has been made for anti-pause enhancers that regulate promoter-proximal pause release. Binding by the histone demethylase JMJD6 and the bromodomain-containing protein Brd4 appears to occur at pre-established enhancer-promoter contacts which are not disrupted by loss of either of these two factors (Figure 2B)45. This suggests that enhancer-promoter contacts can be established without initiating gene expression, and that JMJD6 and Brd4-mediated pause release is an independently regulated event downstream of enhancer-promoter looping. The mechanism of establishment and maintenance of enhancer-promoter contacts in the absence of transcription may rely on the Mediator complex (see below), whose depletion leads to loss of enhancer-promoter looping at anti-pause enhancers45. The function of enhancer-promoter contacts with regards to the initiation of transcription, and additional factors required to initiate transcription from an enhancer-contacted promoter will be important areas for future study.
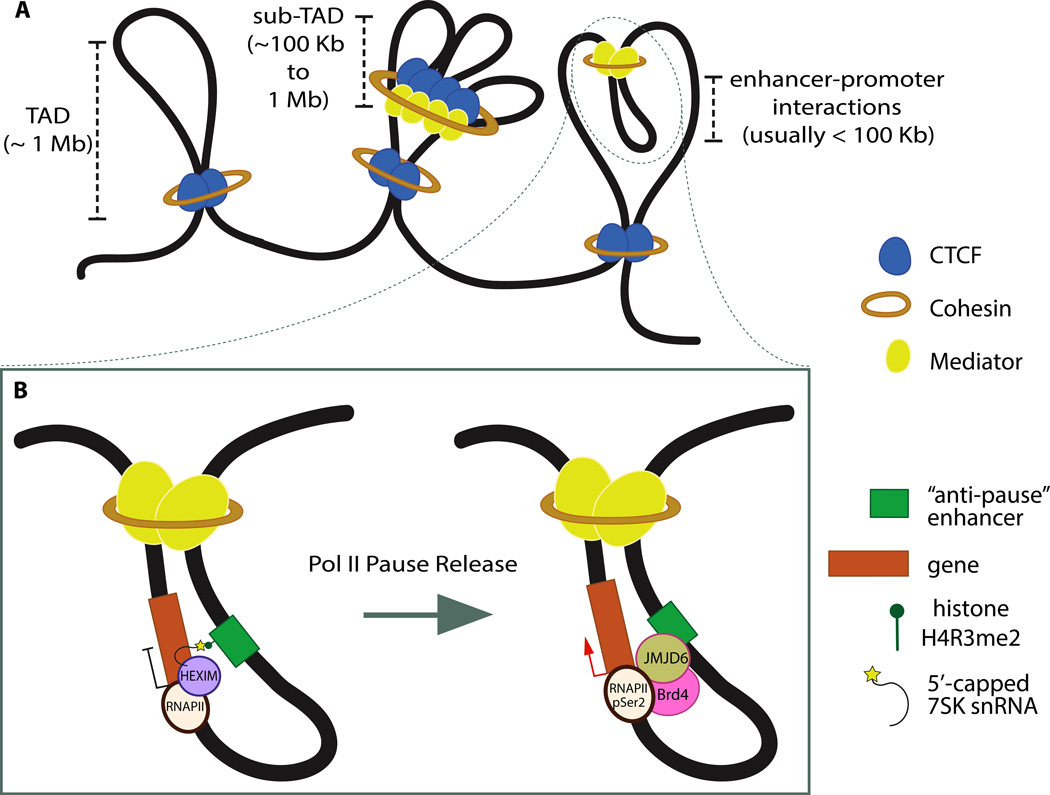
Figure 2. Architectural proteins act combinatorially to organize chromatin at different length-scales.
A) TAD boundaries are enriched for CTCF and Cohesin, but these proteins can also act in combination with other factors, such as Mediator to partition these large Mb-scale TADs into smaller sub-TADs and to facilitate enhancer-promoter interactions.
B) A gene regulatory event involving a constitutive promoter-enhancer interaction. Pre-established Mediator-dependent looping of "anti-pause" enhancers to a target gene promoter is followed by the recruitment of the jumonji C-domain-containing protein 6 (JMJD6) and bromodomain-containing protein 4 (Brd4) complex. Brd4/JMJD6-dependent erasure of H4R3me2 and concomitant decapping/demethylation of 7SK snRNA ensures the release of the 7SK snRNA/HEXIM complex, which inhibits elongation factor P-TEFb, permitting pause release and transcriptional elongation.
The linchpins of looping: architectural proteins and chromatin contacts
The establishment and maintenance of both inter- and intra-TAD chromatin interactions is thought to occur via recruitment of Cohesin, a protein complex that is known for its role in sister chromatid cohesion during mitosis46. Recruitment of Cohesin can occur through transcription factor-mediated recruitment of the Mediator complex and the Cohesin loading factor Nipbl47, allowing for cell type-specific chromatin organization associated with gene-regulatory networks. Cohesin can also be recruited by the insulator protein CTCF48–50, which governs cell type-invariant features of genome organization11,21 and is required for proper Cohesin localization to CTCF-enriched sites51. As such, CTCF, Cohesin, and Mediator act as the “architectural” proteins of the nucleus (Figure 2). In mouse ESCs and neural progenitor cells, CTCF, Cohesin, and Mediator are found at more than 80% of chromatin interactions, as defined by 5C, further supporting the notion that the three proteins play a central role in organizing chromatin28.
Consistent with their role as effectors of cell type invariant features of chromatin organization, TAD boundaries are enriched for CTCF and Cohesin binding17,18. Genes found within chromatin loops anchored by CTCF binding sites often share similar chromatin modifications52, in agreement with the co-regulated nature of genes located within a single TAD20, supporting the idea that gene regulation often acts at the scale of TADs. TAD boundaries are well conserved across mammalian species and cell types17,18, and insulator-binding proteins also serve to delimit distinct chromatin domains in Drosophila53,54 arguing that insulator accumulation at TAD boundaries is an evolutionarily conserved aspect of genome organization. Despite an enrichment at TAD boundaries, CTCF/Cohesin-bound sites are not sufficient to block chromatin interactions18,39, and CTCF binding is not sufficient to demarcate TAD boundaries, as only ~15% of all CTCF binding sites are estimated to be found at TAD boundaries17. Similarly, insulator-binding proteins do not always block chromatin interactions in Drosophila54. Interestingly, CTCF/Cohesin co-occupancy within TADs form chromatin loops at length scales of a few hundred kilobases, leading to the concept of “sub-TADs”, which often form constitutive interactions around developmentally regulated, tissue-specific genes21 (Figure 2A). Together, these results suggest that these architectural proteins can serve as boundaries of interactions of different strength, blocking certain interactions while allowing others dependent on the context.
Knockdown of CTCF not only reduces intra-TAD interactions, but also increases inter-TAD interactions, implying that CTCF depletion results in less well-defined TAD boundaries and more promiscuous short-range chromatin interactions, which are accompanied by alterations in gene expression55. Conversely, disruption of the Cohesin complex via proteolytic cleavage of the Rad21 protein leads to a diminution of intra-TAD interactions, but the TADs themselves remained intact55, demonstrating a role for Cohesin in the maintenance of intra-TAD interactions. In line with this finding, knockdown of a Cohesin subunit in ESCs disrupted an interaction between the Pou5f1 promoter and a neighboring enhancer, causing the loss of self-renewal in pluripotent cells. Extending the functional requirement for Cohesin to inter-TAD interactions, Apostolou and colleagues demonstrated a functional requirement for the Cohesin and Mediator complexes in the re-establishment and maintenance of pluripotency-specific long-range contacts of the Nanog locus upon reprogramming14. Similarly, depletion of Klf4 in ESCs leads to loss of Cohesin loading at the Pou5f1 enhancer, and loss of inter-TAD chromatin contacts that are specific for the pluripotent state22. Supporting a combinatorial role for Cohesin and Mediator in facilitating tissue-specific contacts, Phillips-Cremens and colleagues showed that these two factors act together to facilitate interactions between enhancers and core promoters, mainly within TADs, but also at long-range between TADs21. Altered chromatin conformations and gene expression profiles upon loss of Cohesin do not appear to be due to mitotic defects, as genetic ablation of Cohesin in post-mitotic astrocytes caused decreased intra- and inter-TAD contacts, resulting in profound global architectural changes and extensive misregulation of gene expression. Cohesin deletion did not ablate TAD boundaries, arguing that although Cohesin is required for proper chromatin organization and gene expression, it is not necessary for TAD boundary formation23.
Together, the emerging data suggest that architectural protein-mediated interand intra-TAD chromatin contacts constitute a key mechanism for ensuring the stability of both cell type-specific and cell type-invariant features of mammalian genomic architecture and global gene regulation, and also facilitate changes in genome architecture associated with differentiation (Figure 2).
Completing the loop and looping ahead: future directions
Recent cutting-edge cytological and 3C-based genome-scale research has helped to provide a deeper understanding of the complicated relationship between gene regulation and nuclear architecture in mammalian development. This work has made clear that the linear genomic features that control transcription help to shape the 3D space of the nucleus, and that the 3D organization of chromatin in turn plays a vital role in the regulation of gene expression, and by extension in the maintenance and establishment of cell identity.
Given the strong propensity of genomic regions bound by similar gene regulatory factors to co-localize, it will be important to determine how specific genomic regions locate each other within the space of the nucleus. Complementary work on the mechanisms used to avoid contacts with regions bound by different regulatory factors will also be important. Similarly, defining the molecular events that follow enhancer-promoter contacts and precede initiation of transcription will be important to properly define enhancer action and the relevance of promoter-enhancer interactions to gene expression.
A limitation of 3C-based studies is the requirement for a large population of cells during library preparation, meaning the resulting data represent the average chromatin contacts across the entire ensemble, making it difficult to gauge the relevance and frequency of individual chromatin interactions. Single-cell, genome-wide chromatin contact maps recently recapitulated the domain structure characterized using population-based Hi-C, and showed that inter-TAD and inter-chromosomal contacts are highly variable between individual cells and that active domains were generally found at CT boundaries56. In future studies, it will be important to compare the variability observed for chromosomal interactions with that of gene expression at the single cell level. Matching this work with genome editing approaches able to disrupt and induce specific chromatin interactions35, single cell studies will go a long way towards resolving the direct effect of chromatin organization on gene expression.
Acknowledgements
K.P. is supported by the NIH (P01 GM099134), CIRM (RB3-05080 and RB4-06133), the Jonsson Comprehensive Cancer Center, and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA; G.B. by the Whitcome Pre-doctoral Training Program; and M.D. by pre-doctoral fellowships from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, CIRM, and the UCLA Graduate Division.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 2.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denholtz M, Plath K. Pluripotency in 3D: genome organization in pluripotent cells. Curr Opin Cell Biol. 2012;24:793–801. doi: 10.1016/j.ceb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev. 2014;25C:30–37. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiratani I, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. * Demonstrated a differentiation-dependent, olfactory receptor (OR) gene-exclusive clustering of silenced OR genes in mouse olfactory neurons, required for mono-genic OR expression.
- 13. Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. * Demonstrated that Polycomb-dependent clustering of Polycomb-regulated genomic regions is an evolutionarily conserved aspect of Polycomb-mediated gene regulation and genome organization.
- 14.Apostolou E, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. * Demonstrated that ectopic recruitment of the pluripotency transcription factor Nanog to chromatin was sufficient to induce long-range, inter-TAD chromatin interactions with other genomic regions enriched for binding by Nanog.
- 16.Papantonis A, et al. TNFalpha signals through specialized factories where responsive coding and miRNA genes are transcribed. Embo J. 2012;31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Z, et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. Embo J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem. 2006;50:161–176. [PubMed] [Google Scholar]
- 26.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223–272. [PubMed] [Google Scholar]
- 27.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. ** Used Hi-C in the prokaryote Caulobacter crescentus to demonstrate the existence of TAD-analogous chromosomal interaction domains (CIDs) in this organism and applied chemical and genetic approaches to demonstrate that CIDs are a biophysical property of the prokaryotic genome, dependent upon transcription.
- 29.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 30.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Takebayashi S, et al. Murine esBAF chromatin remodeling complex subunits BAF250a and Brg1 are necessary to maintain and reprogram pluripotency-specific replication timing of select replication domains. Epigenetics Chromatin. 2013;6:42. doi: 10.1186/1756-8935-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13:30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 35. Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155:606–620. doi: 10.1016/j.cell.2013.09.051. ** Demonstrated a requirement for chromatin interactions in the regulation of gene expression by disrupting chromatin contacts between a set of NF-kappaB-coregulated genes, leading to loss of their co-localization and expression.
- 36.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andrey G, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. * Demonstrated that genes encoded within the HoxD locus switch their chromatin interactions between enhancers in contiguous TADs during development, assuring co-linear gene expression during limb specification.
- 41.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. * As part of the NIH Mouse Regulome Project, showed that genes expressed broadly across different cell types utilize cell type-specific enhancers regulated by lineage-specific transcription factors.
- 43. Mercer TR, et al. DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nat Genet. 2013;45:852–859. doi: 10.1038/ng.2677. * Linked chromatin organization to alternative splicing by identifying a subset of exons with DNase hypersensitivity sites that form chromatin loops with their respective promoters. Exons with these features are enriched for alternative splicing events.
- 44. Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. ** Used ultra-deep sequencing of Hi-C data to generate genome-wide contact maps in human cells with enhancer/promoter contact resolution. Surprisingly, they found that many TNF-alpha responsive gene promoters were already in contact with their respective enhancers prior to induction of signaling, and that these pre-existing contacts were predictive of transcriptional activation upon signaling activation.
- 45. Liu W, et al. Brd4 and JMJD6-Associated Anti-Pause Enhancers in Regulation of Transcriptional Pause Release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. ** Demonstrated a mechanism by which paused polymerases could be released from promoters with pre-established promoter-enhancer looping.
- 46.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 52.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Negre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz YB, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. * Developed single-cell Hi-C, using it to demonstrate that the TAD-based structure of the genome is conserved at the single cell level, and that active domains tend to cluster at the surface of chromosome territories.
- 57.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 59.Splinter E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Berkum NL, et al. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp. 2010 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]