Abstract
A sophisticated mechanistic understanding of physiology and disease requires knowledge of how sex-biasing factors cause sex differences in phenotype. In therian mammals, all sex differences are downstream of the unequal effects of XX vs. XY sex chromosomes. Three major categories of sex-biasing factors are activational and organizational effects of gonadal hormones, and sex chromosome effects operating outside of the gonads. These three types of effects can be discriminated from each other with established experimental designs and animal models. Two important mouse models, which allow conclusions regarding the sex-biasing effects of sex chromosome complement, interacting with gonadal hormone effects, are the Four Core Genotypes model and the XY* model. Chromosome Y consomic strains give information about the role of the Y chromosome. An important recent change in sexual differentiation theory is the increasing realization that sex-biasing factors can counteract the effects of each other, reducing rather than producing sex differences in phenotype. This change in viewpoint rationalizes a change in experimental strategies for dissecting sex chromosome effects. The overall goal is to understand the sexome, defined as the sum of effects of sex-biasing factors on gene systems and networks.
Keywords: Sexual differentiation, testosterone, estradiol, sex chromosomes, X chromosome, Y chromosome, X inactivation, sex differences in disease, Four Core Genotypes, XY*, sexome, compensation
Increasing interest in sex differences in physiology and disease
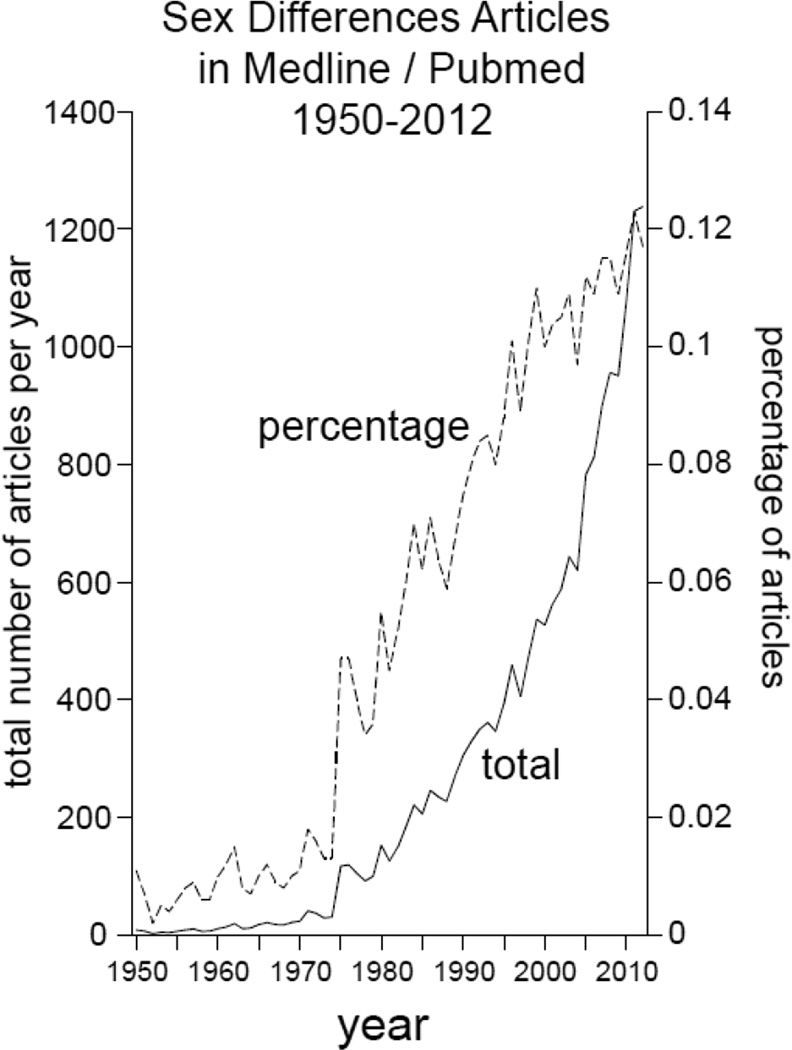
Within the scientific and medical community, there is increasing realization that many disease mechanisms differ in the two sexes. One sex may be affected by a specific disease much more than the other (Karastergiou et al., 2012: Miller et al., 2011: Sandberg and Ji, 2012: Voskuhl, 2011), so that even a basic appreciation of disease mechanisms requires understanding how sex-biased factors influence the disease. The majority of basic science research is performed on males (Beery and Zucker, 2011), but conclusions drawn from those studies may not apply fully to females. Importantly, if one sex is protected from disease, then study of the sex-biased protective mechanisms could lead to discovery of regulatory mechanisms that could be targeted for novel therapies. These ideas have contributed to an increase in the number and percentage of publications on sex differences in the last 15–20 years (Figure 1).
Figure 1.
Pubmed publications on sex differences. A search of Pubmed shows the increasing number of publications on sex differences since 1950. The search was for articles using the phrases “sex difference” or “gender difference” or “sexual dimorphism” or “sexually dimorphic”. See http://dan.corlan.net/medline-trend.html
Although the study of both sexes individually is important to establish the broad validity of scientific principles or therapeutic approaches, direct comparison of the sexes offers even greater advantages. Resolving the reasons for sex differences in disease leads to the discovery of unexpected regulatory mechanisms, and suggests new levels of protection that can be achieved in both sexes. Without reference to the other sex, it is sometimes not clear what aspects of physiology can be regulated by factors that occur already in nature. For example, the discovery that males die at greater rates at most ages across the lifespan, frames questions about what sex-specific social and biological factors are responsible for this sex difference, and whether these factors can be altered to increase lifespan of both sexes.
Fundamentally, we are asking where sex differences come from. Both phylogenetic and ontogenetic viewpoints are helpful in answering that question. Here, we discuss evolutionary reasons why sex-biasing factors might often be in opposition to each other, and review types of ontogenetic factors that can be discriminated by specific experimental designs.
The “big three” causes of sex differences in phenotype
Research between 1916 and 2010 gave rise to a relatively straightforward tripartite classification of categories of proximate (ontogenetic) causes of sex differences in phenotypes: (1) activational effects of gonadal steroid hormones, (2) organizational effects of gonadal steroid hormones, and (3) sex chromosome effects (Arnold, 2009b). These three classes are both conceptual and operational, because specific experimental outcomes define each class. Considering sex differences in adulthood, testicular and ovarian secretions act on many tissues to induce non-gonadal phenotypes to differ in the two sexes. These hormonal effects, predominantly of androgens, estrogens, and progestins, are reversible because they typically disappear in hours to weeks after removal of the gonads. Operationally, therefore, sex differences that are eliminated by adult gonadectomy are classified as activational effects. Some sex differences do not disappear after gonadectomy, but are caused by long-lasting, differentiating, or permanent changes caused by gonadal hormones acting at early stages of development (organizational effects of gonadal hormones, Phoenix et al., 1959). Examples include sexual differentiation of the external and internal genitals, and of specific sexual dimorphisms in the brain and behavior (Arnold and Gorski, 1984: Breedlove et al., 1999: Jost et al., 1973: McCarthy and Arnold, 2011). Classic sexual differentiation theory posits that testicular secretions, especially testosterone and Müllerian Inhibiting Hormone, act to cause masculine patterns of differentiation not found in females. Finally, some sex differences are not explained by either activational or organizational effects of gonadal hormones, but by direct effects of sex chromosome genes acting outside of the gonads. Both X and Y genes, which are differentially present in each XX vs. XY cell, act in a sex-specific or sex-biased manner to cause sex differences in non-gonadal phenotypes (Arnold, 2004: Arnold, 2009b).
This conceptual framework gives rise to a relatively standard strategy (called the A-O-S approach here: activational then organizational then sex chromosome) for discovering sex-biased factors that cause sex difference in tissue function or protection from disease (Becker et al., 2005). In an animal model, the first experiment is often to remove the gonads, preferably of both sexes, to determine whether the sex difference depends on the secretion of gonadal hormones in adulthood (for simplicity we are considering adult phenotypes, and use mice as an example). Adult gonadectomy is the first choice, because the majority of sex differences appear to be caused by activational effects of gonadal hormones (e.g., Van Nas et al., 2009), although this may not always hold (Seney et al., 2013). If the sex difference is eliminated by adult gonadectomy, then the sex difference is classified as caused by activational effects of gonadal hormones, leading to further experiments to investigate which hormones are relevant, and their downstream mechanisms of action. By Occam’s razor, eliminating the sex difference by adult gonadectomy means that there is no reason to invoke sex biasing factors other than activational effects. If the sex difference persists after gonadectomy, however, or is found in adult mice that have the same levels of hormones (for example, in female and male mice gonadectomized and treated with the same levels of sex steroid hormones in adulthood), then it is appropriate to test next for organizational effects. Organizational effects are discovered if females are permanently masculinized by exposure to androgens during an early development stage (in rodents just before or after birth), or if males are demasculinized or feminized when they are deprived of testosterone or androgen receptors at the same early stages of life (or later periods of organizational effects, Juraska et al., 2013; Schulz et al., 2009). If these manipulations of gonadal hormones do not explain the sex difference, then the remaining option is to consider sex chromosome effects, for example by comparing mice with different numbers of X or Y chromosome, under conditions in which the effects of gonadal hormones are similar across groups (Arnold, 2009a). Two relevant mouse models are discussed below.
The A-O-S experimental approach just outlined answers a variety of essential questions that are the first steps for finding the cellular and molecular mechanisms that explain sex-biased protection from disease in an animal model. These experiments are inherently mechanistic, because they establish variables that control the phenotype of interest, and point to other mechanistic experiments to define the hormones, gene, receptor mechanisms, and downstream molecular pathways that cause the sex difference. Nevertheless, the thesis of this paper is that the A-O-S approach is based on an overly simplistic view of the interactions of distinct sex-biasing factors. Several considerations, reviewed in the section on sexual monomorphism below, may justify a different priority of experiments in specific cases.
Mouse models for separating effects of the big three sex-biasing factors
To discover sex-biased factors that protect from disease, we need to generate an accurate catalog of possible sex-biasing factors that could be relevant. In addition, we need animal models and experimental designs that help us manipulate these variables, individually or in combination, to discriminate the sex-biasing factors that are important in specific cases.
Constructing the catalog of sex-biasing factors starts with the idea that all phenotypic sex differences stem from the imbalance in sex chromosome complement (XX vs. XY), which is present from the beginning, in the zygote (Arnold, 2011). This genetic sex difference leads to at least four primary classes of sex bias in the genome: (1) Y genes act only in XY cells. For example, the Y chromosome harbors male-specific genes that induce formation of testes (Sry) or are critical for spermatogenesis (Burgoyne and Mitchell, 2007: Tilmann and Capel, 2002). The Sry-induced formation of testes is enormously important, because it sets up lifelong sex differences in levels of gonadal hormones that cause activational and organizational effects, two dominant classes within A-O-S. In addition, however, the small number of genes on the Y chromosome may act outside of the testes to cause male-specific effects, for example Sry effects on the brain and other tissues (Czech et al., 2012; Dewing et al., 2006; Turner et al., 2011). (2) X genes escaping X inactivation may be expressed constitutively higher in XX than XY cells (Berletch et al., 2010). (3) XX cells receive both a paternal and maternal imprint on the X chromosomes, whereas XY cells receive only a maternal imprint. Differences in imprint could theoretically induce a sex difference in cell function, although no example of this type of phenotypic sex difference has been reported to date. Mouse models that manipulate X chromosome imprint are available (Davies et al., 2005; Isles et al., 2004) but are not discussed further here. (4) The X or Y chromosomes harbor segments of sex-specific heterochromatin that may alter the epigenetic status of autosomes in a sex-specific manner. For example, the presence of a large heterochromatic inactive X chromosome only in XX cells (or the smaller heterochromatic Y chromosome only in XY cells) could alter the availability of factors modulating the euchromatic - heterochromatic status of specific autosomal loci (Wijchers and Festenstein, 2011). This category differs from the first three because the sex chromosome effect is thought not to stem from gene expression from the X or Y chromosomes, but from alterations in the availability of factors regulating the epigenome. Studies of Drosophila support the existence of Y-linked epigenetic effects of this type (Lemos et al., 2008, 2010), but mechanistic evidence is lacking for similar Y effects in mammalian cells (but see Case et al., 2013; Praktiknjo et al., 2013; Spach et al., 2009).
Recent reviews have discussed important mouse models that allow the investigator to manipulate specific sex-biasing factors (Arnold, 2009a: Arnold and Chen, 2009: Majdic and Tobet, 2011; Cox et al., 2014). Here, we summarize briefly two mouse models, Four Core Genotypes (FCG), and XY*, which have been most useful for comparing the effects of the “big three” (A-O-S) sex-biasing factors. We also discuss ChrY substitution strains, which give information on the effects of Y genes.
Four Core Genotypes (FCG) mouse model
The FCG model makes gonadal sex independent of sex chromosome complement (Figure 2A)(Arnold et al., 2009; De Vries et al., 2002). It allows conclusions about which sex differences are affected by gonadal hormones and sex chromosome complement, and the interaction of these factors. Thus, it is often used as the first step in detecting sex chromosome effects (Abel and Rissman, 2011; Chen et al., 2008, 2009; Gatewood et al., 2006; Seney et al., 2013).
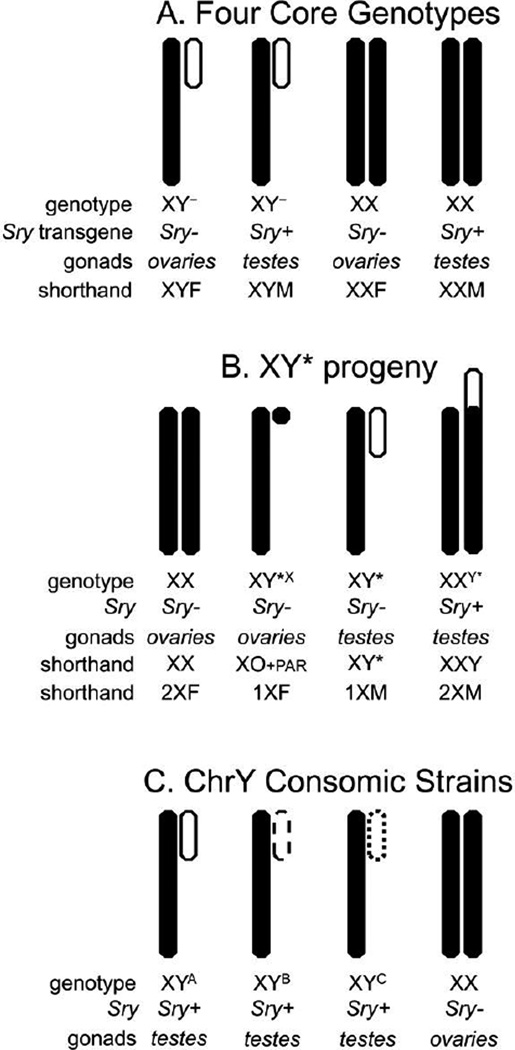
Figure 2.
Simplified schematic models of three mouse models for assessing effects of sex chromosomes. A. The FCG model makes gonad type independent of sex chromosome complement (XX vs. XY), and allows assessment of the independent and interactive effects of gonads and sex chromosome complement on any measurable trait. See text for further discussion (de Vries et al., 2002; Arnold and Chen, 2009). B. The progeny of XY* mice allows comparison of gonadally males mice with one vs. two copies of the non-pseudoautosomal (non-PAR) X chromosome, and gonadally females mice with one vs. two copies of the non-PAR X chromosome. Details of this model are discussed elsewhere (Burgoyne et al., 1998b; Chen et al., 2013; Eicher et al., 1991; Wolstenholme et al., 2012). C. ChrY consomic strains are mice with the same strain background but differing in the strain origin of the Y chromosome. These mice allow measuring the effects of the Y chromosome on traits, assessed by genetic variation of the Y chromosome.
FCG mice comprise four genotypes, XX and XY gonadal males (XXM, XYM), and XX and XY gonadal females (XXF, XYF)(Figure 2A, Figure 3AB). This is possible because of two mutations, deletion of the testis-determining gene Sry from the Y chromosome, and insertion of an Sry transgene onto an autosome (Lovell-Badge and Robertson, 1990: Mahadevaiah et al., 1998). These two changes remove gonadal determination from the sex chromosomes, so that XX and XY mice are generated either with Sry (XXM, XYM) or without Sry (XXF, XYF). Two major comparisons are informative. When XX and XY mice with the same gonadal type differ (XXF vs. XYF, or XXM vs. XYM), the difference is attributable to sex chromosome complement (number of X or Y chromosomes), which is the only genetic difference between the mice. The second comparison is between mice with testes (Sry present) vs. ovaries (Sry absent) (XXM vs. XXF, XYM vs. XYF), which tests for the effects of the type of gonad, or direct effects of Sry outside of the gonads. Importantly, the model allows testing for the interaction of gonadal and sex chromosome effects, for example if testicular hormones have different effects in cells with different sex chromosome complement.
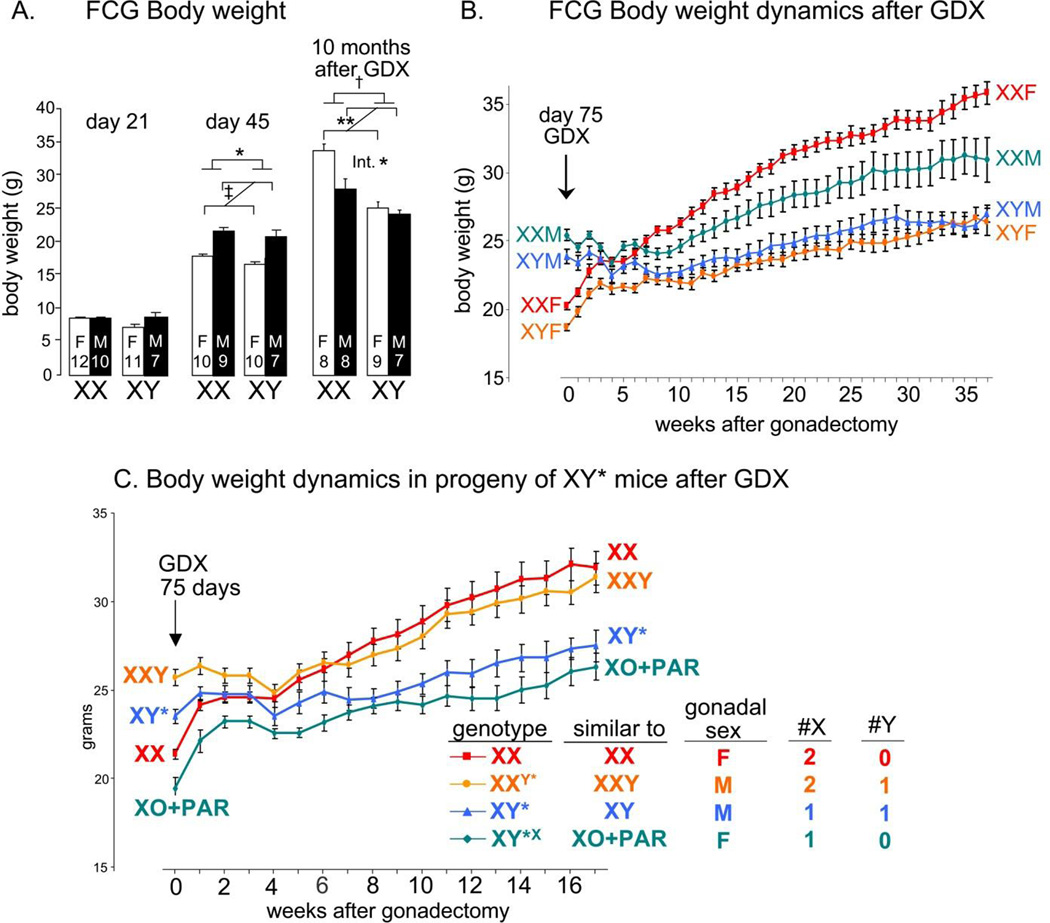
Figure 3.
Using Four Core Genotypes (FCG) and XY* models to dissect sex-biasing factors causing sex differences in body weight. The FCG model varies sex chromosome complement and gonadal type independently, producing four groups: XXM, XX gonadal males; XYM, XY gonadal males; XXF, XX gonadal females; XYF, XY gonadal females. See text and Figure 2 for further discussion of this model. A. FCG mice showed no group differences in body weight at day 21 after birth. By day 45, secretion of gonadal hormones makes gonadal males heavier than gonadal females (‡ p<0.000001), but XX mice weigh slightly more than XY (*p<0.05). Ten months after gonadectomy (GDX at 75 days of age), XX mice are 24% heavier than XY mice († p<0.0001). The effects of sex chromosome complement and gonadal sex interacted significantly (Int, *p<0.05) because XX gonadal females are heavier than gonadal males, but XY gonadal males and females are not different. ** p<0.01 B. Body weight in gonadally intact FCG mice at day 75 and after GDX at day 75. In gonadally intact mice (week 0), when all sex-biasing factors are operating, gonadal males are 25–28% heavier than gonadal females, but XX mice are 6–9% heavier than XY mice. The sex difference caused by activational effects of gonadal secretions disappears in the first month after GDX, after which XX mice gain more weight than XY mice until the sex chromosome effect is nearly as large at 10 months after GDX as the activational effect was prior to GDX. The difference between mice with XX and XY sex chromosome complement is reduced when gonads are present. C. The XY* model varies the number and type of sex chromosomes as shown here and in Figure 2, with two gonadally female groups and two gonadally male groups. Gonadally intact progeny of XY* at 75 days of age (week 0) show greater body weight in the gonadally male groups. After GDX at 75 days of age, the activational effects of gonadal hormones are lost (little difference between gonadal males and females) but mice with two X chromosomes gain weight more than mice with one X chromosome (p<0.000001). The presence of the Y chromosome has little effect. Thus, the sex chromosome effect on body weight is attributable to XX vs. XY differences in the number of X chromosomes. From Chen et al., 2012.
Although several studies have determined that adult XX and XY FCG mice with the same type of gonads have similar levels of circulating gonadal hormones (Gatewood et al., 2006; Palaszynski et al., 2005; Sasidhar et al., 2012), we do not assume that XX and XY mice with the same type of gonad have identical levels of gonadal hormones at all life stages. Specific experimental designs eliminate or reduce potential confounds between sex chromosome complement and levels of gonadal hormones. For example, to eliminate the possibility that an XX vs. XY difference is secondary to different levels of hormones in adulthood, many investigators remove the gonads with or without hormone treatments to make the levels of gonadal hormone equivalent across groups (Chen et al., 2012; De Vries et al., 2002; Seney et al., 2013). If XX and XY mice differ despite having similar hormone levels, the result points more strongly to a direct role of sex chromosome effects outside of the gonads. This is especially convincing when the phenotype is similar in mice that possessed testes vs. ovaries before the age of gonadectomy, a result that implies that large differences in the levels of gonadal hormones before adulthood (organizational effects) are not playing role in causing a sex difference in the phenotype. Ultimately, however, the question of hormonal mediation of a sex chromosome effect is most rigorously tested after identifying the specific X or Y genes (or genetic factors) that cause the sex chromosome effect, and establishing the molecular mechanism of action of the genes or factors.
XY* mouse model
XY* male mice possess the Y* chromosome, which has an aberrant pseudoautosomal region (PAR), allowing abnormal crossing over with the X chromosome during meiosis, which produces abnormal recombination products of the X and Y chromosomes. The detailed genetics of this model are discussed elsewhere (Burgoyne et al., 1998b; Chen et al., 2013; Eicher et al., 1991; Wolstenholme et al., 2012). Mating XY* males to XX females produces progeny that are gonadal males or females, each with one vs. two X chromosomes (Figure 2B, Figure 3C)(Burgoyne et al., 1998a; Eicher et al., 1991). The four main genotypes produced are gonadally female mice with one vs. two copies of the non-PAR region of the X chromosome (containing most of the X genes), and gonadally male mice (with a Y chromosome that contains Sry) with one vs. two copies of the non-PAR X. In the male mice with two copies of the non-PAR X, the second X is fused with the Y chromosome via a disrupted PAR (Figure 2B). If a phenotype has previously been found to differ in XX vs. XY mice in the FCG model, independent of gonadal sex, the XY* model can be used to resolve if the sex chromosome effect is caused by the number of X or Y chromosomes (Chen et al., 2008; Chen et al., 2012, 2013). The XY* model can also be used by itself, to assess if the number of non-PAR X chromosome regions affect a phenotype (Cox and Rissman, 2011; Wistuba et al., 2010; Bonthuis and Rissman, 2013). Because the Y* chromosome contains Sry, the XY* model by itself cannot test if Y-linked factors have effects independent of testicular secretions.
ChrY Substitution (Consomic) Strains
The Y chromosome has long been overlooked as a significant source of factors that cause male-specific effects outside of the testes, because it has few genes that have been thought to act mostly within the testes to regulate testicular differentiation and spermatogenesis (Burgoyne and Mitchell, 2007). Moreover, attempts to test Y chromosome function have been hindered because knocking out Y genes using classic methods has not been successful, although TALEN gene manipulation technology now offers hope for future studies (Wang et al., 2013). Direct Sry effects on the brain have been reported, and may also occur in other tissues such as adrenal and kidney (Dewing et al., 2006; Turner et al., 2011), thus it is clear that the Y chromosome genes can influence traits via actions outside of the testes. A more classic method to study Y chromosome function involves crossing different Y chromosomes (from different mouse strains) onto a single background strain of mice, producing ChrY substitution or consomic strains (Figure 2C). Because genetically identical male mice, differing only in their Y chromosome, show differences in various phenotypes such as embryo size, aggression, susceptibility to autoimmune and other diseases, and androgen effects on the heart (Burgoyne et al., 1995; Monahan and Maxson, 1999; Praktiknjo et al., 2013; Spach et al., 2009), Y genes are implicated in an increasing number of non-testicular functions and diseases. A significant issue in these cases is whether the phenotypic effects of varying the Y chromosome are the results of direct Y gene effects outside the testes, or are mediated by changes in testicular function, for example resulting in different levels of testosterone. The testosterone-mediation of Y chromosome effects cannot usually be answered by showing a lack of effect on testosterone levels at one time of life, because differences at any time of life, especially prenatally, could produce phenotypic effects measured later. In two reported cases of ChrY substitution experiments, substituting the Y chromosome altered plasma levels of testosterone in adulthood, or altered the anogenital distances at birth, an androgen-sensitive phenotype, Thus, the composition of the Y chromosome likely can alter levels of testosterone prenatally (Praktiknjo et al., 2013; Tordjman et al., 1995).
In the context of sex differences in disease, the most extensive study to date of ChrY substitution strains was recently reported (Case et al., 2013). Varying the Y chromosome had dramatic effects on the susceptibility to experimental autoimmune encephalomyelitis (EAE, a mouse model of multiple sclerosis), producing XY mice that were equivalent to, or different from XX mice in the severity of EAE. The effects of different Y chromosomes on EAE did not correlate well with circulating testosterone levels, suggesting that the variability in Y chromosome effects on EAE were not downstream of effects on androgen levels. Instead, the effects of EAE correlated specifically with the Y chromosome content of specific Y alleles, a finding that motivates further studies of the importance of these alleles in EAE. These studies strongly support the idea that Y chromosome variation influences disease, and that Y genes could contribute to the XX vs. XY difference in severity of EAE. The ramifications of these findings for understanding the origin of sex differences are discussed further below.
Sexually dimorphic mechanisms underlying sexual monomorphism
It is fair to say that the A-O-S conceptual framework as outlined above relied on the incorrect assumption that sex-biasing factors operate in each sex only to make that sex different from the other. Activational, organizational, and sex chromosome effects were thought all to “push” in the same direction, increasing sexual dimorphism. It was not considered (or at least, not discussed in studies of phenotypic sex differences) that factors encoded by the Y chromosome, for example, might make males more like females. In contrast, an increasing emphasis of the last 10 years is that compensatory effects of sex-biasing factors may be common (De Vries, 2004). In other words, the lack of a sex difference in a phenotype may not mean sexual equivalence, because various sex-biased factors may cancel each other out. Or, males and females can get to the same place via different underlying mechanisms.
A salient example is X-inactivation, the epigenetic process by which one X chromosome is transcriptionally silenced (to a large extent) in each XX cell (Heard and Disteche, 2006a). X inactivation effectively eliminates a pattern of female-biased expression of X genes that would occur because each X gene is present twice in the XX genome but once in the XY genome (Itoh et al., 2007). In this example, one female-specific process (X-inactivation) has evolved to offset another female-specific condition (presence of a second X chromosome). The female-specific process of X-inactivation makes females more like males than they otherwise would be.
A second example of compensation stems from investigations of sex chromosome and gonadal effects on body weight and adoposity. FCG and XY* mice with two X chromosomes (either XX gonadal females or XX gonadal males) develop greater body weight and adiposity than mice with a single X chromosome (of either gonadal sex), especially if gonadal hormones are removed in adulthood (Figures 3ABC)(Chen et al., 2012, 2013). Gonadal hormones also have potent effects on body weight and adiposity, but in the opposite direction from the effects of sex chromosome complement. Male (testicular) hormones increase body weight (Figure 3A), but a male sex chromosome complement (with a single X chromosome) decreases body weight relative to XX (Figures 3BC). Thus, each of the two sex-biasing factors reduces the impact of the other factor: the effects of gonadal hormones appear to differ in XX vs. XY mice, and the effects of sex chromosome complement are different in mice with testes vs. ovaries.
At a basic level, the A-O-S strategy is based on the idea that if one removes sex-biasing factors, that the sexes become equivalent. The experimental goal is sexual equivalence, or “sex-reversing” one sex to match the phenotype of the other sex, for example by administering hormones. Given that all XX cells perform X inactivation, at the cellular and genomic level sexual equivalence is probably not possible. Preventing X inactivation (for example, by knocking out Xist, the gene that initiates inactivation) is lethal to embryos (Jaenisch et al., 1998), so for phenotypic measurements using live mice the investigator is often constrained to compare XX mice, composed of cells performing X inactivation, with XY mice composed of cells that do not. In live mice XX and XY cells are not equivalent, although this genomic difference may not produce an XX vs. XY difference in many cellular or whole-organ phenotypes. On the contrary, it often reduces phenotypic differences by balancing X gene expression in the two sexes (Heard and Disteche, 2006b: Itoh et al., 2007). It is not clear at present precisely which cellular and whole-organ phenotypes are made sexually dimorphic by the inherent genomic differences between XX and XY cells.
The compensation idea also affects our interpretation of experiments using ChrY consomic strains. In the interesting study of Y chromosome effects on EAE (Case et al., 2013), XYA males, possessing one type of Y chromosome, were found to be are equivalent to XX females, but XYB males with another type of Y chromosome differed from XX females. Based on these results, it might be tempting to conclude that the sex difference in EAE is largely controlled by the Y chromosome. That conclusion rests on the assumption that some Y chromosomes have no effect on the trait (i.e., those variants that make XY males equivalent to XX females). If, instead, all Y chromosomes have some effect, but some variants more perfectly compensate for the lack of a second X chromosome in XY males and make them equivalent to XX females, then the result leaves open a sex-biasing role for one vs. two X chromosomes, in addition to any effect of one vs. zero Y chromosomes. Indeed, among Y genes are a few that have a function similar to a partner gene on the X chromosome (so-called X-Y gene pairs), so that XX and XY mice might be made more similar to each other by the presence of similar proteins encoded by the second sex chromosome, whether it is X or Y. In some mouse strain backgrounds, a second sex chromosome (either X or Y) increases body weight and adiposity relative to XO mice, demonstrating that the second X compensates for the lack of a Y in XX mice, and the Y chromosome in XY mice compensates for the lack of the second X (Chen et al., 2013). ChrY consomic strains, which are valuable for detecting Y chromosome effects on traits, do not directly address the effects of one vs. no Y chromosome, which is the normal male-female difference, which is tested better using the FCG and XY* models together. In future studies, it will be important to vary the number of X and Y chromosomes, independently of the each other, to decide the contributions of X and Y chromosome number to sex difference in EAE. These considerations are an example of the use of sexual differentiation theory to frame experimental questions that would not otherwise be asked, and which are important for a deep understanding of disease processes.
Evolutionary origins of compensation in sex-biasing networks
Why might compensatory sex-biasing effects be common? One answer comes from a consideration of evolutionary forces that generate sex differences. Males and females occupy different biological and social niches, so that mutations can be favored more in one sex than the other. Sexual antagonism occurs in the genome when a mutation has a differential effect on the fitness of the two sexes (Parsch and Ellegren, 2013: Pennell and Morrow, 2013). The differential fitness of the mutation would almost certainly result in sex differences in tissues affected by the mutation. The most extreme case is a mutation that increases fitness of one sex, but decreases the fitness of the other. If the fitness advantage in one sex is sufficiently large, the mutation may become fixed within a population (Ellegren and Parsch, 2007), but the accompanying loss of fitness in the other sex will set up selection pressures that favor mutations that offset the disadvantage. These selection pressures are sex-specific, resulting in a sex-specific change in physiology to protect the disadvantaged sex. For example, the allele might evolve sensitivity to regulatory signals that down-regulate the allele only in the disadvantaged sex, resulting in a sex difference in expression. In any case, the resolution of the sexual conflict involves evolutionary changes in which a sex-biased signal (e.g., hormones or gene products that are present or higher in one sex relative to the other) gains regulatory control over the allele in question. Thus, sexual antagonism may often be resolved when one sex-biased signal creates a sexual dimorphism that counterbalances and offsets another sex-biased process. Within gene networks, the push and pull of sex-biasing forces may therefore reduce sex differences in function. Testosterone could reduce the male-biasing effects of some Y chromosome genes, or the second X chromosome could reduce the effects of ovarian hormones. Because many signals and gene products have pleiotropic effects, it is easy to envision that often individual mutations will result in a variety of changes in function that differ in their degree of sexual antagonism. The separate pleiotropic effects might require different tissue-specific or age-specific countermeasures to reduce the disadvantages of the mutation according to sex, developmental stage, disease state, or tissue type. In the end, a complex array of counterbalancing network interactions may contribute to adaptive tissue function under a wide variety of circumstances.
One implication of these ideas is that in the course of sex-differences research, when one sex-biasing factor is removed, for example by gonadectomy to eliminate sex differences in the effects of gonadal hormones, an opposing sex-biased process (e.g., differential effects of XX vs. XY sex chromosome complement) may be uncovered because it is no longer opposed by compensatory hormonal factors. In the data shown in figure 3B, for example, the effect of sex chromosome complement (differences in body weight of XX vs. XY mice) is much larger after the sex-biasing effects of gonadal hormones are removed. Similarly, a disease process may reduce the effects of one type of sex-biasing factor (for example, by reducing hormone levels, or altering the ability of gene networks to respond to hormones) and lead to sex differences in gene networks that are not obvious in healthy individuals (Seney et al., 2013).
Studying sex differences in animal models for neurology: Where to start?
How does the idea of compensatory sex-biasing factors impact our experimental strategies for investigating sex differences in physiology and disease? Compensation complicates the A-O-S practical approach that we embraced previously (Becker et al., 2005), summarized above. Thus, removing one of the sex-biasing factors (for example, gonadal hormones in adult animals) may be necessary to unmask other sex-biasing factors, and the relative importance of each factor may be dependent on the status of the others. This situation appears to be the case in the example of hormonal and sex chromosomal effects on body weight and adiposity in mice, where gonadectomizing adult mice leads to a large increase in the differential effects of XX vs. XY sex chromosomes (Figure 3B). Although we do not propose that sex differences necessarily lurk behind every sexual monomorphism, the idea of compensation might at least reduce the expectation that the order of investigation of sex-biasing effects is always logically A-O-S. Also, if eliminating one type of sex-bias (e.g., activational effects) eliminates the sex difference, that does not mean that another sex-biasing factor might be important in creating sex differences, especially under other conditions (disease state, age, etc.).
The A-O-S conceptual framework suggests that studying activational effects of hormones is a first reasonable step in discovering the sex-biased mechanisms leading to sex differences in any phenotype, in any organ of the body outside of the gonads. The A-O-S framework also suggests that the investigator investigate sex chromosome effects last, only when the sex difference has been shown not to be fully explained by gonadal hormonal effects, activational or organizational. Following this type of rationale, sex differences in EAE were first studied by manipulations of adult levels of testosterone, which were found to protect females or males from EAE (Palaszynski et al., 2004; Voskuhl, 2011). The potent effects of androgens might have signaled that there was no need to look any further for factors causing the sex difference in EAE. Nevertheless, further studies uncovered a role for sex chromosome complement (FCG XX mice were affected in EAE more than XY mice, (Smith-Bouvier et al., 2008), and study of ChrY consomic strains showed that the Y chromosome influences severity and progression of EAE (Case et al., 2013; Spach et al., 2009). Based on the compensatory effects of sex chromosome and hormonal effects, there is really no longer any reason not to test the sex chromosome effects even if a hormonal effect is robust or strong. Under some conditions (disease states, or hypogonadal conditions such as occur in aging humans), the sex chromosome effects may be more significant than can be predicted by measurement of hormonal effects. Indeed, Seney et al (2013) used FCG mice to vary all three variables (A-O-S) in the context of a single study. Thus, adult FCG mice were gonadectomized and treated with testosterone or not, producing eight groups that allowed comparisons of mice with and without testosterone (activational effects), gonadal females vs. males (a comparison that addresses long term organizational effects of gonadal hormones acting prior to gonadectomy), and XX vs. XY (sex chromosome complement). The authors asked which sex-biasing factors exert control of frontal cortex expression of candidate genes implicated in mood disorders, under the specific experimental condition of chronic unpredictable stress. Under those conditions, sex chromosome effects were more salient than hormone effects, either organizational or activational. That result is surprising because of the historical conclusion that nearly all sex differences in the brain are explained by hormonal effects.
In summary, the compensation concept indicates that effects of A, O and S are not exclusive. If a sex difference in a trait is found to be controlled by activational effects of hormones, for example, the result does not mean that organizational effects or sex chromosome effects are not significant or important, especially under other experimental conditions. Inconveniently, sex differences research has become more complicated. Varying sex chromosome complement, independent of type of gonad, is so far possible only in a few mouse models such as FCG and XY*. These models offer striking advantages discussed above. Nevertheless, the models must be used and results interpreted carefully, cognizant of potential confounds of sex hormone levels with sex chromosome complement (e.g., XX and XY mice with the same type of gonads are probably not precisely equivalent in their levels of gonadal hormones throughout life), and with concern that the presence of genetic rearrangements (e.g., insertion of the Sry transgene) may not mimic the endogenous situation (the endogenous Sry gene on the Y chromosome) in all cases (e.g., De Vries et al., 2002). These issues have generally not limited the utility of the models, for example because XX-XY differences are found whether the Sry transgene is present or not, and use of two completely different models (FXG and XY*) show that results are generalizable and not an artefact of a single genetic model (e.g., Chen et al., 2012, 2013). Thus, use of FCG and XY* models has uncovered convincing and striking differential effects of sex chromosome complement. These discoveries now motivate the search for the genes causing sex chromosome effects. Once the genes are found, it will be possible to investigate the molecular mechanisms that differ in XX vs. XY mice, which might mimic similar differences in humans.
Understanding the Sexome
Conceptualizing gene networks has transformed modern systems biology (Barabasi and Oltvai, 2004). A dominant idea is that genes are nodes in large interacting networks that are mathematically tractable. Genes drive and are driven by each other, and an increasing number of algorithms allow the investigator to discover modules of interacting genes that influence specific phenotypes or are disrupted during disease. Pleiotrophic effects of genes are seen as multiple edges in the network connecting each gene with a few or many other genes. In this conceptual framework, the sex-biasing factors, which are more potent in one sex than the other, push and pull specific parts of the network, and have relatively little effect on other parts. The sexome is the aggregate of all sex-biasing effects on gene networks (Arnold and Lusis, 2012). We are at very early stages of understanding the sexome, because relatively few studies compare gene networks in males and females, or compute how the networks change in response to specific sex-biasing factors (Cvitic et al., 2013: Seney et al., 2013: Van Nas et al., 2009). As more information is gathered, it is likely that we will find instances in which individual sex-biasing factors counteract each other when they act at specific nodes of the network. Unraveling these complex interactions is part of the overall goal of discovering which factors protect from disease, how the protection is exerted within specific pathways, and which protective pathways might respond to novel therapeutic interventions.
Highlights.
-
-
A sophisticated mechanistic understanding of physiology and disease requires knowledge of how sex-biasing factors cause sex differences in phenotype.
-
-
In therian mammals, all sex differences are downstream of the unequal effects of XX vs. XY sex chromosomes.
-
-
Three major categories of sex-biasing factors are activational, organizational, and sex chromosome effects.
-
-
Sex- biasing factors can counteract the effects of each other, reducing rather than producing sex differences in phenotype.
-
-
The Four Core Genotypes and XY* mouse models offer advantages for dissecting sex-biasing effects of gonadal hormones and sex chromosomes.
Acknowledgements
Supported by NIH grants NS043196 and DK083561. I am indebted to many collaborators for providing insight on the issues discussed here, including Paul Burgoyne, Geert de Vries, Emilie Rissman, Karen Reue, Eric Vilain, Mansoureh Eghbali, Xuqi Chen, Yuichiro Itoh, and Shayna Williams-Burris.
Abbreviations
- A-O-S
an experimental strategy for discovering specific categories of sex-biasing factors that cause disease, explained in the text.
- FCG
Four Core Genotypes mouse model
- Sry
Sex-determining region of the Y chromosome, a protein-coding gene causing differentiation of testes
- XXF
Gonadal female mice with XX sex chromosome complement
- XYF
Gonadal female mice with XY sex chromosome complement
- XXM
Gonadal male mice with XX sex chromosome complement
- XYM
Gonadal male mice with XY sex chromosome complement
- Xist
X-inactive specific transcript, an RNA gene on the X chromosome expressed from the inactive X chromosome, which initiates X inactivation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel JL, Rissman EF. Location, location, location: Genetic regulation of neural sex differences. Rev. Endocr. Metab Disord. 2011 doi: 10.1007/s11154-011-9186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol. 2009a;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Hormones and Behavior. 2009b;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2011;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Chen X. What does the "four core genotypes" mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;30:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Gorski RA. Gonadal steroid induction of structural sex differences in the CNS. Annual Review of Neuroscience. 1984;7:413–442. doi: 10.1146/annurev.ne.07.030184.002213. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Lusis AJ. Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology. 2012;153:2551–2555. doi: 10.1210/en.2011-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Becker JB, Arnold AP, Berkley KJ, Blaustein JD, Eckel LA, Hampson E, Herman JP, Marts S, Sadee W, Steiner M, Taylor J, Young E. Strategies and methods for research on sex differences in brain and behavior. Endocrinology. 2005;146:1650–1673. doi: 10.1210/en.2004-1142. [DOI] [PubMed] [Google Scholar]
- Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neuroscience Biobehavior Review. 2011;35:565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthuis PJ, Rissman EF. Neural growth hormone implicated in body weight sex differences. Endocrinology. 2013;154:3826–3835. doi: 10.1210/en.2013-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedlove SM, Cooke BM, Jordan CL. The orthodox view of brain sexual differentiation. Brain, Behavior and Evolution. 1999;54:8–14. doi: 10.1159/000006607. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. The Y* rearrangement in mice: new insights into a perplexing PAR. Cytogenetics and Cell Genetics. 1998a;80:37–40. doi: 10.1159/000014954. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. The Y* rearrangement in mice: new insights into a perplexing PAR. Cytogenet Cell Genet. 1998b;80:37–40. doi: 10.1159/000014954. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mitchell MJ. The role of mouse Y chromosome genes in spermatogenesis. In: Lau YFC, Chan WY, editors. Y Chromosome and Male Germ Cell Biology. Hackensack NJ: World Scientific Publisher; 2007. pp. 27–45. [Google Scholar]
- Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos. TransRSoc. Lond B Biol. Sci. 1995;350:253–260. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- Case LK, Wall EH, Dragon JA, Saligrama N, Krementsov DN, Moussawi M, Zachary JF, Huber SA, Blankenhorn EP, Teuscher C. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 2013;23:1474–1485. doi: 10.1101/gr.156703.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Grisham W, Arnold AP. X chromosome number causes sex differences in gene expression in adult mouse striatum. Eur. J. Neurosci. 2009;29:768–776. doi: 10.1111/j.1460-9568.2009.06610.x. [DOI] [PubMed] [Google Scholar]
- Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, Reue K. The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8:e1002709. doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, McClusky R, Itoh Y, Reue K, Arnold AP. X and Y chromosome complement influence adiposity and metabolism in mice. Endocrinology. 2013;154:1092–1104. doi: 10.1210/en.2012-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Watkins R, Delot E, Reliene R, Schiestl RH, Burgoyne PS, Arnold AP. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev. Neurobiol. 2008;68:265–273. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- Cox KH, Bonthuis PJ, Rissman EF. Mouse model systems to study sex chromosome genes and behavior: Relevance to humans. Front Neuroendocrinol. 2014 doi: 10.1016/j.yfrne.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011;10:465–472. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitic S, Longtine MS, Hackl H, Wagner K, Nelson MD, Desoye G, Hiden U. The Human Placental Sexome Differs between Trophoblast Epithelium and Villous Vessel Endothelium. PLoS ONE. 2013;8:e79233. doi: 10.1371/journal.pone.0079233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech DP, Lee J, Sim H, Parish CL, Vilain E, Harley VR. The human testis- determining factor SRY localizes in midbrain dopamine neurons and regulates multiple components of catecholamine synthesis and metabolism. Journal of Neurochemistry. 2012 doi: 10.1111/j.1471-4159.2012.07782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- De Vries GJ. Minireview: Sex differences in adult and developing brains: compensation, compensation, compensation. Endocrinology. 2004;145:1063–1068. doi: 10.1210/en.2003-1504. [DOI] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. Journal of Neuroscience. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewing P, Chiang CWK, Sinchak K, Sim H, Fernagut PO, Kelly S, Chesselet MF, Micevych PE, Albrecht KH, Harley VR, Vilain E. Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Hale DW, Hunt PA, Lee BK, Tucker PK, King TR, Eppig JT, Washburn lL. The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenetics and Cell Genetics. 1991;57:221–230. doi: 10.1159/000133152. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. Journal of Neuroscience. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes and Development. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Isles AR, Davies W, Burrmann D, Burgoyne PS, Wilkinson LS. Effects on fear reactivity in XO mice are due to haploinsufficiency of a non-PAR X gene: implications for emotional function in Turner's syndrome. Human Molecular Genetics. 2004;13:1849–1855. doi: 10.1093/hmg/ddh203. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. Dosage compensation is less effective in birds than in mammals. J. BIol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Beard C, Lee J, Marahrens Y, Panning B. Mammalian X chromosome inactivation. Novartis. Found. Symp. 1998;214:200–209. doi: 10.1002/9780470515501.ch12. [DOI] [PubMed] [Google Scholar]
- Jost A, Vigier B, Prepin J, Perchellet JP. Studies on sex differentiation in mammals. Recent Progress in Hormone Research. 1973;29:1–41. doi: 10.1016/b978-0-12-571129-6.50004-x. [DOI] [PubMed] [Google Scholar]
- Juraska JM, Sisk CL, DonCarlos LL. Sexual differentiation of the adolescent rodent brain: hormonal influences and developmental mechanisms. Hormones and Behavior. 2013;64:203–210. doi: 10.1016/j.yhbeh.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Karastergiou K, Smith SR, Greenberg AS, Fried SK. Sex differences in human adipose tissues - the biology of pear shape. Biol. Sex Differ. 2012;3:13. doi: 10.1186/2042-6410-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development. 1990;109:635–646. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn lL, McCarrey JR, Cattanach BM, Lovell-Badge R, Burgoyne PS. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Human Molecular Genetics. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- Majdic G, Tobet S. Cooperation of sex chromosomal genes and endocrine influences for hypothalamic sexual differentiation. Front Neuroendocrinol. 2011;32:137–145. doi: 10.1016/j.yfrne.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat. Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Kaplan JR, Schork NJ, Ouyang P, Berga SL, Wenger NK, Shaw LJ, Webb RC, Mallampalli M, Steiner M, Taylor DA, Merz CN, Reckelhoff JF. Strategies and methods to study sex differences in cardiovascular structure and function: a guide for basic scientists. Biol. Sex Differ. 2011;2:14. doi: 10.1186/2042-6410-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan EJ, Maxson SC. Y chromosome, urinary chemosignals, and an agonistic behavior (offense) of mice. Physiology & Behavior. 1999;64:123–132. doi: 10.1016/s0031-9384(98)00041-9. [DOI] [PubMed] [Google Scholar]
- Palaszynski KM, Loo KK, Ashouri JF, Liu HB, Voskuhl RR. Androgens are protective in experimental autoimmune encephalomyelitis: implications for multiple sclerosis. Journal of Neuroimmunology. 2004;146:144–152. doi: 10.1016/j.jneuroim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR. A Yin-Yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinology. 2005;146:3280–3285. doi: 10.1210/en.2005-0284. [DOI] [PubMed] [Google Scholar]
- Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- Pennell TM, Morrow EH. Two sexes, one genome: the evolutionary dynamics of intralocus sexual conflict. Ecol. Evol. 2013;3:1819–1834. doi: 10.1002/ece3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- Praktiknjo SD, Llamas B, Scott-Boyer MP, Picard S, Robert F, Langlais D, Haibe- Kains B, Faubert D, Silversides DW, Deschepper CF. Novel Effects of Chromosome Y on Cardiac Regulation, Chromatin Remodeling, and Neonatal Programming in Male Mice. Endocrinology. 2013;154:4746–4756. doi: 10.1210/en.2013-1699. [DOI] [PubMed] [Google Scholar]
- Sandberg K, Ji H. Sex differences in primary hypertension. Biol. Sex Differ. 2012;3:7. doi: 10.1186/2042-6410-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis. 2012;71(8):1418–22. doi: 10.1136/annrheumdis-2011-201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KM, Molenda-Figueira HA, Sisk CL. Back to the future: The organizational- activational hypothesis adapted to puberty and adolescence. Hormones and Behavior. 2009;55:597–604. doi: 10.1016/j.yhbeh.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seney ML, Ekong KI, Ding Y, Tseng GC, Sibille E. Sex chromosome complement regulates expression of mood-related genes. Biol. Sex Differ. 2013;4:20. doi: 10.1186/2042-6410-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR, Voskuhl RR. A role for sex chromosome complement in the female bias in autoimmune disease. Journal of Experimental Medicine. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spach KM, Blake M, Bunn JY, McElvany B, Noubade R, Blankenhorn EP, Teuscher C. Cutting edge: the Y chromosome controls the age-dependent experimental allergic encephalomyelitis sexual dimorphism in SJL/J mice. Journal of Immunology. 2009;182:1789–1793. doi: 10.4049/jimmunol.0803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilmann C, Capel B. Cellular and molecular pathways regulating mammalian sex determination. Recent Prog. Horm. Res. 2002;57:1–18. doi: 10.1210/rp.57.1.1. [DOI] [PubMed] [Google Scholar]
- Tordjman S, Roubertoux PL, Carlier M, Moutier R, Anderson G, Launay M, Degrelle H. Linkage between brain serotonin concentration and the sex-specific part of the Y-chromosome in mice. Neuroscience Letters. 1995;183:190–192. doi: 10.1016/0304-3940(94)11148-c. [DOI] [PubMed] [Google Scholar]
- Turner ME, Ely DL, Prokop J, Milsted A. Sry, more than testis determination? Am. J. Physiol Regul. Integr. Comp Physiol. 2011;301:R561–R571. doi: 10.1152/ajpregu.00645.2010. [DOI] [PubMed] [Google Scholar]
- Van Nas A, GuhaThakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, Arnold AP, Lusis AJ. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuhl R. Sex differences in autoimmune diseases. Biol. Sex Differ. 2011;2:1. doi: 10.1186/2042-6410-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. TALEN-mediated editing of the mouse Y chromosome. Nat. Biotechnol. 2013;31:530–532. doi: 10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Wistuba J, Luetjens CM, Stukenborg JB, Poplinski A, Werler S, Dittmann M, Damm OS, Hamalainen T, Simoni M, Gromoll J. Male 41, XXY* mice as a model for klinefelter syndrome: hyperactivation of leydig cells. Endocrinology. 2010;151:2898–2910. doi: 10.1210/en.2009-1396. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Rissman EF, Bekiranov S. Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 2012;12:166–180. doi: 10.1111/gbb.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]