Abstract
Growth on several carbon sources is dependent on the catabolite repressor/activator (Cra) protein although a Cra consensus DNA binding site is not present in the control regions of the relevant catabolic operons. We show that Cra regulates growth by activating expression of the crp gene. It thereby mediates catabolite repression of catabolic operons by an indirect mechanism.
Keywords: Transcription factors, genetic pleiotropy, carbon catabolite repression
Introduction
Several mechanisms of carbon catabolite repression are operative in large genome organisms such as Escherichia coli (Saier, 1996) and Bacillus subtilis (Stulke et al., 1998; Stulke and Hillen, 1998; Bruckner and Titgemeyer, 2002). In E. coli, two dominant mechanisms involve transcriptional regulation by the cyclic AMP receptor protein (Crp) encoded by the crp gene, and by the catabolite repressor/activator (Cra) protein, encoded by the fruR gene (Ramseier et al., 1993). Crp generally regulates the initiation of carbon metabolism (Kolb et al., 1993) while Cra more frequently regulates carbon flux through the dominant metabolic pathways (Ramseier et al., 1995). For example, the free (unliganded) form of Cra serves as an activator of catabolite repressible, oxidative Krebs cycle and gluconeogenic enzyme genes, but as a repressor of catabolite activatable, anaerobic glycolytic enzyme genes (Chin et al., 1987). By contrast, the cyclic AMP-complexed form of Crp serves as an activator of almost all operons that initiate the metabolism of the many carbon sources that E. coli is capable of utilizing (Xu and Johnson, 1997; Busby and Ebright, 1999). However, there is extensive overlap as Crp, for example, regulates transcription of most Krebs cycle enzymes while Cra directly regulates the initiation of the metabolism of some sugars (e.g., fructose, allose, etc.) (Ramseier et al., 1993; Ramseier et al., 1995).
We have recently found that Cra influences the expression of many cyclic AMP-Crp activatable genes that initiate carbon metabolism even though no Cra binding site could be identified in the control regions of the operons encoding the relevant transport systems and metabolic enzymes. We here document this phenomenon and provide compelling evidence that it is due to the fact that Cra positively controls expression of the crp gene. Although extensive evidence shows that Crp autoregulates expression of its own structural gene (Hanamura and Aiba, 1992; Ishizuka et al., 1993; Tagami et al., 1995), this is the first evidence that Cra cross controls crp gene expression. It therefore affects catabolite repression of these operons by an indirect secondary rather than a direct primary mechanism.
Materials and methods
The strains used in this study are described in Table 1. All strains were derived from E. coli K12 strain MG1655. The fruR gene was deleted using the method of Datsenko and Wanner (Datsenko and Wanner, 2000), yielding MG1655ΔfruR. To make the chromosomal construct in which the native lacZ gene is driven by the crp promoter (Pcrp), Pcrp was cloned into the SalI and BamHI sites of pKDT (Klumpp et al., 2009), yielding pKDT-Pcrp. Using a site-directed mutagenesis approach, the Cra-binding operator in Pcrp was altered by changing GCTGAAGCGAGACACC to GCTCCTGTTAGACACC, yielding pKDT_PcrpOCra. The Pcrp or PcrpOCra promoter plus the upstream rrnB terminator and the Kmr (kanamycin-resistance) gene (Kmr:rrnBT:Pcrp or Kmr:rrnBT:PcrpOCra) in pKDT was integrated into the chromosome of MG1655ΔlacY (Klumpp et al., 2009) to replace the lacI gene and the native lacZYA promoter, yielding strain MG1655 Pcrp-lacZ or MG1655 PcrpOCra-lacZ. In each of these chromosomal constructs, there are a Kmr gene and an rrnB terminator upstream of the promoter. The chromosomal Pcrp-lacZ and PcrpOCra-lacZ in MG1655 were transferred to MG1655lacYΔfruR by P1 transduction, yielding MG1655ΔfruR Pcrp-lacZ and MG1655ΔfruR PcrpOCra-lacZ, respectively. The oligonucleotides used in this study are listed in Table 1.
Table 1.
Strains and oligonucleotides used in this study
| Strain | Relevant genotype or characteristic | Source or reference |
|---|---|---|
| MG1655 | Wild type | Blattner et al (3) |
| ΔfruR | ΔfruR in MG1655 | This study |
| MG1655ΔlacY | ΔlacY in MG1655 | Klumpp et al (11) |
| MG1655ΔlacY ΔfruR | ΔlacY ΔfruR in MG1655 | This study |
| MG1655 (Pcrp-lacZ) | MG1655ΔlacY carrying Pcrp-lacZ at the lac locus | This study |
| MG1655 ΔfruR (Pcrp-lacZ) | ΔlacY ΔfruR carrying Pcrp-lacZ at the lac locus | This study |
| MG1655 (PcrpOCra-lacZ) | ΔlacY carrying PcrpOCra-lacZ at the lac locus | This study |
| MG1655 ΔfruR (PcrpOCra-lacZ) | ΔlacY ΔfruR carrying PcrpOCra-lacZ at the lac locus | This study |
| Oligonucleotide | Sequence | Use |
| fruR1-P1 | gtgaaactggatgaaatcgctcggctggcgggagtgtcgcggaccactgcgtgtaggctggagctgcttcg | fruR mutation |
| fruR2-P2 | gctacggctgagcacgccgcggcgatagagattacgtttaatgcgcgttacatatgaatatcctccttag | fruR mutation |
| Pcrp-Xho-F | atactcgagcttgcatttttgctactccactg | Cloning Pcrp into pKDT |
| Pcrp-Bam-R | ttaggatccctggtgaataagcgtgctcttggatg | Cloning Pcrp into pKDT |
| Pcrp-Z-P1 | gcatttacgttgacaccatcgaatggcgcaaaacctttcgcggtatgtgtaggctggagctgcttc | Integration of Pcrp-lacZ at the lac locus |
| Pcrp-Z-P2 | gattaagttgggtaacgccagggttttcccagtcacgacgttgtaaaacgacctggtgaataagcgtgctcttggatg | Integration of Pcrp-lacZ at the lac locus |
| Pcrp-F | gttttagcatagctttcgctttgtgtctcctggtgtctaacaggagcatg | Cra operator mutation in Pcrp region |
| Pcrp-R | caccaggagacacaaagcgaaagc | Cra operator mutation in Pcrp region |
Miller’s minimal medium 63 (M63) (Miller, 1972), supplemented with 0.5% (w/v) of various carbon sources, was used for measuring growth rates and promoter activities. Growth experiments were conducted following the method of Scott et al (Scott et al., 2000). Seed cultures were grown in LB broth and used to inoculate appropriate pre-culture media. After overnight growth, pre-cultures were used to inoculate 5-ml of experimental media (at a 1000x dilution) contained in test tubes (20 mm × 25 cm). The tubes were shaken in a 37 °C water bath at 250 rpm. Growth rate was monitored by measuring OD600 in a Bio-rad SmartSpec3000 spectrophotometer over time. At least 5 measurements were made during the exponential growth phase. The slope of the plot of logOD600 versus time (min) was used to derive the growth rate in min/generation.
For Pcrp activity determination, 300 ul of culture samples were taken at least 4 times during exponential growth. β-Galactosidase assays were carried out using the method of Miller (Miller, 1972). The slope of the plot of activity (U/ml) versus the sample OD600 yielded the activity in Miller units (U/ml/OD600). To test if a similar activation effect occurred during the stationary growth phase, samples were also taken to determine Pcrp activity.
To determine cAMP-receptor protein (Crp) levels, a freshly isolated colony from an LB plate of each isolate (MG1655 and MG1655ΔfruR) was used to inoculate 5 ml M63 supplemented with 0.5% (w/v) of xylose. The bacteria were incubated overnight in a shaking water bath at 37°C. The culture obtained was used for inoculation of 1 L of the same medium in a 2 L conical flask. The flasks were incubated on a rotary shaker at 37°C at 275 rpm until an OD600 of about 0.7 was achieved. The cells were harvested by centrifugation in a Sorvall SS-34 rotor at 4°C at 6000 rpm for 20 min, and washed twice with and resuspended in M63. The pellet suspension (15 ml) was lysed by passage 3X through a French Pressure cell at 0°C and 16,000 psi. The lysate produced was centrifuged at 40,000 rpm for 2 h at 4°C using a Beckman Ti70 rotor. The high-speed supernatant (HSS) produced was collected, and its total protein content was determined as described by Aboulwafa and Saier (2008) using the Bio-Rad colorimetric protein reagent (Cat. #500–0006) and bovine serum albumin as the standard protein.
SDS-PAGE was conducted as described by Aboulwafa and Saier (2008) using a stacking gel of 5% acrylamide, and a separating gel of 10% acrylamide. Equal volumes of protein solution and Laemmli sample buffer were placed in a boiling water bath for 3 min, cooled to room temperature, and applied in 30 μl aliquots to a 10 × 7 cm gel. The samples were electrophoresed at 75 V. Proteins were transferred from the gel to nitrocellulose membranes (0.22 μM) as described by Towbin et al. (1979) using tris·glycine buffer/20% methanol, pH 8.3, no SDS, and a voltage of 100 V for 1 h. After electroblotting, the membrane was blocked in a tris buffered saline (TBS) solution containing 5% non fat milk (BioRad) and 0.1% tween 20 for 4 h at 4°C with gentle shaking. The first antibody was rabbit polyclonal antibody raised against a peptide mapping within an internal region of E. coli Crp (Crp (T-14): sc-136636, Santa Cruz Biotechnology, Inc.) and used at a dilution of 1:50 in the same solution for 2 h at room temperature. The second antibody was mouse anti-rabbit IgG-HRP (sc-2357, Santa Cruz Biotechnology, Inc.) and used at a dilution of 1:500 in the same solution for 1 h at room temperature. Detection was performed using SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific Pierce Cat # 34075). Following the protocols described in Cheng et al., (2012), Chua et al., (2009), Sharma et al., (2008) and Sujobert et al., (2005), the Crp protein levels were determined by using the ImageJ software (National Institute of Health, Bethesda, MD) after scanning the film. The amount of Crp from MG1655ΔfruR was normalized to 1.
In order to identify Cra binding sites in the E. coli genome, experimentally-determined 16-base Cra binding site sequences from the following genes were used as input for the GRASP DNA program; fruB, icdA, aceB, epd, ppsA, mtlA, pykF, ptsH, nirB, fbp, pckA, adhE and edd (Schilling et al., 2000). The consensus-weighted matrix used to screen the genome was determined using this program.
Results
Table 2 shows the growth rates of a wild-type and an isogenic fruR mutant in M63 minimal medium containing various carbon sources (0.5%, w/v). As can be seen, the mutant grew at a much slower rate than the isogenic wild-type strain on xylose. The same behavior was observed for several other carbon sources including glycerol, casamino acids (CAA) and lactose. When D-sorbitol or succinate was used as the carbon source, very little or no growth of the ΔfruR mutant was detected. The utilization of gluconate as well as glycerol+CAA was slightly decreased; glucose utilization was only slightly decreased, and fructose utilization was substantially increased by the loss of Cra (Table 2).
Table 2.
Growth rates of E. coli wild type and the isogenic fruR deletion mutant in M63 minimal media with 0.5% (w/v) of various carbon sources.
| Carbon source | Wild type (min/generation) | ΔfruR (min/generation) |
|---|---|---|
| Xylose | 80±3 | 95±4 |
| Glycerol | 89±2 | 130±6 |
| Casamino Acids (CAA) | 84±5 | 129±5 |
| Glycerol + CAA | 58±3 | 63±3 |
| Lactose | 56±3 | 78±4 |
| Sorbitol | 88±3 | >500 |
| Succinate | 119±5 | 468±12 |
| Gluconate | 67±3 | 72±3 |
| Glucose | 58±2 | 61±2 |
| Fructose | 85±2 | 63±4 |
These last observations can be explained because growth of wild type cells in the presence of fructose yields fructose-1-phosphate, the high affinity ligand for Cra, while growth on glucose, the most rapidly utilized sugar in E. coli, and the primary substrate of both the glucose and mannose permeases, should produce the highest level of cytoplasmic fructose-1,6-bisphosphate, the primary low affinity ligand for Cra (Ramseier et al., 1993; Ramseier et al., 1995).
The utilization of xylose, glycerol, lactose, succinate and sorbitol are subject to stringent Crp control as well as weak Cra control (Table 2), but there is no Cra binding site upstream of their catabolic enzyme-encoding operons. It appeared that these regulatory effects might be due to an indirect effect of the loss of Cra on an unidentified regulatory process.
A search for Cra binding sites in the E. coli genome using the GRASP-DNA program (Schilling et al., 2000) revealed genes that could potentially be regulated by this protein. The Cra-like sequence G C T G A A G C G A G A C A C C was found at positions −53 to −37 relative to the primary transcriptional start site of the crp gene. Fig. 1 presents a schematic depiction of the control region of the E. coli crp gene. The Cra binding site overlaps one of the two Crp binding sites, the upstream CrpI site, by two nucleotides. While CrpI activates crp gene expression, CrpII represses expression (Hanamura and Aiba, 1992; Ishizuka et al., 1993; Tagami et al., 1995).
Figure 1.
Control region preceding the crp gene in E. coli. The two Crp binding sites (CrpI and CrpII) are indicated by lines above the sequence while the Cra binding site is underlined. Mutations introduced in the Cra binding site are labeled with letters above the sequence. For the primary promoter (crpP1), the −35 and −10 hexamers as well as the transcriptional start site (+1) are shaded. Two subsequent weak promoters (that are functional under specific conditions) are also indicated +1 (Gonzalez-Gil et al., 1998).
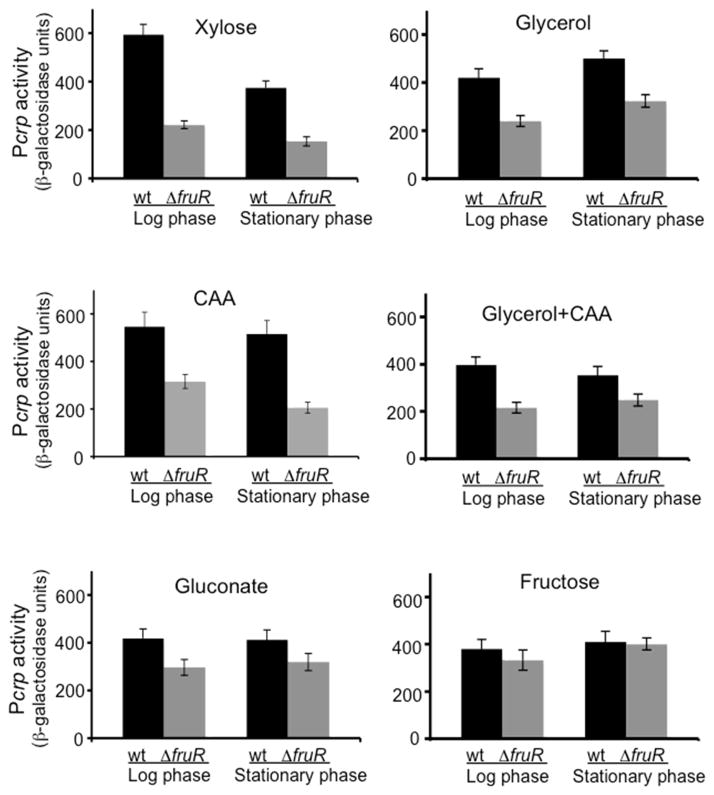
In order to test the possibility that Cra controls transcription of the crp gene, we constructed a chromosomal Pcrp-lacZ fusion in wild type and fruR genetic backgrounds. The results are presented in Fig. 2. Cra activates expression of the crp gene when xylose, glycerol ± CAA or gluconate was used as the sole carbon source. Similar activation effects were observed in cells from the stationary phase. Thus, the loss of this protein gives rise to reduced crp expression. As expected, the Pcrp activity was unaffected by the loss of Cra when cells were grown with fructose since fructose, when converted to fructose-1-P, releases Cra from its operator (Fig. 2; Ramseier et al., 1993).
Figure 2.
The activation of the crp promoter by Cra. The chromosomal Pcrp-lacZ fusion at the lac locus was made. MG1655ΔlacY and MG1655ΔlacYΔfruR cells containing the Pcrp-lacZ fusion were cultured in M63 with 0.5% (w/v) of various carbon sources. For β-galactosidase assays, samples were collected during the exponential growth phase (first two columns) and the stationary phase (last two columns). For brevity, wt = MG1655ΔlacY; ΔfruR = MG1655ΔlacYΔfruR.
These results were confirmed using Western blotting. As shown in Fig. 3, the wild type strain contained higher levels of Crp (lanes 1–4) than did the isogenic fruR mutants (lanes 5–8). The figure shows the results for three independently conducted experiments all of which gave the same results. The relative amounts of the Crp protein in the Western blots were quantified using the ImageJ software (NIH). The ratio of the wild type to the fruR mutant was 1.4 ± 0.1.
Figure 3.
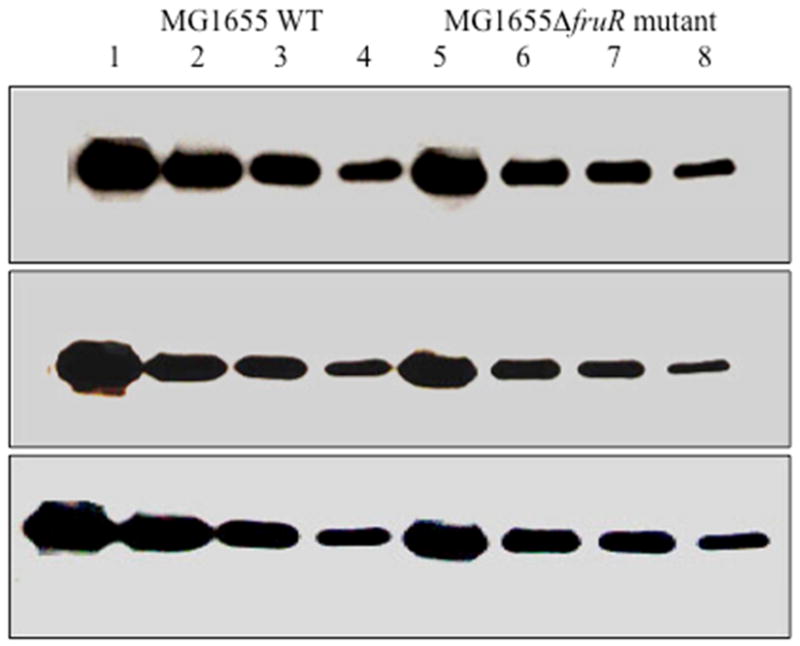
Western blot of Crp in high-speed supernatants prepared from log phase cells of E. coli MG1655 wild type and its fruR deletion mutant MG1655ΔfruR grown in M63 containing 0.5% xylose (see Methods). Lanes 1 – 4 (MG1655 WT) and 5–8 (MG1655ΔfruR) contained 34, 17, 8.5 and 4.25 μg of total protein of the 2 h HSS of the corresponding strain. The amount of the Crp protein in each band was quantified using the ImageJ software (NIH) after scanning (see methods).
Activation of the crp gene by Crp, but not by Cra, had been reported previously (Hanamura and Aiba, 1992; Ishizuka et al., 1993; Tagami et al., 1995). Interestingly, these effects proved not to be additive as the crp fruR double mutant showed the same basal activity as the crp mutant in LB medium (data not shown). These results are in accordance with expectation that gene expression is dependent on a complex of σ70 RNA polymerase with both Crp and Cra. Neither Crp nor Cra alone can fully activate gene expression. Because of the close proximity of the CrpI and Cra binding sites, these two proteins might form a complex without DNA looping. Alternatively or in addition, Cra binding could antagonize the repressive effect of Crp bound to CrpII.
The Cra binding site in the upstream crp operon regulatory region was altered by site specific mutagenesis (see Fig. 1). As shown in Fig. 4, this alteration resulted in decreased expression of the Pcrp-lacZ fusion construct. However, loss of Cra resulted in an even greater loss (Fig. 4). These results are consistent with the conclusion that Cra mediates its effect, at least in part, by binding to the proposed Cra binding site.
Figure 4.
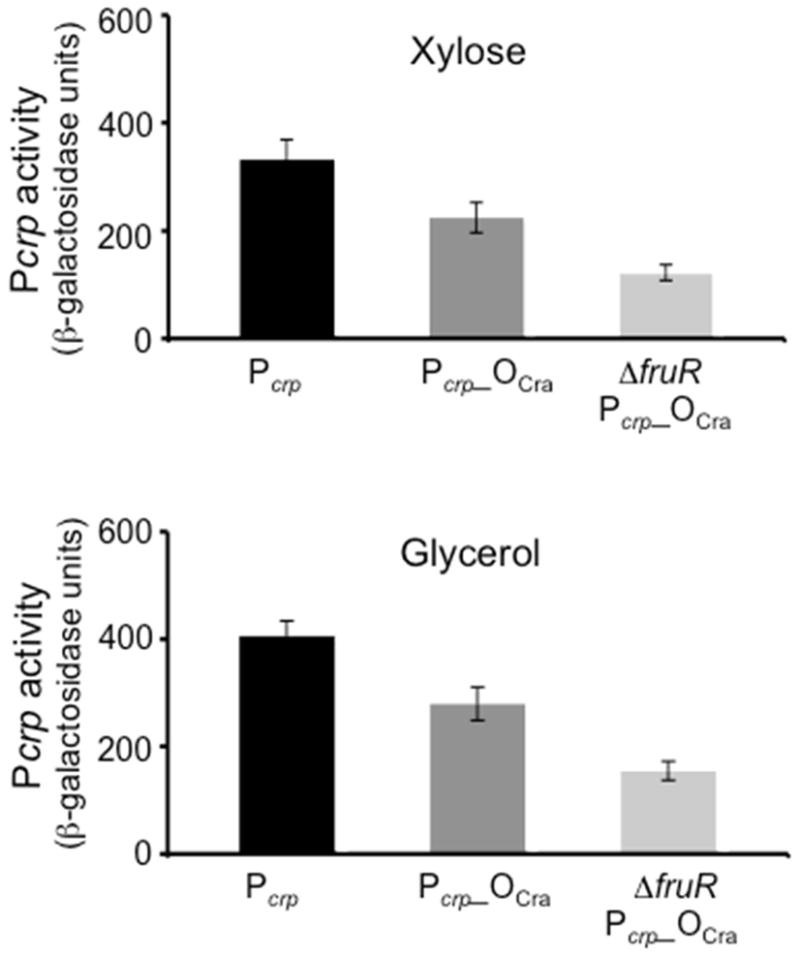
Effect of a Cra operator mutation on Cra activation of Pcrp. The Cra operator was mutated by changing GCTGAAGCGAGACACC to GCTCCTGTTAGACACC (see Fig. 1). The effect of such an alteration on the Pcrp activity was determined using the altered promoter-lacZ fusion in the MG1655ΔlacY and MG1655ΔlacYΔfruR strains in both the presence and absence of Cra.
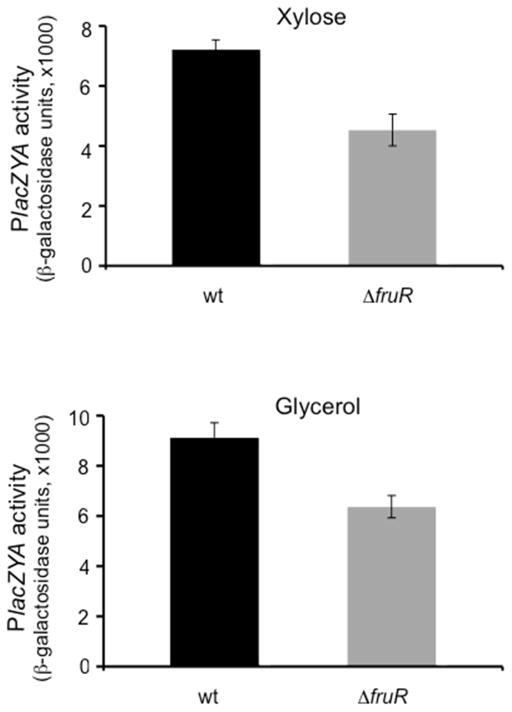
The wild type chromosomal lacZYA operon is known to be positively controlled by Crp. This operon was similarly examined in wild type and a ΔfruR mutant strain. As shown in Fig. 5, activity was decreased by the loss of Cra during growth in minimal medium containing 0.5% Dxylose (A) or 0.5% glycerol (B). All of the results presented in Figs. 2, 3, 4 and 5 as well as Table 2 are consistent with the conclusion that Cra regulates crp gene expression and thereby indirectly controls expression of numerous operons involved in the initiation of carbon metabolism.
Figure 5.
Expression of lacZ from the native E. coli lac operon promoter showing the effect of the loss of Cra. lacZ expression was measured in MG1655ΔlacY and MG1655ΔlacYΔfruR cells grown in M63 medium plus xylose or glycerol.
Discussion
The results reported in this communication indicate a novel level of complexity in the regulation of carbon metabolism in E. coli. Cra and Crp both activate expression of the crp gene, apparently by a mechanism that requires the simultaneous presence of both proteins. The cytoplasmic concentration of Crp thus generated then determines the degree of Crp-regulon gene activation by the cyclic AMP-Crp complex. Those operons that bind the cyclic AMP-Crp complex with low affinity will be particularly sensitive to the Crp concentration. Regulation of crp gene expression by Cra and Crp is therefore suggested to be part of a single regulatory mechanism. The details of this proposed regulatory mechanism are currently under investigation. It is anticipated that interrelated networks of regulatory interactions such as those described here will provide bacteria with mechanisms for fine-tuning and coordinating the different aspects of their metabolism and physiology (Amar et al., 2002; Babu and Teichmann, 2003).
Acknowledgments
We thank Carl Welliver and Maksim Shlykov for assistance in the preparation of this manuscript. This work was supported by NIH grants GM64368 and GM077402 from the National Institutes of General Medical Sciences.
Contributor Information
Zhongge Zhang, Email: zzhang@biomail.ucsd.edu.
Mohammad Aboulwafa, Email: maboulwafa@yahoo.com.
Milton H. Saier, Jr., Email: msaier@ucsd.edu.
References
- Aboulwafa M, Saier MH., Jr Characterization of the e. Coli glucose permease fused to the maltose-binding protein. J Basic Microbiol. 2008;48:3–9. doi: 10.1002/jobm.200700263. [DOI] [PubMed] [Google Scholar]
- Amar P, Ballet P, Barlovatz-Meimon G, Benecke A, Bernot G, Bouligand Y, Bourguine P, Delaplace F, Delosme JM, Demarty M, Fishov I, Fourmentin-Guilbert J, Fralick J, Giavitto JL, Gleyse B, Godin C, Incitti R, Kepes F, Lange C, Le Sceller L, Loutellier C, Michel O, Molina F, Monnier C, Natowicz R, Norris V, Orange N, Pollard H, Raine D, Ripoll C, Rouviere-Yaniv J, Saier M, Jr, Soler P, Tambourin P, Thellier M, Tracqui P, Ussery D, Vincent JC, Vannier JP, Wiggins P, Zemirline A. Hyperstructures, genome analysis and i-cells. Acta Biotheor. 2002;50:357–373. doi: 10.1023/a:1022629004589. [DOI] [PubMed] [Google Scholar]
- Babu M, Teichmann S. Evolution of transcription factors and the gene regulatory network in escherichia coli. Nucleic Acids Res. 2003;31:11. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner R, Titgemeyer F. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (cap) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Cheng X, Liu Y, Chu H, Kao HY. Promyelocytic leukemia protein (pml) regulates endothelial cell network formation and migration in response to tumor necrosis factor alpha (tnfalpha) and interferon alpha (ifnalpha) J Biol Chem. 2012;287:23356–23367. doi: 10.1074/jbc.M112.340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AM, Feucht BU, Saier MH., Jr Evidence for regulation of gluconeogenesis by the fructose phosphotransferase system in salmonella typhimurium. J Bacteriol. 1987;169:897–899. doi: 10.1128/jb.169.2.897-899.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua BT, Gallego-Ortega D, Ramirez de Molina A, Ullrich A, Lacal JC, Downward J. Regulation of akt(ser473) phosphorylation by choline kinase in breast carcinoma cells. Mol Cancer. 2009;8:131. doi: 10.1186/1476-4598-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in escherichia coli k-12 using pcr products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil G, Kahmann R, Muskhelishvili G. Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. Embo J. 1998;17:2877–2885. doi: 10.1093/emboj/17.10.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Aiba H. A new aspect of transcriptional control of the escherichia coli crp gene: Positive autoregulation. Mol Microbiol. 1992;6:2489–2497. doi: 10.1111/j.1365-2958.1992.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Ishizuka H, Hanamura A, Kunimura T, Aiba H. A lowered concentration of camp receptor protein caused by glucose is an important determinant for catabolite repression in escherichia coli. Mol Microbiol. 1993;10:341–350. doi: 10.1111/j.1365-2958.1993.tb01960.x. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by camp and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. xvi Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Ramseier TM, Bledig S, Michotey V, Feghali R, Saier MH., Jr The global regulatory protein frur modulates the direction of carbon flow in escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Ramseier TM, Negre D, Cortay JC, Scarabel M, Cozzone AJ, Saier MH., Jr In vitro binding of the pleiotropic transcriptional regulatory protein, frur, to the fru, pps, ace, pts and icd operons of escherichia coli and salmonella typhimurium. J Mol Biol. 1993;234:28–44. doi: 10.1006/jmbi.1993.1561. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Phylogenetic approaches to the identification and characterization of protein families and superfamilies. Microb Comp Genomics. 1996;1:129–150. doi: 10.1089/mcg.1996.1.129. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Held L, Torre M, Saier MH., Jr Grasp-DNA: A web application to screen prokaryotic genomes for specific DNA-binding sites and repeat motifs. J Mol Microbiol Biotechnol. 2000;2:495–500. [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: Origins and consequences. Science. 2000:1109–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Sharma V, Dhillon P, Wambolt R, Parsons H, Brownsey R, Allard MF, McNeill JH. Metoprolol improves cardiac function and modulates cardiac metabolism in the streptozotocin-diabetic rat. Am J Physiol Heart Circ Physiol. 2008;294:H1609–1620. doi: 10.1152/ajpheart.00949.2007. [DOI] [PubMed] [Google Scholar]
- Stulke J, Arnaud M, Rapoport G, Martin-Verstraete I. Prd--a protein domain involved in pts-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- Stulke J, Hillen W. Coupling physiology and gene regulation in bacteria: The phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften. 1998;85:583–592. doi: 10.1007/s001140050555. [DOI] [PubMed] [Google Scholar]
- Sujobert P, Bardet V, Cornillet-Lefebvre P, Hayflick JS, Prie N, Verdier F, Vanhaesebroeck B, Muller O, Pesce F, Ifrah N, Hunault-Berger M, Berthou C, Villemagne B, Jourdan E, Audhuy B, Solary E, Witz B, Harousseau JL, Himberlin C, Lamy T, Lioure B, Cahn JY, Dreyfus F, Mayeux P, Lacombe C, Bouscary D. Essential role for the p110delta isoform in phosphoinositide 3- kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106:1063–1066. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- Tagami H, Inada T, Kunimura T, Aiba H. Glucose lowers crp* levels resulting in repression of the lac operon in cells lacking camp. Mol Microbiol. 1995;17:251–258. doi: 10.1111/j.1365-2958.1995.mmi_17020251.x. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Johnson RC. Cyclic amp receptor protein functions as a repressor of the osmotically inducible promoter prop p1 in escherichia coli. J Bacteriol. 1997;179:2410–2417. doi: 10.1128/jb.179.7.2410-2417.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]