Abstract
Ubiquitin-dependent proteosome-mediated proteolysis is an important pathway of degradation that controls the timed destruction of cellular proteins in all tissues. All intracellular proteins and many extracellular proteins are continually being hydrolyzed to their constituent amino acids as a result of their recognition by E3 ligases for specific targeting of ubiquitination. Gustavus is a member of an ECS-type E3 ligase which interacts with Vasa, a DEAD-box RNA helicase, to regulate its localization during sea urchin embryonic development, and Gustavus mRNA accumulation is highly localized and dynamic during development. We tested if the core complex for Gustavus function was present in the embryo and if other SOCS box proteins also had restricted expression profiles that would inform future research. Expression patterns of the key members of the proteasomal function, such as the E3 core complex which interacts with Gustavus, and other E3-SOCS box proteins, are widely spread and dynamic in early development of the embryo suggesting broad core complex availability in the proteasome degradation pathway and temporal/spatial enrichments of various E3 ligase dependent targeting mechanisms.
Keywords: Ubiquitination, proteolysis, E3 ligases, proteasome
1. INTRODUCTION
The ubiquitin proteasome pathway (UPP) is integral to the normal function of eukaryotic cells (Joazeiro and Weissman 2000; Tanaka et al., 2001; Pan et al., 2004; Dohmen, 2004; Pickard and Eddins 2004; Aragon, 2005; Mani and Gelmann, 2005; Denison et al., 2005). It is ATP dependent and involves the covalent attachment of chains of ubiquitin molecules to target substrates. Proteins modified in this manner are recognized by the proteasome, a 26S multiprotein complex that catalyzes the breakdown of poly-ubiquitinated proteins. The major functions of the pathway are rapid removal of cell cycle proteins, regulation of gene transcription by degradation of transcription factors, quality control mechanism to degrade abnormally folded or damage proteins, generate amino acids for new protein synthesis, and others (Lecker et al., 2006). Several protein functions are also regulated by mono-ubiquitin attachment, and are independent of degradation, including, trafficking, chromatin re-structuring, and the modulation of protein function. Ubiquitin attachment to proteins is referred to as ubiquitination (or ubiquitylation; Sawasdikosol et al., 2000; Tyers and Jorgensen, 2000; Marmor and Yarden, 2004). Ubiquitination of target proteins proceeds in a stepwise format involving E1, E2 and E3 enzymes. E1, an ubiquitin-activating enzyme, uses ATP to catalyze the covalent transfer of ubiquitin (Ub) to the active site cysteine of an E2 Ub-conjugating enzyme. The E2 enzyme then interacts with an E3 Ub-protein ligase, which is the key enzyme in the process because it recognizes a specific protein substrate and catalyzes the transfer of activated ubiquitin to it, resulting in the ubiquitination of target proteins on specific lysine residues (Pickart, 2001; Burger and Seth, 2004; Fang and Weissman, 2004; Canning et al., 2013). Generally, the addition of one to four ubiquitin molecules to a target protein leads to a change in its localization and/or function. The addition of many ubiquitin molecules (poly-ubiquitination) leads to protein degradation by the 26S proteosome. Specificity in targeting proteins for ubiquitination lies mostly in the E3 enzyme (Amemiya et al., 2008).
Cullin-RING E3 Ub-ligases (CRLs) comprise the largest class of E3 Ub-ligases (Petroski and Deshaies, 2005). CRLs contain a substrate specificity receptor that binds the ubiquitinated target and a RING (Really Interesting New Gene) protein that is involved in recruiting an E2-conjugating enzyme. RING proteins and particular substrate specificity receptors are brought together by scaffold Cullin proteins. The two best characterized subfamilies are the SCF (Skp1/Cullin/F-box) and ECS (Elongin B/C-Cullin-SOCS box) E3 Ub-ligase families, the latter of which includes the von Hippel-Lindau (VHL) tumour suppressor and the wider SOCS box-containing protein families (Linossi and Nicholson, 2012). SCF Ub-ligases are multiprotein complexes, and every protein in a SCF complex is homologous to a component found in ECS E3 Ub-ligases. In SCF complexes, the F-box has been shown to play a comparable role to SOCS box in ECS complexes (Kibel et al., 1995; Kamura et al., 1998; Patton et al., 1998; Zhang et al., 1999). In the SCF complex, the F-box of Skp2 binds to the adaptor protein Skp1 which in turn, binds to the cullin family member, and forms a bridge between the Cullin and the F-box (Carrano et al., 1999; Kugler et al., 2010). This is similar to the connectivity in ECS complexes, with the SOCS box or VHL box binding to elongin C, which in turn, binds to the cullin family member Cul-5 in the case of SOCS box proteins and Cul-2 in the case of VHL box proteins (Kamura et al., 1998; Zhang et al., 1999; Kile et al., 2002). For SOCS families of proteins, assembly with the E2-Ub-conjugating machinery Elongin B/C-Cullin 5 complex is predicted through the SOCS box LPXP motif, which confers Cul-5 selection (Bullock et al., 2006).
The SOCS box is a conserved domain that was initially discovered in SH2 domain-containing proteins of the suppressor of cytokine signaling (SOCS) family (Starr et al., 1997). SOCS-1 was identified simultaneously on the basis of its interaction with JAKs, its antigenic cross-reactivity to STATs and its ability to inhibit cytokine signaling. SOCS-1 has a central SH2 (Src homology 2) domain and is most similar to cytokine-inducible SH2-containing protein (CIS) and has a high degree of sequence similarity in a 40 amino acid C-terminal region that was named the SOCS box. Since then, other members of SOCS family (SOCS-2 to SOCS-7) have been identified in mouse and human. All these proteins contain SH2 domains in their N-termini and SOCS box domains in their C-termini (Hilton, 1999).
In addition to the canonical SOCS proteins, several additional SOCS protein families have been identified. Rather than containing a SH2 domain upstream of the SOCS box, these proteins contain other domains implicated in protein-protein interactions. The major families of proteins that contain a SOCS box domain are the ankyrin-repeat proteins (ASBs), the SPRY domain-containing proteins (SSBs), the WD40 repeat-containing proteins (WSBs), the Neuralized family of proteins and a previously described family of small GTPases, Rar and its relatives (Hilton, 1999). Gustavus is an E3 Ub-ligase identified in Drosophila and involved in the balance of Vasa ubiquitination controlling pole plasm accumulation (Styhler et al., 2002; Kugler et al., 2010). It contains a B30.2/SPRY sequence comprising a single domain that biochemically interacts with Vasa protein in vitro and in vivo (Woo et al., 2006a; Woo et al., 2006b; Styhler et al., 2002; Kugler et al., 2010) and a SOCS box that interacts with Elongin B/C-Cullin 5 complex (Woo et al., 2006a; Kugler et al., 2010).Vasa is a conserved DEAD-box RNA helicase associated with germ-line development and is expressed in multipotent cells in many animal species (Lasko and Ashburner, 1988; Raz, 2000; Gustafson and Wessel, 2010). During embryogenesis of the sea urchin, vasa transcripts are uniformly distributed through blastula formation, followed by specific expression in the small micromere lineage during gastrulation, but the Vasa protein remains uniformly distributed through only the first three cleavage divisions. In the fourth cleavage division it is enriched in the four micromeres and subsequently in the small micromeres (Juliano et al., 2006; Voronina et al., 2008). Gustafson et al. (2011) provided evidence that gustavus appears to degrade Vasa in all cells except the small micromeres thereby limiting vasa accumulation in a general background of Vasa mRNA translation (Gustafson et al., 2011).
Expression of some proteasomal components and members of the proteasome-dependent degradation system were described previously in specified adult tissues from the sea urchin (Loram and Bodnar, 2012). However, the members of the ubiquitination machinery complex in the embryo have not been documented. We were intrigued by the expression of Gus, and hypothesized that other SOCS-box proteins would have restricted, and perhaps informative expression profiles that might lead to their functional analysis. We also felt compelled to test if the other machinery for Gus function was present in the embryo – machinery that would be essential for the proposed functions of Gus in vasa regulation and of widespread function for general SOCS box protein functions.
2. RESULTS AND DISCUSSION
We obtained sequences of D. melanogaster, human and mouse proteins of all tested genes from NCBI (http://www.ncbi.nlm.nih.gov/). Orthologous protein sequences from sea urchins were found by BLAST analysis against the published Sea Urchin Genome Database (Spbase.org). The top hits were used for reciprocal BLAST analysis to the nonredundant NCBI database to test orthology and to identify specific protein domains. S. purpuratus Gene Expression Database (Spbase.org) was used to reflect their abundance (an estimate of copy numbers per embryo as calculated by normalization to specific mRNAs at 48hrs) and differential expression (Wei et al., 2006). Abundant mRNAs show signal intensities between 25,000 and 200,000 AU (Arbitrary Units) whereas those which are expressed in only a few cells during development are correspondingly lower (between 200 and 300 AU). These data provided us rough estimates of mRNA abundance that were helpful in identifying interesting candidate genes. The list of primers used for polymerase chain reaction (PCR) amplification of each gene in sea urchin and the length of RNA in situ probes are shown (Table 1).
Table 1.
Members of the protein degradation system in the sea urchin.
| Genes (Spbase reference number) | Orthologs (organism) | % Identities | NCBI reference number | Highest signal intensity during development (AU*) | Primer sequences | Length of RNA in situ probe (nucleotid |
|---|---|---|---|---|---|---|
| Sp-Elongin B (SPU_011920) | Mm-EloB | 68.64 | NP_080581. | 4,184 | F: TCAAGCGTCAGAAGACGACC | 760 |
| Dm-EloB | 61.02 | NP_524416.1 | R: taatacgactcactatagggTGAAACGCCCCCTGGTATTC | |||
| Sp-Elongin C (SPU_001062) | Mm-EloC | 82.29 | P83940.1; NP_080732.1 | 7,673 | F: TCCTTCTAGACAACCGAACGC | 491 |
| Dm-EloC_A | 86.32 | NP_725894. | R: taatacgactcactatagggAGGCCTATAGTTTCTGACACGC | |||
| Sp-Cullin 5 (SPU_006755) | Dm-Cul5 | 72.59 | NP_651665.2 | 882 | F: AAACAATGCCCAGCTCCAGA | 783 |
| R: taatacgactcactatagggACAAAGGTTGGAGGGCACA | ||||||
| Sp-Gustavus (SPU_004717) | Dm-Gus_G | 62.5 | NP_001246140.1 | 9,277 | F: GAAAGTGTCCGGAGGCATGA | 651 |
| R: taatacgactcactatagggTCACAATGTCCCCAGACTGC | ||||||
| Sp-NeurL2 (SPU_007486) | Mm-NeurL2 | 36.84 | NP_001076443.1 | 201 | F: TGAGAACCATGGGGTGAACG | 751 |
| R: taatacgactcactatagggGCATTCCTGGAGTGTGGGAA | ||||||
| Sp-Socs4/5 (SPU_026496) | Mm-Socs5 | 47.2 | NP_062628. | 2,717 | F: TGCAAAGAACAACAGAGCCAG | 633 |
| R: taatacgactcactatagggCTTTCAGACCGTTTGGCAGC | ||||||
| Sp-Socs6L (SPU_011298) | Mm-Socs6 | 27.66 | AAH85245.1 | 2,750 | F: CGGAAAACTCGGGGGATGAA | 872 |
| R: taatacgactcactatagggGCAAGACCACCACTCTCGAA | ||||||
| Sp-Asb5 (SPU_023368) | Hs-Asb5 | 39.78 | NP_543150 | 746 | F: CAACCACCCGTACTCAGCAT | 596 |
| R: taatacgactcactatagggCCGACAGGTCAAGAGACGAG | ||||||
| Sp-Rab40 (SPU_006006) | Mm-Rab40B | 48.72 | NP_631886.2 | 1,898 | F: AGAGGAGGCCCAGGACTATG | 505 |
| R: taatacgactcactatagggTAAAACTGGATGGCGCTCTT |
Signal intensity as per Wei et al., 2006, an approximate measure of transcripts per embryo.
2.1. Multiprotein complex ECS-type Ub-ligase components are broadly present during early development of embryos
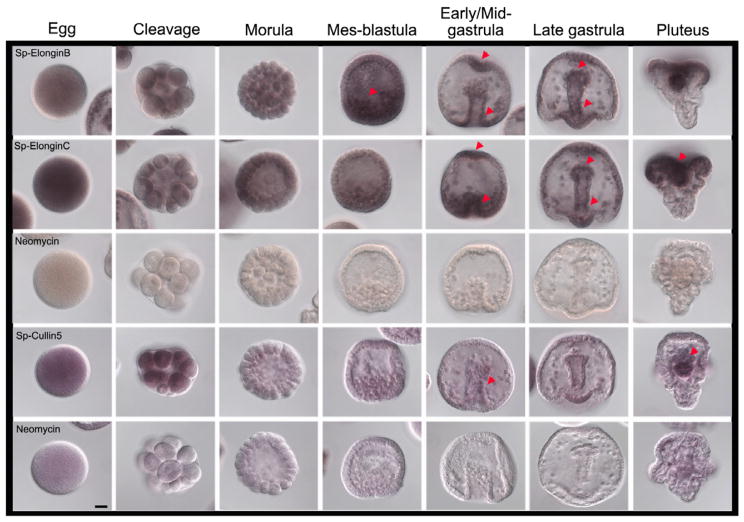
The SOCS box motifs interact specifically with Elongin C (Kamura et al., 1998; Zhang et al., 1999) whereas Elongin B acts to stabilize the complex and has only minimal interaction with the SOCS box itself (Bullock et al., 2006). Further, one might predict that the Elongin B, Elongin C, and Cullin 5 expression profiles overlap the cells expressing Gustavus. The S. purpuratus Gene Expression database (Spbase.org) showed that Elongin B transcripts are present throughout development but less abundant than Gustavus transcripts (Table 1). It is clear that Elongin B transcripts accumulate throughout the early embryo. At mid-gastrula, Elongin B transcripts are enriched in the animal pole and in the blastopore (Figure 1, red arrows). In late gastrulae, transcripts are present in the archenteron of the embryo, and within the highly prolific ciliary band (Figure 1, red arrows). Elongin C transcripts are more abundant than Elongin B mRNAs in eggs and early cleavage divisions. During gastrulation, the areas of mRNA signal are similar to those observed for Elongin B transcripts, mainly in highly proliferating cells of the late gastrulae and in plutei (Figure 1, red arrows). Cullin 5 mRNA is the least detectable of the mRNAs that contribute to the Elongin B/C-Cullin 5 protein complex; transcript signal is present throughout the egg and early embryos, but it decreased in mes-blastula stage. In early/mid-gastrula, a slight enrichment is seen in the middle region of the archenteron (Figure 1, red arrow). In plutei, some transcript enrichment is seen for Cullin 5 in the gut of the larva (Figure 1, red arrow), as also observed with Elongin B and Elongin C transcripts suggesting an important role for the expression of these components during this specific stage in the embryo. The data on expression of these genes supports the hypothesis that these gene products are widely present to target proteins for proteasome-mediated degradation.
Figure 1.
Expression of members of the multiprotein complex ECS-type E3 Ub-ligase which interacts with Gustavus protein. Line 1: Elongin B transcripts are widely spread in embryos during development and become slightly enriched at the bottom and at the tip of the archenteron. Line 2: Elongin C transcripts shown areas of enrichment similar to Elongin B transcripts, but Elongin C mRNAs are more enriched in eggs and early cleavage divisions than Elongin B transcripts. Staining reactions of Elongin B and Elongin C were stopped at 14.5 h. Line 3: Neomycin control stopped at 14.5 h. Line 4: Cullin 5 mRNAs are less enriched during development than Elongin B and C. They are spread from egg to morula stage but they decreased during later developmental stages, increasing again at pluteus stage. Line 5: Neomycin control. Cullin 5 and Neomycin staining reactions were stopped at 29h. Red arrows show areas of emphasis for transcript detection. Scale bar=20μm.
2.2. Members of SOCS box protein families are present broadly in sea urchin embryos
Orthologous sequences for SOCS box proteins were found through a BLAST search in the Sea Urchin Genome Database (Spbase.org) and our screening showed 14 protein sequences besides Gustavus, containing a SOCS box domain in their C-terminus (Figure 2). Four sequences contained SH2 motifs (CIS/SOCS family), one sequence contained a neuralized homology repeat (NHR) domain that belongs to the Neuralized and Neuralized-like family of proteins, NeurL2, seven sequences containing ankyrin-repeats (ASBs family), one with a WD40-repeat motif (WSBs family) and a Rab GTPase domain-containing protein.
Figure 2.
Schematic representation of SOCS box proteins found in sea urchin. The SOCS box motif is shown in green. The SPRY domain of the SPRY-domain proteins with a SOCS box (SSBs) is shown in aqua. The Neuralized domain of the Neuralized-like protein is shown in red. The Src-homology (SH2) domains are shown in orange. The ankyrin repeats in the ankyrin-repeat proteins with a SOCS box (ASBs) are shown in pink. The WD40 repeats in WD40-repeat proteins with a SOCS box (WSBs) are shown in blue. The Rab GTPase domain is shown in purple.
Canonical members of the SOCS protein family are inhibitors of cytokine signaling pathway and physiological regulators of both innate and adaptive immune systems in vertebrates. In mammals, members of the SOCS protein family have been shown to regulate growth hormone (GH) signaling pathway in vitro through multiple mechanisms (Flores-Morales et al., 2006; Metcalf et al., 2000). SOCS-1 targets for degradation members of the Src family of tyrosine kinases (Venkitachalam et al., 2011; Whiting et al., 2012). Thus, testing appearance in the embryo may help inform as to the mechanism used in for example, the egg-to-embryo transition, various differentiation steps, and cell cycle transitions.
Gustavus transcripts were analyzed by in situ hybridization during embryonic development. Of particular note is a uniform and strong signal in eggs, which remains uniform during early cleavage divisions. In mesenchyme blastula stage embryos, Gustavus transcripts are most readily detectable at the vegetal pole, but are also present in the ingressing primary mesenchyme cells (Figure 3, red arrow in mes-blastula stage). Some transcript enrichment is observed in the blastopore area and in some of the remaining mesenchyme cells during early/mid-gastrula stage embryos relative to other cells (Figure 3, red arrow). These observations are consistent with Gustavus mRNA accumulation described previously by Gustafson et al (2011).
Figure 3.
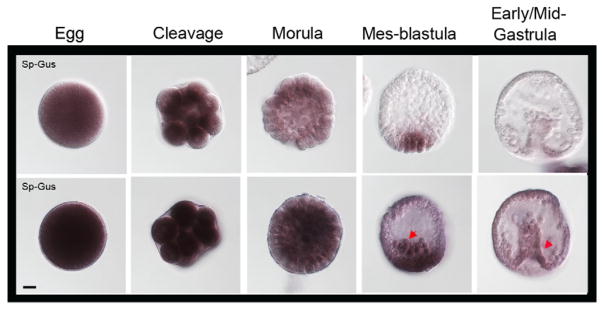
Expression pattern of Gustavus mRNAs. Gus transcripts are enriched in egg and early cleavage division stages and become restricted to the vegetal pole and in the ingressing primary mesenchyme cells during mes-blastula stage. A slight enrichment is present in the blastopore area and in some of the remaining mesenchyme cells during gastrulation. Line 1: Staining reaction was stopped at 10.5h. Line 2: Staining reaction was stopped at 19.5 h. Red arrows show areas of emphasis for transcript detection. Scale bar=20 μm.
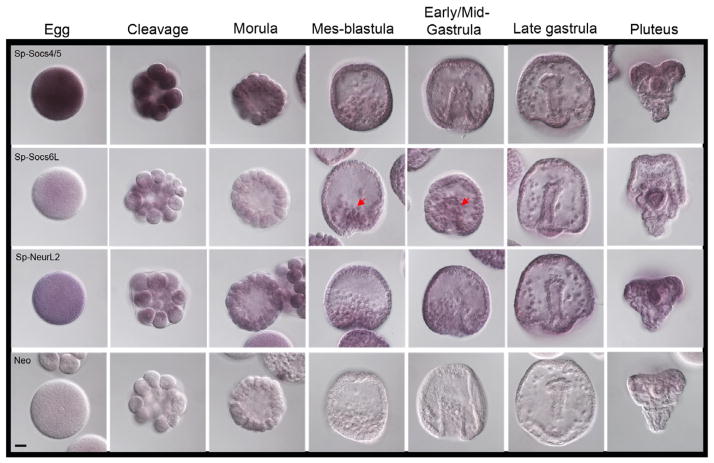
In general, we find that the mRNAs of the SOCS family of proteins are broadly expressed in the sea urchin. As might be expected, the elements of the core machinery (Elongin B, C, Cul-5) are present generally whereas one of the E3-ligase proteins, Gus, is restricted in its presence. Thus, the Gus pattern of expression is unique among these protein modifying activities. Socs4/5 transcripts are enriched only in the egg and early cleavage stages, and then decreased during blastulation and before mesenchyme cell migration (Figure 4). Socs6L transcripts are not more apparent in the egg and early cleavage divisions but they became restricted to the ingressing mesenchyme cells during mes-blastula and early/mid-gastrula stages (Figure 4, red arrows).
Figure 4.
Expression profiles of two members of the CIS/SOCS box family of proteins, Sp-Socs 4/5 and Sp-Soc6L, and one member of the Neuralized-like family, Sp-NeurL2. Line 1: Socs4/5 transcripts are enriched in the egg and early cleavage divisions but they decreased after morula stage and they remained in low concentration until pluteus. Line 2: Sp-Socs6L has a low transcript level in egg and early cleavage but it enrichment slightly increasedduring gastrulation in the mesenchyme cells. Line 3: Sp-NeurL2 mRNAs are slightly expressed and they are shown spread in the egg and in the whole embryo during early cleavage divisions and a slight enrichment after gastrula stage which remained until pluteus stage. Staining reactions were stopped at 29h. Red arrows show areas of emphasis for transcript detection. Scale bar=20 μm.
In both Drosophila and Xenopus, the Neuralized protein has been shown to target Delta, the ligand for the Notch receptor, for internalization, ubiquitination and degradation (Lai et al., 2001; Deblandre et al., 2001; Daskalaki et al., 2011). In Drosophila, the neuralized protein (Neur) is required in a subset of Notch pathway-mediated cell fate decisions during development of the nervous system. Neur binds to the Notch receptor ligand Delta through its first NHR1 domain and mediates its ubiquitination for endocytosis. However, during embryonic development in Drosophila, Neur is also required for the remodeling of the midgut epithelium via Brd proteins and this function of Neur in the regulation of epithelial polarity is independent of its known activity in Notch signaling (Chanet and Schweisguth, 2012). In mice, Neurl1 expression promotes lysosomal degradation of Jagged, a Notch ligand, in vitro (Koutelou et al., 2008). In sea urchin, NeurL2 showed low accumulation and spread broadly in the egg and embryo during early cleavage divisions, with only a slight signal increase after gastrulation (Figure 4).
To date, 18 Asb genes have been identified in mouse and human (Asb-1 to Asb-18) and one has been identified in C. elegans (ceASBa). Members of this large family of proteins are ubiquitously expressed in mammalian tissues, but their roles and function during development have been not reported. Some Asb family members, such as Asb-4, Asb-9and Asb-17, are expressed in male germ cells, suggesting their role in mammalian testis development and spermatogenesis (Kim et al., 2004; Kim et al., 2008; Lee et al., 2008). However, only the Asb-1 gene has been deleted in mice and this had a little effect on their development, health, or fertility (Kile et al., 2001). In sea urchin, Sp-Asb5 transcripts are not detectable in eggs and early embryos but increase and maintain signal levels in early/mid gastrulae (Figure 5).
Figure 5.
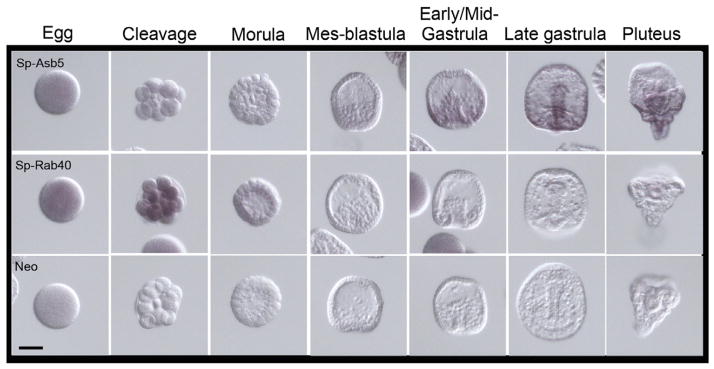
Expression patterns of Sp-Asb5 and Sp-Rab40. Line 1: Sp-Asb5 transcripts are not present in the egg and during early cleavage divisions. Its enrichment is observed at early/mid-gastrula stage, restricted to the forming gut of the embryo. Line 2: Sp-Rab40 is slightly enriched in the egg and it increases during early cleavage divisions. In morula stage, Sp-Rab40 mRNA decreases dramatically and cannot longer be perceived. Staining reactions were stopped at 20h. Scale bar=50 μm.
The Rab40 subfamily contains the paralogs Rab40a, Rab40b, and Rab40c. Rab small GTPases are key regulators in membrane trafficking and are regulated by GTPase activating proteins (GAPs), Guanine nucleotide exchange factors (GEFs) and Guanine nucleotide dissociation inhibitors (GDIs). Lipid binding is essential for membrane attachment, a key feature of most Rab proteins. In the sea urchin, Sp-Rab40 mRNA is present in the egg and its level increases during early cleavage divisions (Figure 5). In morula stage, Sp-Rab40 transcript signals decrease and can no longer be detected following gastrulation.
Since some of the transcripts detected in this study were broadly distributed and at low abundance, we conducted control experiments to test if the procedure used was faithfully representing transcript accumulation. For this control, we used Sp-Nanos, which accumulates selectively in the small micromeres of the sea urchin (Juliano et al., 2006). As reported, we found highly specific Sp-nanos accumulation with low background signal (Figure 6). Thus, we believe the signals detected for the ubiquitination machinery reported herein are true representations of their transcript accumulations.
Figure 6.
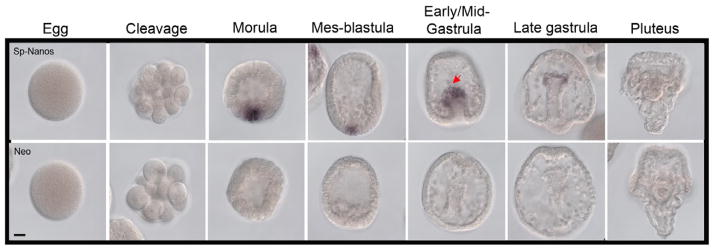
Expression pattern of Sp-Nanos. Transcripts are undetectable in cleavage stage embryos but they are locally enriched in morula and in a group of cells at the vegetal plate in blastula stage embryos and in the small micromere descendents at the tip of the archenteron in gastrula stage. Staining reaction was stopped at 8h. Scale bar=20 μm.
3. CONCLUSIONS
Overall, the results of this work suggest that the key members of the proteasomal function are present broadly in the early embryo. It supports the functional capability of Gustavus, Socs4/5 and other Socs box proteins in their functions, and suggests that the ubiquitination machinery is regulated by more than just presence and absence. Likely in this embryo the ubiquitination activity is biochemically regulated, to make uniform machinery be selectively functional. The reagents generated herein will assist in such activity identifications, and should be helpful in strategizing on harnessing the proteosomal machinery for engineered protein degradation in response to light (optogenetic) or small molecule regulation.
4. EXPERIMENTAL PROCEDURES
4.1. Animals and Embryo culture
Strongylocentrotus purpuratus were collected in Long Beach, CA, USA, and housed in aquaria cooled to 16°C in artificial sea water (ASW; Coral Life Scientific Grade Marine Salt; Energy Savers Unlimited, Carson, CA, USA). Animals were shed by KCl (0.5 M) injection. Eggs were collected in ASW and sperm were collected dry. Eggs were fertilized with a dilute sperm suspension in ASW supplemented with 1 mM 3-amino-triazol to waken and remove fertilization envelops, and embryos were cultured in filtered sea water and incubated at 16°C with rotation. Samples from different developmental stages (Eggs; Cleavage, 4.5 hr post-fertilization (hpf); Morula, 8.5 hpf; Mes-blastula 20 hpf; Early/Mid-gastrula, 27 hpf; Late gastrula 45 hpf; Pluteus, 4.5-days post-fertilization) were collected, fixed and stored in 70% ethanol at −20°C as described (Arenas-Mena et al., 2000).
4.2. RNA analysis
Whole-mount in situ RNA hybridizations were performed using digoxigenin-labeled RNA probes as previously described (Arenas-Mena et al., 2000). cDNAs from egg and 2-day embryo stages were used as templates for PCR reactions. Primers designed to amplify each gene of interest included a T7 RNA polymerase sequence in the 5′ end of reverse primers. The resultant PCR products were used as templates for transcription by T7 RNA polymerase to yield an antisense RNA probe with DIG RNA Labeling Kit (SP6/T7) (Roche Applied Science, IN). Eggs and embryos were fixed, hybridized with 0.1 ng/μl final concentration of the RNA probe diluted in hybridization buffer containing 70% formamide for one week at 50°C, and the signals were detected as described (Arenas-Mena et al., 2000). In the case of Sp-Elongin B, Sp-Elongin C and Sp-Cullin 5, complete Open Reading Frames (ORF) and a small part of 3′UTR were used to design RNA probes. Sequences of the ORFs from Sp-Gustavus, Sp-Socs4/5, Sp-Asb5, Sp-Socs6L, and Sp-NeurL2 were used to synthesize their RNA probes. In the case of Sp-Rab, the 5′UTR, ORF and 3′UTR sequences were used to design its RNA probe. Negative controls for these experiments included the use of a non-relevant transcript probe (Neomycin-resistance sequence). A positive control was achieved using a RNA probe specific for sea urchin (Sp-Nanos). Eggs and embryos were visualized on a Zeiss Axioplan microscope.
Highlights.
post-transcriptional regulation of genes is an essential part of germ line development in animals.
sea urchins rely on selective ubiquitin-dependent protein turnover to restrict germ line factors to the primordial germ cells.
here we learn that the core ubiquitination machinery is present broadly during development.
the sea urchin embryo contains multiple SOCs box continaing E3-ligases, each with dynamic localizations during development.
Gustavus mRNA accumulation is uniquely selective in the E-3 family of ligases tested.
Acknowledgments
We are grateful to other members of PRIMO for helpful discussion and feedback, and especially to S. Zachary Swartz by his kind donation of Sp-Nanos RNA probe.
Grant sponsor: NIH2R01HD028152; CONACyT
ABBREVIATIONS
- UPP
Ubiquitin proteasome pathway
- SOCS
Suppressor of cytokine signaling
- RING
Really interesting new gene
- Cul-5
Cullin 5
- Ub
ubiquitin
- ECS
ElonginB/C–Cullin–SOCS box
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6:1385–1396. doi: 10.1158/1541-7786.MCR-08-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon L. Sumoylation: a new wrestler in the DNA repair ring. Proc Natl Acad Sci USA. 2005;102:4661–2. doi: 10.1073/pnas.0501342102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–4643. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci USA. 2006;103:7637–7642. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer. 2004;40:2217–29. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: the sea urchin genome database and web site. Nucleic Acids Research. 2009:D750–754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Marsden BD, Gileadi O, Knapp S, von Delft F, Bullock AN. Structural basis for Cul3 assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288:7803–7814. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chanet S, Schweisguth F. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nature Cell Biology. 2012;14:467–477. doi: 10.1038/ncb2481. [DOI] [PubMed] [Google Scholar]
- Daskalaki A, Shalaby NA, Kux K, Tsoumpekos G, Tsibidis GD, Muskavitch MA, Delidakis C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J Cell Biol. 2011;195:1017–31. doi: 10.1083/jcb.201105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus Neuralized is an ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- deHostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein β-subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison C, Kirkpatrick DS, Gygi SP. Proteomic insights into ubiquitin and ubiquitin- like proteins. Curr Opin Chem Biol. 2005;9:69–75. doi: 10.1016/j.cbpa.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695:113–31. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–61. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Greenhalgh CJ, Norstedt G, Rico-Bautista E. Negative regulation of growth hormone receptor signaling. Mol Endocrinol. 2006;20:241–253. doi: 10.1210/me.2005-0170. [DOI] [PubMed] [Google Scholar]
- Fong HK, Hurley JB, Hopkins RS, Miake-Lye R, Johnson MS, Doolittle RF, Simon MI. Repetitive segmental structure of the transducin β-subunit: Homology with the CDC4 gene and identification of related mRNAs. Proc Nat Acad Sci. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Vasa genes: Emerging roles in the germ line and in multipotent cells. Bioessays. 2010;32:626–637. doi: 10.1002/bies.201000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Jualiano CE, Wessel GM. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol. 2011;349:440–450. doi: 10.1016/j.ydbio.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton DJ. Negative regulators of cytokine signal transduction. Cell Mol Life Sci. 1999;55:1568–1577. doi: 10.1007/s000180050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Weinzierl RO, Gill G, Chen JL, Dynlacht BD, Tjian R. Molecular cloning and functional analysisof Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–52. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in the sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, Conaway JW. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, Ras, WD-40repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibel A, Ilopoulos O, DeCarpio JA, Kaelin WG., Jr Binding of the vonHippel–Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- Kile BT, Metcalf D, Mifsud S, DiRago L, Nicola NA, Hilton DJ, Alexander WS. Functional analysis of Asb-1 using genetic modification in mice. Mol Cell Biol. 2001;21:6189–6197. doi: 10.1128/MCB.21.18.6189-6197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HME, Hilton DJ. The SOCS box: a tale of destruction and degradation. TRENDS in Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- Kim KS, Kim MS, Kim SK, Baek KH. Murine Asb-17 expression during mouse testis development and spermatogenesis. Zygote. 2004;12:151–6. doi: 10.1017/s0967199404002722. [DOI] [PubMed] [Google Scholar]
- Kim SK, Rhim SY, Lee MR, Kim JS, Kim HJ, Lee DR, Kim KS. Stage-specific expression of ankyrin and SOCS box protein-4 (Asb-4) during spermatogenesis. Mol Cells. 2008;25:317–21. [PubMed] [Google Scholar]
- Kugler JM, Woo JS, Oh BH, Lasko P. Regulation of Drosophila Vasa in vivo through paralogous Cullin-RING E3 ligase specificity receptors. Mol Cell Biol. 2010;30:1769–82. doi: 10.1128/MCB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Sato S, Tomomori-Sato C, Florens L, Swanson SK, Washburn MP, Kokkinaki M, Conaway RC, Conaway JW, Moschonas NK. Neuralized-like 1 (Neurl1) Targeted to the Plasma Membrane by N-Myristoylation Regulates the Notch Ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila Neuralized is an ubiquitin ligase that promotes the internalization and degradation of Delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor -4A. Nature. 1988;335:611–7. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Lee MR, Kim SK, Kim JS, Rhim SY, Kim KS. Expression of murine Asb-9 during mouse spermatogenesis. Mol Cells. 2008;26:621–4. [PubMed] [Google Scholar]
- Loram J, Bodnar A. Age-related changes in gene expression in tissues of the sea urchin Strongylocentrotus purpuratus. Mech Ageing Dev. 2012;133:338–47. doi: 10.1016/j.mad.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–70. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an express way to protein destruction. Oncogene. 2004;23:1985–97. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control inubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formationfrom the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasdikosol S, Pratt JC, Meng W, Eck MJ, Burakoff SJ. Adapting to multiple personalities: Cbl is also a RING finger ubiquitin ligase. Biochim Biophys Acta. 2000;1471:M1–12. doi: 10.1016/s0304-419x(00)00013-5. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008;22:1265–1268. doi: 10.1101/gad.1676208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kawakami T, Tateishi K, Yashiroda H, Chiba T. Control of IKBA proteolysis by the ubiquitin-proteasome pathway. Biochimie. 2001;83:351–6. doi: 10.1016/s0300-9084(01)01237-8. [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Vaisman N, Tsouladze A, Robzyk K, Ben-Yehuda S, Kupiec M, Kassir Y. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol Gen Genet. 1995;247:123–136. doi: 10.1007/BF00705642. [DOI] [PubMed] [Google Scholar]
- Venkitachalam S, Chueh F, Leong K, Pabich S, Yu C. Suppressor of cytokine signaling 1 interacts with oncogenic lymphocyte-specific protein tyrosine kinase. Oncology reports. 2011;25:677–683. doi: 10.3892/or.2011.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer C, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Dev Biol. 2006;300:476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting RJ, Payne CJ, Satiaputra J, Kucera N, Qiu TW, Irtegun S, Gunn NJ, Lavova-Azmanova NS, Mulhern TD, Ingley E. Targeting Lyn tyrosine kinase through protein fusions encompassing motifs of Cbp (Csk-binding protein) and the SOCS box of SOCS1. Biochem J. 2012;442:611–620. doi: 10.1042/BJ20111485. [DOI] [PubMed] [Google Scholar]
- Williams FE, Varanasi U, Trumbly RJ. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiaeare associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SB, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]