Highlights
-
•
Coronaviruses and arteriviruses encode papain-like protease domains that process the replicase polyprotein and are required for viral replication.
-
•
Many viral papain-like proteases are multifunctional and have protease, deubiquitinating and deISGylating activity.
-
•
Structural and enzymatic studies revealed the multifunctional nature of coronavirus and arterivirus papain-like proteases.
-
•
Viral DUB and deISGylating activity is proposed to modulate the innate immune response.
-
•
An arterivirus papain-like protease has been shown to modulate the innate immune response to viral infection.
Keywords: Coronavirus, Arterivirus, Papain-like protease, Deubiquitinating enzyme, DeISGylating activity, MERS-CoV
Abstract
Coronaviruses and arteriviruses, members of the order Nidovirales, are positive strand RNA viruses that encode large replicase polyproteins that are processed by viral proteases to generate the nonstructural proteins which mediate viral RNA synthesis. The viral papain-like proteases (PLPs) are critical for processing the amino-terminal end of the replicase and are attractive targets for antiviral therapies. With the analysis of the papain-like protease of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), came the realization of the multifunctional nature of these enzymes. Structural and enzymatic studies revealed that SARS-CoV PLpro can act as both a protease to cleave peptide bonds and also as a deubiquitinating (DUB) enzyme to cleave the isopeptide bonds found in polyubiquitin chains. Furthermore, viral DUBs can also remove the protective effect of conjugated ubiquitin-like molecules such as interferon stimulated gene 15 (ISG15). Extension of these studies to other coronaviruses and arteriviruses led to the realization that viral protease/DUB activity is conserved in many family members. Overexpression studies revealed that viral protease/DUB activity can modulate or block activation of the innate immune response pathway. Importantly, mutations that alter DUB activity but not viral protease activity have been identified and arteriviruses expressing DUB mutants stimulated higher levels of acute inflammatory cytokines after infection. Further understanding of the multifunctional nature of the Nidovirus PLP/DUBs may facilitate vaccine development. Here, we review studies describing the PLPs’ enzymatic activity and their role in virus pathogenesis.
1. Introduction
How do you pack a lot of information into a small space? This is the challenge for all microbes and analysis of viral genomes reveals interesting strategies of genetic economy. Positive strand RNA viruses employ genetic economy by encoding polyproteins that are processed by viral proteases during replication. This strategy minimizes genome size by allowing for the expression of multiple protein products from a single open reading frame. An additional twist to this genetic economy is that the viral protease itself may be multifunctional, i.e. the protease may act on both viral and host cell proteins. Hepatitis C virus encodes a polyprotein that is processed by a viral protease, NS3/4a, that also cleaves host cell mitochondrial associated viral sensor (MAVS), thus inactivating the innate immune response to viral infection (Li et al., 2005). Poliovirus 2A protease processes the viral polyprotein and host cell factor eIF4G, which shifts the ribosomes from cap-dependent to cap-independent translation, whereas 3C protease cleaves poly(A)-binding protein to facilitate the complete host translation shutoff (Kuyumcu-Martinez et al., 2004, Gradi et al., 1998). Here, we focus on coronaviruses and arteriviruses, two families of positive strand RNA viruses in the order Nidovirales, and review recent findings illuminating the mutifuctionality of the papain-like proteases (PLPs) encoded in the replicase polyproteins. For these viruses, PLPs play a critical role in processing the amino-terminal portion of the replicase polyprotein and are attractive targets for antiviral drug development. In addition, structural studies have revealed a striking similarity of the viral PLPs to cellular deubiquitinating enzymes (DUBs), which are involved in regulation of the innate immune response to viral infection. This raises the question of the multifunctional potential of Nidovirus PLPs and their role in antagonism of the innate immune response to viral replication. In this review we discuss current knowledge on the multifunctionality of PLPs encoded within coronaviruses and arteriviruses genomes and their potential role in viral pathogenesis (summarized in Table 1 ).
Table 1.
Coronavirus and arterivirus papain-like protease characteristics.
| Virus family | Virus | Protease characteristics | Reference |
|---|---|---|---|
| Coronaviridae | Betacoronavirus | ||
| SARS-CoV PLpro | Proteolytic activity | Harcourt et al., J Virol. (2004) | |
| Crystal structure | Ratia et al., PNAS (2006) | ||
| DUB activity in vitro toward K-48 Ub2 and Ub7, and K-63 Ub7 | Barretto et al., 2005, Lindner et al., 2005 | ||
| DUB activity in cell culture | Frieman et al., J Virol (2009) | ||
| Blocks IFNβ induction | Devaraj et al., J Biol Chem. (2007) | ||
| DeISGylating activity in vitro | Lindner et al., ABB (2007) | ||
| MERS-CoV PLpro | Proteolytic activity | Kilianski et al., J Virol. (2013) | |
| DUB and deISGylating activities in cell culture | Mielech et al., 2014, Yang et al., 2013 | ||
| Blocks IFNβ induction | |||
| MHV PLP2 | Proteolytic activity | Kanjanahaluethai and Baker, 2000, Kanjanahaluethai et al., 2003 | |
| DUB activity in cell culture | Zheng et al., Cell Res. (2008) | ||
| Blocks IFNβ induction | |||
| Deubiquitinates TBK-1 and IRF3 in cell culture | Wang et al., PLoS ONE (2011) | ||
| Alphacoronavirus | |||
| HCoV-NL63 PLP2 | Proteolytic activity and DUB activity in vitro toward K-48 Ub6 | Chen et al., J Virol. (2007) | |
| DUB activity in vitro toward K-48 and K-63 Ub6, and in cell culture | Clementz et al., J Virol. (2010) | ||
| DeISGylating activity in cell culture | |||
| Blocks RIG-I mediated IFNβ | |||
| TGEV PLP1 | Proteolytic activity | Wojdyla et al., J Virol. (2010) | |
| Crystal structure | |||
| DUB activity in vitro toward K-48 and K-63 polyubiquitin chains | |||
| PEDV PLP2 | DUB activity in cell culture | Xing et al., J Gen Virol. (2013) | |
| Blocks IFNβ induction | |||
| Deubiquitinates RIG-I | |||
| Arteriviridae | EAV PLP2 | Proteolytic Activity | Snijder et al., J Biol Chem. (1995) |
| DUB and deISGylating activity in cell culture | Frias-Staheli et al., CHM (2007) | ||
| DUB activity in vitro toward K-48 and K-63 Ub7 chains | van Kasteren et al., J Virol. (2012) | ||
| Blocks IFNβ induction | |||
| DUB activity in cell culture; Deubiquitinates RIG-I | |||
| Crystal structure with ubiquitin | van Kasteren et al., PNAS (2013) | ||
| DUB activity is required for innate immune inhibition in infected cells | |||
| PRRSV PLP2 | Proteolytic activity | Han et al., 2009, Han et al., 2010 | |
| DUB and deISGylating activity in cell culture | Frias-Staheli et al., CHM (2007) | ||
| Blocks IFNβ induction | Sun et al., J Virol. (2010) | ||
| DeISGylating activity in cell culture | Sun et al., J Virol. (2012) | ||
The Coronaviridae is a family of enveloped, positive strand RNA viruses with very large genomes ranging in size from 28 to 32 kilobases. Coronaviruses were identified as etiologic agents of respiratory, gastrointestinal and neurologic diseases in humans and other animals. Coronaviruses are now notorious for emerging from animal reservoirs into the human population, sometimes with pandemic potential. In 2002, the coronavirus responsible for the pandemic of Severe Acute Respiratory Syndrome (SARS) emerged from Chinese horseshoe bats, and through an intermediate host (likely civet cats), into humans. This resulted in over 8000 infections with 10% mortality (Perlman and Netland, 2009). The SARS pandemic was controlled by public health measures of isolating infected individuals and their contacts which disrupted the chain of human to human transmission. Since 2012, a coronavirus designated Middle East Respiratory Syndrome Coronavirus (MERS-CoV) has been detected in 178 patients with a reported mortality of 43% (www.who.int, 2014). MERS-CoV-like sequences have been detected in bats suggesting bats as a potential reservoir of the virus (Memish et al., 2013). In addition, camels have been implicated as a potential intermediate host since the discovery of MERS-CoV neutralizing antibodies and infectious virus in dromedary camels (Reusken et al., 2013, Haagmans et al., 2014, Meyer et al., 2014). Additional endemic human coronaviruses that cause respiratory tract disease include: HCoV-229E and HCoV-OC43, which cause common colds; HCoV-NL63, which has been associated with croup in children; and HCoV-HKU1, associated with lower respiratory tract infection and pneumonia in the elderly. Coronaviruses are also important pathogens of livestock and pets including transmissible gastroenteritis virus (TGEV) (pigs), porcine epidemic diarrhea virus (PEDV) (pigs); bovine coronavirus (cows); infectious bronchitis virus (chickens); feline coronavirus (cats); and canine coronavirus (dogs). Interestingly, we now recognize that bats harbor diverse strains of coronaviruses, and these bat viruses may be the ancestors of the “species specific” viruses (Lau et al., 2013). The identification of a common therapeutic target in the genomes of coronaviruses may allow for the development of a broad spectrum antiviral therapy to existing and potentially emerging coronaviruses.
2. Coronavirus papain-like proteases and their role in virus replication
All coronaviruses replicate in the cytoplasm of infected cells through the action of the viral replicase complex. The coronavirus replicase is produced upon translation of the incoming RNA genome. The replicase gene contains two open reading frames, ORF1a and ORF1b, which are connected by a frame shift region allowing for translation of the ORF1ab polyprotein. The replicase polyproteins, designated pp1a and pp1ab, are processed into 16 non-structural proteins (nsps) by two or three, depending on the coronavirus, viral proteases. The PLPs are responsible for the cleavage of the amino-terminal portion of the polyprotein. Coronavirus PLP activity was identified by in vitro transcription/translation studies of genomic RNA and the recognition that the polyprotein product was processed by an encoded protease domain (Denison and Perlman, 1986). Site-directed mutagenesis and deletion studies reveal that for the model coronavirus, mouse hepatitis virus (MHV), there are two PLP domains with PLP1 processing the polyprotein between nsp1/nsp2 and nsp2/nsp3 (Denison et al., 1992, Baker et al., 1989, Baker et al., 1993, Teng et al., 1999). Further expression studies of MHV ORF1a revealed that the PLP2 domain processes the polyprotein at the nsp3/nsp4 site using a recognition site of LXGG (Kanjanahaluethai and Baker, 2000). Similar studies of other coronaviruses revealed the existence of either one or two PLP domains that were required for processing the amino-terminal region of the replicase polyprotein (Fig. 1 ). SARS-CoV encodes only one PLP domain termed PLpro within nsp3 which cleaves the replicase polyprotein at the junctions between nsp1/nsp2, nsp2/nsp3, and nsp3/nsp4 through recognition of a LXGG motif (Harcourt et al., 2004). For the recently emerged MERS-CoV the cleavage sites recognized by the PLpro are predicted and await experimental validation (van Boheemen et al., 2012). However, Kilianski et al. (2013) showed that MERS-CoV PLpro can recognize an LXGG motif similarly to SARS-CoV PLpro. It has been shown that when two PLP domains are encoded within the coronavirus replicase polyprotein, the PLP2 has similar characteristics to SARS-CoV PLpro and recognizes the LXGG motif (Fig. 1). Coronavirus PLP2 domains are known to or predicted to cleave between nsp3 and nsp4, whereas the cleavage between nsp1/nsp2 and nsp2/nsp3 is usually mediated by the PLP1 domain. Interestingly, gamma- and deltacoronaviruses encode one PLpro domain within nsp2 that is predicted to cleave between nsp1/nsp2 and nsp2/nsp3.
Fig. 1.
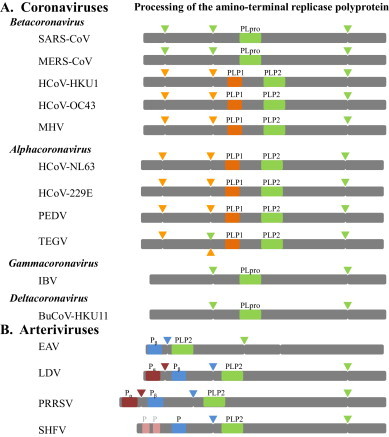
Coronavirus and arterivirus papain-like proteases. Schematic depiction of the Nterminal region of the replicase polyprotein of selected coronaviruses (A) and arteiviruses (B). The papain-like protease domains, termed PLpro, PLP1 or PLP2 for CoV, or Pα, Pβ or PLP2 for arteriviruses are depicted with correspondingly colored arrowheads indicating predicted or confirmed cleavage sites.
3. From viral protease to viral deubiquitinase
The multifunctionality of coronavirus PLPs was first proposed by Sulea et al. (2005) from their study of molecular modeling of the SARS-CoV PLpro domain. In their study, the authors predicted PLpro DUB activity based on the observations that PLpro could be modeled onto the structure of the herpesvirus-associated ubiquitin-specific protease (HAUSP/USP18), which was a known cellular DUB.
DUBs are enzymes that can remove ubiquitin modifications from target proteins. Ubiquitination is a post translational modification of proteins that allows for the addition of ubiquitin molecules to a specific lysine residue within the target protein in an ATP-dependent reaction. This system is mediated by three enzymes: an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase. These enzymes might add one (monoubiquitination) or several (polyubiquitination) ubiquitin moieties to a target protein. The ubiquitin molecules in the polyubiquitin chain can be covalently linked via any of the seven lysine residues present within ubiquitin. Several types of polyubiquitination have been characterized [reviewed in Komander and Rape, 2012]. The form of ubiquitin linkage determines the fate of the modified protein and influences its function. Three types of polyubiquitination are known to be involved in the regulation of the innate immune signaling pathways: Lys48-linked Ub (K-48-Ub), Lys63-linked Ub (K-63-Ub), and linear polyubiquitination. K-63 linked ubiquitin and linear ubiquitin modifications are associated with activation of proteins in certain innate immune signaling cascades. In contrast, K-48 linked polyubiquitination is a signal that directs proteins for proteosomal degradation. Removal of ubiquitin from cellular targets is mediated by cellular enzymes called deubiquitinases (DUBs) (Komander et al., 2009). DUBs recognize the RLRGG motif that links ubiquitin chains and remove ubiquitin conjugates from cellular proteins.
The SARS-CoV PLpro DUB activity predicted by Sulea et al. was tested in vitro and in transfected cells by several groups. Using purified enzyme, two groups showed SARS-CoV PLpro DUB activity in vitro and that catalytic activity of the protein is required for DUB activity. They showed that mutation of the predicted catalytic cysteine or aspartic acid residue to alanine leads to loss of DUB activity (Lindner et al., 2005, Barretto et al., 2005). Further studies revealed that PLpro is capable of processing both K-48-linked diubiquitin and heptaubiquitin, as well as K-63-linked heptaubiquitin chains in vitro (Lindner et al., 2007). These in vitro studies were the first to demonstrate the multifunctional nature of the coronavirus PLPs.
Structural studies have contributed to a detailed understanding of SARS-CoV PLpro (Ratia et al., 2006). The structure of SARS-CoV PLpro displays a thumb-palm-fingers architecture resembling cellular ubiquitin specific proteases (USPs) (Fig. 2A) (Ratia et al., 2006). Structural superposition of SARS-CoV PLpro with USP14 resulted in a pairwise root-mean-square-deviation (RMSD) of 3.6 Å over 198 aligned backbone Cα, although the sequence identity between these two proteins is only around 10%. The active site of SARS-CoV PLpro consists of a canonical Cys-His-Asp catalytic triad, which is in a similar position to that of USP14 (Fig. 2A). In contrast to some USPs, including USP14 and HAUSP, whose fingers domains consist of a Cys4-type-like zinc ribbon without zinc (Hu et al., 2005), the fingers domain of SARS-CoV PLpro contains a zinc finger with four cysteines tetrahedrally coordinating a zinc atom (Ratia et al., 2006). Moreover, the zinc-binding ability seems to be essential for SARS-CoV PLpro activity, as supported by mutagenesis studies (Barretto et al., 2005). Such dependence on zinc seems to be conserved among coronaviral PLPs as zinc binding has also been observed for other PLPs, including TGEV PLP1 and HCoV-229E PLP1 (Herold et al., 1999, Wojdyla et al., 2010).
Fig. 2.
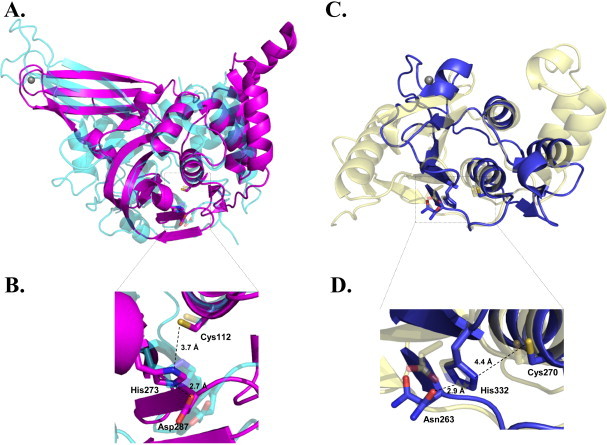
Structures of SARS-CoV PLpro and EAV PLP2 compared to USP14 and yeast OTU1. (A) Structural superposition of SARS-CoV PLpro (magenta, PDB code 2FE8) to USP14 (cyan, PDB code 2AYO). The zinc atom from the zinc finger of SARS-CoV PLpro is shown as a gray sphere. (B) Close-up view of the active site in SARS-CoV PLpro with the catalytic triad residues shown as sticks. Numbering of the residues is based on (26) (C) Structural overlay of EAV PLP2 (blue, PDB code 4IUM) with yeast OTU1 (yellow, PDB code 3BY4) bound to ubiquitin. The ubiquitin molecules have been omitted for clarity. The zinc atom from the zinc finger of EAV PLP2 is shown in gray sphere. (D) Close-up view of the active site in EAV PLP2. Numbering of the residues is based on (49). Asn263 in EAV PLP2 catalytic triad has two alternative conformations. Images were generated using PyMOL.
The structure of the PLP1 domain of TEGV is similar to SARS-CoV PLpro and several cellular USPs including USP 2, 7, 8, 14, and 21 (Wojdyla et al., 2010). Purified TGEV PLP1 has DUB activity in vitro. In addition, the authors showed that TEGV PLP1 has a slight preference for cleavage of K-48-linked over K-63-linked polyubiquitin chains. The role of DUB activity of TEGV PLP1 in cell culture or in infected animals remains to be determined.
HCoV-NL63 PLP2 which has only 22% homology to SARS-CoV PLpro has also been shown to act as a DUB in vitro and in cell culture. Expression of PLP2, but not PLP2 catalytic mutant (C1678A) leads to a decrease in the level of ubiquitinated proteins in transfected cells. In addition, purified PLP2 can process both K-48-linked and K-63-linked hexaubiquitin chains (Chen et al., 2007, Clementz et al., 2010).
Similarly to HCoV-NL63, PEDV encodes two PLPs in the genome; however, only PLP2 has been shown to have the ability to deconjugate ubiquitin from cellular substrates (Xing et al., 2013). Moreover, the authors showed that alanine mutants of the active site residues (cysteine, histidine, or aspartic acid) led to the loss of DUB activity in PEDV PLP2-transfected cells. In addition, the authors showed that PEDV PLP2 can efficiently remove both K-48-linked and K-63-linked polyubiquitin conjugates from cellular proteins. Specifically, the authors showed that PLP2 can remove K-63-linked polyubiquitin chains from RIG-I and STING, two molecules important for induction of innate immunity. These results suggest a possible mechanism by which coronavirus PLP2 DUB activity can block induction of interferon β (IFNβ) (Xing et al., 2013).
DUB activity of MHV PLP2 has also been reported. Frieman et al. (2009) showed that MHV can deubiquitinate multiple cellular proteins conjugated with ubiquitin in infected cells. In addition, Zheng et al. (2008) showed that MHV PLP2 is capable of processing both K-48-linked and K-63-linked polyubiquitin chains from cellular substrates. Further, Wang et al. (2011) determined that MHV PLP2 can deubiquitinate K-63-linked TBK1, a kinase required to activate transcription factor IRF3, which activates expression of proinflammatory cytokines such as IFNβ. These results suggest that MHV PLP2 DUB activity may have a role in blocking induction of the antiviral state.
Of note, the PLpro domain from the recently emerged, highly pathogenic MERS-CoV has been characterized as a protease, DUB and inhibitor of innate immune responses, including IFNβ (Kilianski et al., 2013, Mielech et al., 2014, Yang et al., 2013). Further work is required to determine if the DUB activity detected during overexpression of the PLpro in transfected cells is also critical for viral replication or pathogenesis.
4. From viral protease to deISGylating enzyme
The recognition that coronavirus PLPs had the ability to recognize the LXGG motif and process both polyproteins and ubiquitin chains led to the hypothesis that PLP domains might also process ubiquitin-like molecules such as interferon stimulated gene 15 (ISG15). ISG15 is an interferon stimulated di-ubiquitin-like molecule that can be linked to cellular targets via a mechanism termed ISGylation (Jeon et al., 2010). Similar to ubiquitination, ISGylation requires the activity of E1, E2, and E3 enzymes, and the process is reversible (deISGylation). Several screens have identified cellular targets of ISGylation including molecules important for the activation of innate immune response, particularly RIG-I, JAK1, STAT1, PKR, and MxA [reviewed in Lenschow, 2010]. The exact mechanism of how modification with ISG15 influences these proteins’ activity is not well characterized; however, ISGylation is important for protection and clearance of viral infections. It has been shown that ISG15 knock-out mice are more susceptible to infection with influenza, herpes and Sindbis viruses, than are wild-type mice (Lenschow et al., 2005, Lenschow et al., 2007). Coronavirus encoded PLPs, in addition to DUB activity, have also been shown to be able to remove IGS15 conjugates from cellular targets, an activity which might also contribute to virus pathogenesis.
DeISGylating activity has been demonstrated for several coronavirus PLPs. DeISGylating activity of SARS-CoV PLpro and HCoV-NL63 PLP2 was demonstrated in vitro (Nicholson et al., 2008). In addition, Clementz et al. (2010) showed that HCoV-NL63 PLP2 has the ability to remove ISG15 conjugates from cellular proteins in a catalytically dependent manner. Moreover, the PLpro domain from MERS-CoV has similar properties (Mielech et al., 2014, Yang et al., 2013) suggesting that deISGylating activity of PLPs might be conserved between PLPs from different coronavirus species.
5. Papain-like proteases as antagonists of the innate immune response
DUB and deISGylating activities of the coronavirus PLPs have been proposed as a mechanism to block induction of the cellular response to viral infection. Devaraj et al. determined that SARS-CoV PLpro can inhibit polyI:C and Sendai virus induced IFNβ, and that PLpro catalytic activity is important for antagonism. Co-expression of PLpro with stimulators of interferon (IFN) activation such as RIG-I, MAVS, TRIF, TBK1 or IKKɛ reduced IFNβ induction. In contrast, PLpro was not able to inhibit IFNβ induced by expression of a dominant active form of IRF3. The authors showed that PLpro can down-regulate phosphorylation of IRF3 and prevent IRF3 dimer formation. Furthermore, the authors were able to co-immunoprecipitate IRF3 with transiently expressed PLpro, and co-localization of IRF3 and nsp3/PLpro during SARS-CoV infection (Devaraj et al., 2007). Subsequently, Frieman et al. also showed that PLpro inhibits the IFN response. In addition, the authors showed that PLpro can inhibit NF-κB-luciferase activity by stabilizing IκBα, a molecule that has to be degraded in order to release the transcription factor NF-κB, which activates proinflammatory responses (Frieman et al., 2009). Later studies showed that PLpro can block TNFα-induced NF-κB activation, and further that this block can be removed when the cells are treated with a PLpro inhibitor, revealing a possible antiviral strategy (Clementz et al., 2010). Furthermore, a recent analysis showed that expression of SARS-CoV or MERS-CoV PLpro decreases endogenous RNA levels of proinflammatory cytokines and chemokines in activated cells (Mielech et al., 2014).
6. Multifunctional arterivirus papain-like proteases
The arterivirus family of positive strand RNA viruses contains genomes ranging from 10 to 14 kilobases. The arterivirus family includes important pathogens such as Equine Arteritis virus (EAV) that causes disease in horses; and Porcine Reproductive and Respiratory Syndrome virus (PRRSV) that infects pigs and is lethal to piglets. These diseases can lead to significant economic loss and there are currently no effective vaccines or antiviral drugs available to treat infected animals.
Arteriviruses encode at least one PLP that is responsible for proteolytic processing of the virus polyprotein (Fig. 1). The arterivirus genome has a similar organization to the coronavirus genome and is composed of 2 large ORFs that encode polyproteins pp1a and pp1ab, and 8–11 downstream ORFs that encode accessory and structural proteins. The amino-terminal region of the ORF1 is processed by two or three PLP domains. The PLP encoded within the amino-terminal region of nsp2, termed PLP2 or P2, has a similar function as SARS-CoV PLpro [Snijder et al., 1995 and reviewed in Snijder and Kikkert, 2013]. The sequence recognized by this protease domain is only partially conserved across the arterivirus species. For example, EAV PLP2 recognizes the sequence LIGG, whereas PRRSV P2 can recognize cleavage sites containing either TTGG or PSGG. The partial conservation of the site cleaved by the arterivirus PLP2s, and its similarity to motifs recognized by cellular DUBs and deISGylating enzymes, led to the hypothesis that arterivirus P2s might be multifunctional enzymes.
Expression of the EAV PLP2 domain revealed that it indeed exhibits DUB activity in cell culture (Frias-Staheli et al., 2007). DUB activity of the amino-terminal region of nsp2 (Nsp2(N)) was confirmed in vitro. EAV Nsp2(N) can deubiquitinate K-48-linked and K-63-linked polyubiquitin chains in a catalytic dependent manner, as alanine mutants of the three predicted catalytic residues lost DUB activity (van Kasteren et al., 2012). To address the possible role for nsp2(N) DUB activity in the inhibition of innate immunity, the authors determined the ubiquitination status of RIG-I which requires K-63-linked polyubiquitination for activation. Studies have shown that DUB activity of nsp2(N) from EAV can block RIG-I ubiquitination in a catalytic dependent manner (van Kasteren et al., 2012). This led to the hypothesis that EAV DUB activity might be important for blocking the innate immune response to viral infection. To test this hypothesis, it was critically important to obtain structural information on how the enzyme interacts with ubiquitin. With this information in hand, researchers can attempt to generate PLP2 mutants that retain protease activity but are impaired for interaction with ubiquitin.
The co-crystal structure of EAV PLP2 with ubiquitin was solved by van Kasteren et al. (2013). EAV PLP2 adopts a two-domain fold and shares a β-sheet core and two central helices with eukaryotic OTUs (Fig. 2C). Structural overlay of EAV PLP2 with yeast OTU1 resulted in a RMSD of 3.3 Å over 81 aligned Cα with only around 9% sequence identity. The Cys-His-Asn catalytic triad of EAV PLP2 resides in a structurally conserved region of OTU DUBs (Fig. 2B). What is unique about the structure of EAV PLP2 is the presence of a zinc finger as a part of the OTU domain. This zinc finger is critical for the structural integrity and catalytic activity of EAV PLP2. The presence of a zinc finger has not been observed in other known OTU-like proteases. Thus, it has been proposed that EAV PLP2 may represent the first member of a new class of zinc-dependent OTUs (van Kasteren et al., 2013).
The co-crystal structure of EAV PLP2 with ubiquitin revealed interaction sites between the enzyme and ubiquitin that are distal to the active site (van Kasteren et al., 2013). Using this structural information, the authors were able to successfully separate PLP2 protease and DUB activities. Mutation of the ubiquitin-interacting residues of EAV PLP2 had no evident effect on protease activity but significantly reduced DUB activity in overexpression studies. To study the effect of the DUB mutant on virus replication and innate immunity, the authors used reverse genetics to recover two replication competent mutant viruses (a single mutant I353R and a triple mutant T312A/I313V/I353R in the EAV PLP2 domain) and showed that replication kinetics of these viruses were essentially identical to the wild-type virus. However, the viruses with the mutant PLP2 domain had lost the ability to deconjugate ubiquitin from cellular targets in virus-infected cells. In addition, cells infected with the EAV DUB mutant viruses generated an elevated innate immune response, particularly expression of IFNβ, IL8, and MX1, suggesting that DUB activity is important for inhibition of the innate immune response (van Kasteren et al., 2013). The EAV DUB mutant study was the first study to demonstrate that viral protease and deubiquitinase functions can be separated. This is an important contribution because it opens the door for the study of viral DUB activity in vaccines and as a target for antiviral therapeutics.
Interestingly, EAV is not the only member of arterivirus family for which multifunctional protease/DUB activity has been shown. PRRSV is a well-studied member of the family which causes disease of high economic importance in pigs. The proteolytic activity of PRRSV P2 has been well-studied (Han et al., 2009, Han et al., 2010) and P2 has the ability to cleave ubiquitin conjugates from cellular targets (Frias-Staheli et al., 2007). Sun et al. showed that P2 had deubiquitinating activity in vitro and in cell culture. The authors showed that PRRSV P2 can block Sendai virus induced IFNβ and inhibit NF-κB by preventing IκBα degradation by its deubiquitination in cell culture. In addition, they generated several mutant versions of P2 that had reduced ability to inhibit NF-κB-activation. To test if P2 can block NF-κB activation in the context of the virus, the authors introduced these mutations into PRRSV. Although some of the mutations that altered P2 activity in vitro could not be recovered as viable virus, the authors did report two single amino acid P2 mutant viruses that showed decreased ability to inhibit NF-κB-reporter activity and a decrease in the level of IκBα in infected cells (Sun et al., 2010). However, the authors did not address whether protease or DUB activity was responsible for the effect they observed in infected cells. The authors noticed that the mutant viruses had severe replication defects suggesting that P2 protease activity, as well as DUB activity, may have been impaired (Sun et al., 2010).
Similar to studies on coronavirus PLP activities, groups characterizing arterivirus PLP domains investigated their deISGylating activity. Frias-Staheli et al. (2007) showed that EAV P2/PLP2 is capable of deconjugating IGS15 from cellular proteins in transfected cells. Furthermore, a report by Sun et al. (2012) showed that PRRSV P2 decreases endogenous ISG15 protein levels upon Sendai virus stimulation, and that it has deISGylating activity in cells culture. Furthermore, the authors generated PRRSV P2 mutants that had reduced deISGylating activity. The first mutant had a 23 aa deletion in nsp2 (aa 402–424), the second mutant had a 19 aa deletion in nsp2 (aa 402–420), and the third mutant had the 19 aa deletion and additional point mutation (S462A). When cells were transfected with those mutants the endogenous levels of IGS15 was increased compared to cells transfected with wild-type version of P2. Interestingly, the processing of nsp2/nsp3 was not affected by the deletion or deletion/point mutations in the P2 domain of the virus. The authors were able to recover two viruses (19 aa deletion, and 19 aa deletion/S462A). The growth kinetics analysis showed that those two viruses do not replicate as efficiently as wild-type virus, which may be due to the inability to efficiently process the nsp2/nsp3 site in the replicase polyprotein (Sun et al., 2012). Further studies including solving the X-ray crystal structure of the PRRSV P2 domain and determining to role of P2 DUB/deISGylating mutants on viral pathogenesis in animal models are needed.
7. Final remarks
Viruses have evolved many different mechanisms to down-regulate innate immune response upon infection. The ability of a virus to block or delay the induction of the antiviral state in an infected cell is likely to be an important contributor to pathogenesis. Nidoviruses encode in their genomes several proteins that are capable of blocking or delaying innate immune signaling. One of them is the papain-like protease, a multifunctional enzyme that can act as a potent protease, DUB, deISGylating enzyme, and antagonist of the innate immune response. Separating protease and DUB activities is needed to fully understand the role of these proteins during virus replication. Thus far, separating protease activity from DUB activity has been shown only for the arterivirus EAV (van Kasteren et al., 2013). The specific mechanisms and the contributions of arteri- and coronavirus PLPs to interferon antagonism during virus infection and pathogenesis are an exciting new direction for research that may lead to the development of effective vaccines and novel antiviral therapeutics.
Acknowledgements
We thank Xufang Deng, Andy Kilianski and Rob Mettleman for their suggestions. This work was supported by funding from NIH R01 AI085089 (to SCB and ADM). AMM was supported in part by the Arthur J. Schmitt Dissertation Fellowship from Loyola University Chicago.
References
- Baker S., Shieh C., Soe L., Chang M., Vannier D., Lai M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 1989;63(September (9)):3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Yokomori K., Dong S., Carlisle R., Gorbalenya A., Koonin E. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 1993;67(October (10)):6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(December (24)):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A.D., Wilkinson K.D., Baker S.C. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 2007;81(June (11)):6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84(May (9)):4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M., Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 1986;60(October (1)):12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M., Zoltick P., Hughes S., Giangreco B., Olson A., Perlman S. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189(July (1)):274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282(November (44)):32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2(December (6)):404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83(July (13)):6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A., Svitkin Y.V., Imataka H., Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U. S. A. 1998;95(September (19)):11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B., Al Dhahiry S., Reusken C., Raj S., Galiano M., Myers R. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Rutherford M.S., Faaberg K.S. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 2009;83(September (18)):9449–9463. doi: 10.1128/JVI.00834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Rutherford M.S., Faaberg K.S. Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein. J. Virol. 2010;84(October (19)):10102–10112. doi: 10.1128/JVI.01208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(December (24)):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274(May (21)):14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24(November (21)):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.J., Yoo H.M., Chung C.H. ISG15 and immune diseases. Biochim. Biophys. Acta. 2010;1802(May (5)):485–496. doi: 10.1016/j.bbadis.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74(September (17)):7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Jukneliene D., Baker S.C. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 2003;77(Jul (13)):7376–7382. doi: 10.1128/JVI.77.13.7376-7382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A., Mielech A., Deng X., Baker S.C. Assessing activity and inhibition of MERS-CoV papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 2013;87(August (21)):11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10(August (8)):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez N.M., Van Eden M.E., Younan P., Lloyd R.E. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 2004;24(February (4)):1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Li K., Tsang A., Lam C., Ahmed S., Chen H. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in japanese pipistrelle: implications for the origin of the novel middle east respiratory syndrome coronavirus. J. Virol. 2013;87(August (15)):8638–8650. doi: 10.1128/JVI.01055-13. Epub 29.05.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow D.J. Antiviral properties of ISG15. Viruses. 2010;2(October (10)):2154–2168. doi: 10.3390/v2102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow D.J., Giannakopoulos N.V., Gunn L.J., Johnston C., O’Guin A.K., Schmidt R.E. Identification of interferon-stimulated gene 15 as an antiviral molecule during sindbis virus infection in vivo. J. Virol. 2005;79(November (22)):13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 2007;104(January (4)):1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2005;102(December (49)):17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(December (24)):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466(October (1)):8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z., Mishra N., Olival K., Fagbo S., Kapoor V., Epstein J. Middle east respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;(November) doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Müller M.A., Corman V.M., Reusken C.B.E.M., Ritz D., Godeke G. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg. Infect. Dis. 2014;20(4) doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B., Leach C.A., Goldenberg S.J., Francis D.M., Kodrasov M.P., Tian X. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17(June (6)):1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(June (6)):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103(April (15)):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Müller, Gutierrez C., Godeke G., Meyer B. Middle east respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013 doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. Arteriviridae. [Google Scholar]
- Snijder E., Wassenaar A., Spaan W., Gorbalenya A. The arterivirus Nsp2 protease. an unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995;270(July (28)):16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79(April (7)):4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Chen Z., Lawson S.R., Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 2010;84(August (15)):7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Li Y., Ransburgh R., Snijder E.J., Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J. Virol. 2012;86(April (7)):3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng H., Pinon J.D., Weiss S. Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J. Virol. 1999;73(April (4)):2658–2666. doi: 10.1128/jvi.73.4.2658-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(November (6)) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Beugeling C., Ninaber D.K., Frias-Staheli N., van Boheemen S., Garcia-Sastre A. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 2012;86(January (2)):773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110(February (9)):E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS ONE. 2011;6(2):e17192. doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., van Kasteren P.B., Kikkert M., Snijder E.J., Gorbalenya A.E. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J. Virol. 2010;84(October (19)):10063–10073. doi: 10.1128/JVI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.who.int/csr/disease/coronavirus_infections/MERS_CoV_Update_20_Jan_2014.pdf (accessed 22.01.14).
- Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94(July (Pt 7)):1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y. Proteolytic processing, deubiquitinase and interferon antagonist activities of middle east respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2013 doi: 10.1099/vir.0.059014-0. [Epub ahead of print] doi:10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18(November (11)):1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]


