Abstract
RNAs, more than ever before, are increasingly viewed as biomolecules of the future, in the versatility of their functions and intricate three-dimensional folding. To effectively study them by nuclear magnetic resonance (NMR) spectroscopy, structural biologists need to tackle two critical challenges of spectral overcrowding and fast signal decay for large RNAs. Stable-isotope nucleotide labeling is one attractive solution to the overlap problem. Hence, developing effective methods for nucleotide labeling is highly desirable. In this work, we have developed a facile and streamlined source of recombinant enzymes from the pentose phosphate pathway for making such labeled nucleotides. The Escherichia coli (E. coli) genes encoding ribokinase (RK), adenine phosphoribosyltranferase (APRT), xanthine/guanine phosphoribosyltransferase (XGPRT), and uracil phosphoribosyltranferase (UPRT) were subcloned into pET15b vectors. All 4 constructs together with cytidine triphosphate synthetase (CTPS) and human phosphoribosyl pyrophosphate synthetase isoform 1 (PRPPS) were transformed into the E. coli BL21 (AI) strain for protein overexpression. The enzyme preparations were purified to >90% homogeneity by a one-step Ni-NTA affinity chromatography, without the need of a further size-exclusion chromatography step. We obtained yields of 1530, 22, 482, 3120, 2120 and 2280 units of activity per liter of culture for RK, PRPPS, APRT, XGPRT, UPRT and CTPS, respectively; the specific activities were found to be 70, 22, 21, 128, 144 and 113 U/mg, respectively. These specific activities of these enzyme constructs are comparable to or higher than those previously reported. In addition, both the growth conditions and purification protocols have been streamlined so that all the recombinant proteins can be expressed, purified and characterized in at most two days. The availability and reliability of these constructs should make production of fully and site-specific labeled nucleotides for making labeled RNA accessible and straightforward, to facilitate high-resolution NMR spectroscopic and other biophysical studies.
Keywords: His-tagged recombinant E. coli ribokinase, Human PRPP synthetase, Phosphoribosyltransferases, CTP synthetase, E. coli BL21(AI) strains, Affinity chromatography
Introduction
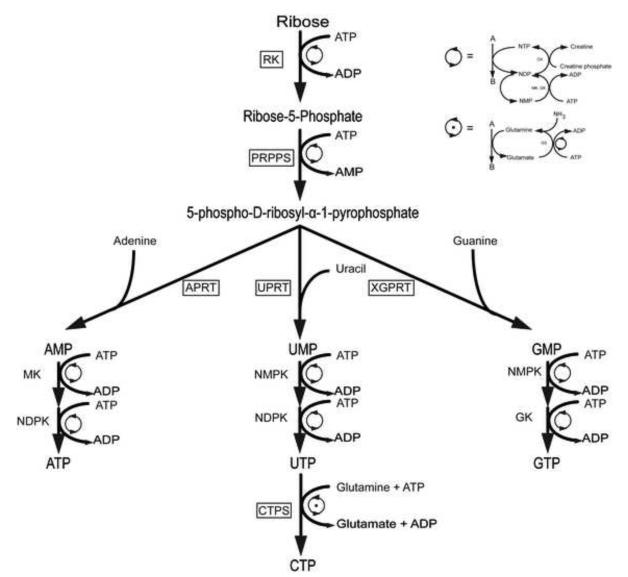
RNA molecules have taken center stage as fundamental transactors of catalysis and gene regulation making them valuable targets for drug discovery efforts and biophysical characterizations [1-16]. Advances in methods for synthesizing labeled RNA [17-20] have fueled the development of new nuclear magnetic resonance (NMR) experiments specifically tailored for RNA biophysical measurements [13-15, 21-23]. Unfortunately, two challenges facing RNA NMR structural biology become even more acute for large RNAs: extensive spectral crowding and increased resonance linewidths. Overcoming both challenges necessitates the development of alternate enzymatic synthetic labeling strategies using the enzymes of the E. coli pentose phosphate pathway, as depicted in figure 1 [17-19]. This method enables the synthesis of all four nucleoside triphosphates (ATP, CTP, GTP, and UTP) using three phosphoribosyltransferases and CTP synthetase, and ribokinase (if using ribose), or hexokinase and three other enzymes if starting with glucose. Ribose-5-phosphate (R5P), synthesized from glucose or D-ribose, is then converted to 5-phospho-D-ribosyl-α-1-pyrophosphate (PRPP) by PRPP synthetase.
Figure 1.
Nucleotide salvage branch of the pentose phosphate pathway. This scheme of reactions is proposed to enzymatically synthesize site-specific labeled nucleoside triphosphates (NTPs), where the boxed enzymes were expressed, purified and characterized in this work, the rest are commercially available. Starting from ribose, ribokinase (RK) would make ribose 5-phosphate. Then, phosphoribosyl pyrophosphate synthetase (PRPPS) would convert it to the activated ribose phosphate, 5-phospho-D-ribosyl-α-1-pyrophosphate. Following that, the respective adenine, uracil or xanthine/guanine phosphoribosyltransferases (APRT, UPRT, XGPRT) would synthesize AMP, UMP or GMP. The nucleoside monophosphates would then be phosphorylated by myokinase (MK), nucleoside monophosphate kinase (NMPK), nucleoside diphosphate kinase (NDPK) or guanylate kinase (GK) to the nucleoside triphosphate stage. Finally, CTP would be synthesized from UTP by CTP synthase (CTPS). Here the ATP and glutamine regeneration schemes use creatine kinase (CK) and glutamine synthase (GS), respectively.
A key advantage of this labeling scheme is the ability to differentially combine labeled glucose or ribose with labeled bases. Using this general strategy, the Williamson group has produced several isotopic labeling patterns [17, 24-28] valuable for NMR spectroscopic analysis of RNA structure and dynamics. For example ribose specifically labeled at C1′ and/or C5′ can be combined with uracil base specifically labeled at C5 or C6 to produce a label that will greatly simplifiy RNA signal crowding in NMR spectra. Note that CTP synthetase easily provides an identically labeled CTP. Unfortunately, one limitation of this enzymatic approach is that 6 out of the 18 enzymes required for synthesis are not commercially available and must be produced from over-expressing E.coli strains. This hurdle must be overcome to make this method widely adopted by the biophysics and chemical biology communities. Of these six enzymes, only ribokinase and adenosine phosphoribosyltransferase (APRT) have robust activities of 350-700 U (U is the unit of activity, defined as μmol of substrate turned over per min) per liter of bacterial culture [29]. The other five are only moderately over-expressed with activities of 28-40 U, making the enzyme preparation labor intensive [17, 24, 26]. By finding constructs that highly over-express the commercially unavailable enzymes, the cost and labor required to synthesize isotopically labeled NTPs could be significantly reduced. Once these constructs are produced, it is expected that the process for obtaining specifically labeled NTPs will be facile and streamlined. Ribokinase, RK (E.C. 2.7.1.15), catalyzes the phosphorylation at the ribose C5′ position to produce ribose-5-phosphate [30]. Phosphoribosyl pyrophosphate synthetase, PRPPS (E.C. 2.7.6.1), catalyzes the addition of a β,γ-diphosphate from ATP to the C1′ position of ribose-5-phosphate, activating it for the addition of a nucleobase [31]. The phosphoribosyl transferases catalyze the formation of a β-substituted ribose-5-phosphate with a specific base starting from PRPP [32]. Adenine and uracil phosphoribosyl transferases, APRT (E.C. 2.4.2.7) and UPRT (E.C. 2.4.2.9), catalyze the reaction between adenine or uracil and PRPP to form AMP or UMP, respectively [33,34]. Xanthine/Guanine phosphoribosyltransferase, XGPRT (E.C. 2.4.2.22), catalyzes not only the transfer of guanine to PRPP to form GMP, but also the transfer of xanthine or hypoxanthine to form XMP or IMP, respectively [35]. For this study, the relevant reaction is that with guanine to form GMP. In the proposed nucleotide synthesis pathway utilizing the pentose phosphate pathway (Fig. 1), cytosine triphosphate is not synthesized by a phosphoribosyl transferase and a kinase as is the case for ATP, GTP and UTP; there is no known cytidine phosphoribosyltransferase. Rather, CTP synthethase, CTPS (E.C. 6.3.4.2), is utilized in a separate reaction as the catalyst in the ATP-dependent production of CTP from UTP, using glutamine or ammonia as the source of nitrogen [26, 36].
In this study, we subcloned the E.coli genes encoding RK, APRT, UPRT, and XGPRT into pET15b vectors. Then we transformed these four constructs and CTPS and PRPPS in E. coli BL21(AI) strains for enzyme overexpression. Subsequently, we purified all six proteins by a one-step Nickel Nitrilotriacetic acid (Ni-NTA) affinity chromatography yielding enzymes with high purity and activity for use in NTP enzymatic synthesis. The production of these enzymes should make it seamless and straightforward to produce labeled nucleotides for biophysical applications.
Materials and Methods
Cloning of RK, UPRT, APRT and XGPRT into His-tagged expression vector
We designed the following primers to contain both XhoI and BamHI restriction sites which allowed the amplified DNA fragments to be ligated into a pET15b vector (Novagen).
RK forward primer
dCGCCTCGAGATGCAAAACGCAGGCAGCCTCGTTGT
RK reverse primer
dGCGGGATCCTCACCTCTGCCTGTCTAAAAATGCGT
UPRT forward primer
dCCGCGCCTCGAGATGAAGATCGTGGAAGTCAAACAC
UPRT reverse primer
dGCGGGATCCTTATTTCGTACCAAAGATTTTGTCACC
APRT forward primer
dCGCCTCGAGATGACCGCGACTGCACAGCAGCTTG
APRT reverse primer
dGCGGGATCCTTAATGGCCCGGGAACGGGACAAGGC
XGPRT forward primer
dCGCCTCGAGATGAGCGAAAAATACATCGTCACCTG
XGPRT reverse primer
dGCGGGATCCTTAGCGACCGGAGATTGGCGGGACGA
The corresponding sequences of the E.coli genes were all amplified from plasmids (kindly provided by Professor Williamson at The Scripps Research Institute, LaJolla, CA) by PCR using Pfu polymerase (Stratagene). The PCR products for the different genes were gel purified using Qiagen kit and ligated into either pCR4-TOPO (Invitrogen) or pGEMT (Promega) vectors.
The vectors were transformed into DH5α cells (Invitrogen), and then plated on LB/agar plates containing 100 μg/mL ampicillin and 1 μg/mL IPTG and Xgal for blue/white screening, and at least three positive white colonies were cultured in 20 mL LB media containing 100 μg/mL ampicillin. Plasmids were isolated using Qiagen Miniprep spin columns and restriction digest reactions were performed using XhoI and BamHI (NEB). The positive plasmids were digested in a stepwise manner with XhoI for 2 h at 37°C followed by DNA purification using Qiagen kit and then digested by BamHI for 2 h at 37°C. Insert fragments as well as the digested pET15b were extracted from 1.5% agarose gel using Qiagen gel extraction kit. Ligation reactions with T4 DNA ligase (NEB) were supplemented with 4% PEG, and the ligated constructs were transformed into TOP10 cells (Invitrogen). The nucleotide sequences of cloned fragments were verified by sequencing. Plasmids from positive colonies, identified by digesting isolated plasmids with XhoI/BamHI, were transformed into E. coli BL21(AI) for protein over-expression.
Protein expression
RK, UPRT, APRT, XGPRT
All four E. coli enzymes not obtained commercially were overexpressed and purified using similar procedures. The pET15b plasmid constructs prepared in this work, were all transformed into E.coli strain BL21(AI) (Invitrogen) for protein overexpression under arabinose regulation and IPTG induction. The expected molecular weights for RK, APRT, XGPRT and UPRT were 32, 20, 17 and 23 KDa, respectively. All protein expressions and purifications were carried out for RK as described previously with very little modification [30,37]. Cells transformed with the RK construct were grown onto LB plates supplemented with 100 μg/mL ampicillin. A single colony was grown at 37°C in a 5-mL starter culture of LB media with 100 μg/mL ampicillin to OD600=0.60 (ca. 4h). The cells were harvested by centrifugation at 3800 rpm for 15 min, resuspended in fresh LB, and added to 100 mL LB media with 100 μg/mL ampicillin. The cell culture was grown at 37°C to OD600=0.60 (ca. 2h); then the cells were harvested by centrifugation at 3800 rpm for 15 min and resuspended in fresh LB and added to 1 L LB media with 100 μg/mL ampicillin. Expression was induced with 0.05% L-(+)-arabinose at 37°C for 2 h followed by addition of 1mM IPTG for 3 h at 37°C. The cells were harvested by centrifugation at 4500 rpm for 25 min at 4°C, and stored at −80°C.
PRPP Synthethase
The plasmid used for expression of full length human PRPP synthetase, 34 KDa, was cloned into the NdeI and XhoI restriction sites of pET22b(+) expression plasmid (a generous gift from Professor Sheng Li, Graduate school of the Chinese Academy of Sciences, Shanghai) and then it was transformed into E.coli strain BL21(AI) (Invitrogen) to produce an overexpressing strain of isoform 1 of human PRPPS. PRPPS expression and purification were carried out as described previously with some modifications [31]. A single colony was grown at 37°C in a 5-mL starter culture of LB media with 100 μg/mL ampicillin to OD600=0.60 (ca. 4h). The cells were harvested by centrifugation at 3800 rpm for 15 min, resuspended in fresh LB, and added to 100 mL LB media with 100 μg/mL ampicillin. The cell culture was grown at 37°C to OD600=0.60 (ca. 2h); then the cells were harvested by centrifugation at 3800 rpm for 15 min and resuspended in fresh LB and added to 1 L LB media with 100 μg/mL ampicillin. Expression was induced with 0.05% L-(+)-arabinose at 37°C for 2 h followed by addition of 1mM IPTG for 3 h at 37°C. The cells were harvested by centrifugation at 4500 rpm for 25 min at 4°C, and stored at −80°C.
CTP Synthetase
The E. coli CTP synthetase gene, 60 KDa protein product, previously sub-cloned into pET15b (a kind gift of Professor Stephen L. Bearne, Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, Nova Scotia) was transformed into E.coli strain BL21(AI) for protein overexpression. A single colony was grown at 37°C in a 5-mL starter culture of LB media with 100 μg/mL ampicillin to OD600=0.60 (ca. 4h). The cells were harvested by centrifugation at 3800 rpm for 15 min, resuspended in fresh LB, and added to 100 mL LB media with 100 μg/mL ampicillin. The cell culture was grown at 37°C to OD600=0.60 (ca. 2h); then cells were harvested by centrifugation at 3800 rpm for 15 min and resuspended in fresh LB and added to 1 L LB media with 100 μg/mL ampicillin. Expression was induced with 0.05% L-(+)-arabinose at 37°C for 2 h followed by addition of 1mM IPTG for 3 h at 37°C. The cells were harvested by centrifugation at 4500 rpm for 25 min at 4°C, and stored at −80°C.
Protein purification
Conditions for the purification of all the recombinant His-tagged enzymes were optimized for maximal yield and purity by nickel affinity chromatography, and, as an optional step, size-exclusion chromatography could be performed using Sephadex S75 gel filtration column for all the proteins. The frozen cell pellet was resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) supplemented with 1 mg/mL lysozyme and placed on ice for 30 min without addition of nucleases. The cells were disrupted by sonication at 4°C for 1 min with 1 min resting periods. The cellular debris was pelleted by centrifugation at 46000 g and 4°C for 30 min and the viscous nucleic acids were manually removed using a pipette. The purifications were carried out following the QIAexpressionist handbook (Qiagen) with very little modification. The supernatant was applied to pre-packed Ni-NTA beads in column mode, previously equilibrated with lysis buffer.
Following incubation, the Ni-NTA column was washed with 6 bed volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole, pH 8). Recombinant proteins were then eluted from the Ni-NTA column with elution buffer (wash buffer with 400 mM imidazole). The volume of the two eluates were reduced using Amicon ultra tubes (Milipore), typically to 200 μL and then buffer-exchanged to its final storage buffer (50 mM NaH2PO4, 150 mM NaCl or 300 mM NaCl for PRPPS only) to a final volume of 1 mL. Equal volume of 100% glycerol was added and the proteins were stored at −20°C. The different stages of purification were monitored by SDS-PAGE. Protein concentrations were determined by Bradford assay employing BSA as the standard.
Enzyme activity assays
Biochemical Assays
Enzymes were assayed using known procedures [17,26,37,38,39,40,41]
Ribokinase
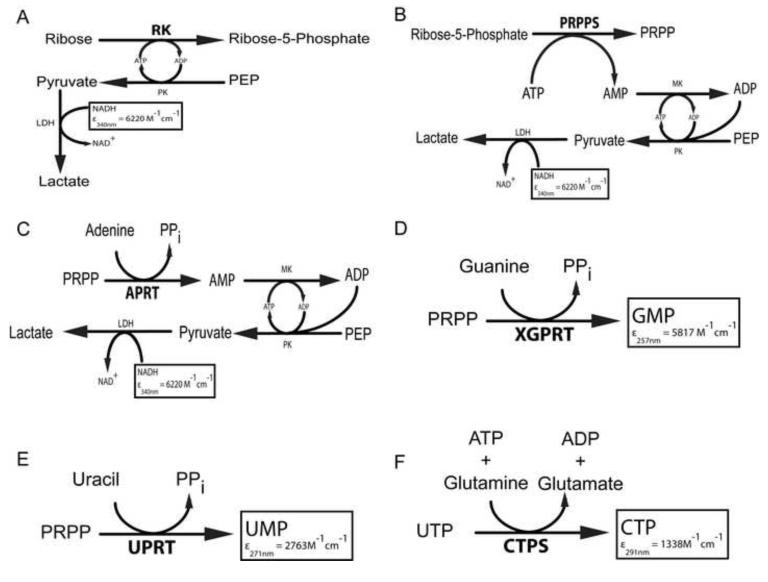
The spectrophotometric assay is based on the coupled enzyme system [17,39] as shown in Figure 3a. The assay mixture (1 mL) contained 50 mM Tris-HCl buffer at pH 7.8, 5 mM Ribose, 3mM ATP, 1mM PEP, 100 mM KCl, 10 mM MgCl2, 0.2mM NADH, 2U of lactate dehydrogenase, and 2U of pyruvate kinase. The mixture was incubated for 5 min until a steady baseline was obtained and then a 2-μL aliquot of ribokinase was added to initiate the reaction. The absorbance change at 340 nm (ΔA340) was monitored as a function of time using the linear range of the kinetic trace. Units of activity were calculated as a function of the total reaction volume (V in L), path length (l in cm), time (t in min), change in extinction coefficient of 6220 M−1 cm−1 at 340nm for oxidation of NADH to NAD+ and K = 1:
| Equation(1) |
Figure 3.
a-f. Direct and indirect continuous activity assays used for the six enzymes. (a) RK (b) PRPPS (c) APRT (d) XGPRT (e) UPRT (f) CTPS. The boxed molecule corresponds to the one being monitored during the assay at the specific wavelength with the corresponding molar extinction coefficient in M−1cm−1. MK: myokinase, PK: pyruvate kinase, LDH: lactate dehydrogenase, PEP: phopshoenolpyruvate, PPi= inorganic pyrophosphate.
PRPP Synthetase
The spectrophotometric assay is based on the coupled enzyme system [39] as depicted in Figure 3b. The assay mixture (1 mL) contained 50 mM Tris-HCl buffer at pH 7.5, 5 mM R5P , 3mM ATP , 1mM PEP , 10 mM MgCl2, 0.2mM NADH, 2U of lactate dehydrogenase, 2U of pyruvate kinase, and 2U of adenylate kinase. The mixture was incubated for 5 min until a steady baseline was obtained and then a 2-μL aliquot of PRPP synthetase was added to initiate the reaction. The absorbance change at 340 nm (ΔA340) was monitored as a function of time using the linear range of the kinetic trace. Units of activity were calculated using equation (1), where ε = 6220 M−1 cm−1 at 340nm and K = 2.
APRT
The spectrophotometric assay is based on the coupled enzyme system [17,39] as shown in Figure 3c. The assay mixture (1 mL) contained 50 mM Tris-HCl buffer at pH 7.8, 1.5 mM PRPP, 3mM ATP, 1mM PEP, 1.5 mM adenine hydrochloride, 10 mM MgCl2, 0.2mM NADH, 2U of lactate dehydrogenase, 2U of pyruvate kinase and 2U of adenosine kinase. The mixture was incubated for 5 min until a steady baseline was obtained and then a 2-μL aliquot of APRT was added to initiate the reaction. The absorbance change at 340 nm (ΔA340) was monitored as a function of time using the linear range of the kinetic trace. Units of activity were calculated using equation (1), where ε = 6220 M−1 cm−1 at 340nm and K = 2.
XGPRT
The spectrophotometric assay is based on monitoring the conversion of guanine (246 nm) to GMP at 257.5 nm using the change in extinction coefficient of ε=5817 M−1 cm−1 [41-43] as shown in Figure 3d. The assay mixture (1 mL) contained 100 mM Tris-HCl buffer at pH 7.5, 1 mM PRPP, 50 μM of guanine, 100 mM MgCl2 and a 2-μL aliquot of XGPRT. The reaction mixture was incubated without guanine for 5 min. Then, the reaction was initiated by the addition of guanine, and the formation of GMP was monitored at 257.5 nm at 25°C until it reached saturation after 5-7 min [42]. Units of activity were calculated using equation (1), where ε = 5817 M−1 cm−1 at 257.5 nm and K = 1.
UPRT
The spectrophotometric assay is based on monitoring the conversion of uracil (271 nm) to UMP using the change in extinction coefficient of ε=2763 M−1 cm−1 [17] as shown in Figure 3e. The assay mixture (1 mL) contained 50 mM Tris-HCl (pH 7.5), 1.5 mM PRPP, 0.1 mM of uracil, 5 mM MgCl2 and a 2-μL aliquot of UPRT. The reaction was incubated without uracil for 5 min. Then the reaction was initiated by addition of uracil, and the formation of UMP was monitored at 271 nm at 25 or 37°C until it reached saturation after 5-7 min. Units of activity were calculated using equation (1), where ε = 2763 M−1 cm−1 at 271nm and K = 1.
CTP Synthetase
The spectrophotometric assay is based on the increase in absorbance at 291nm following conversion of UTP to CTP using the change in extinction coefficient of ε=1338 M−1 cm−1 [17,44] as shown in Figure 3f. The assay mixture (1 mL) contained 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM UTP, 1 mM ATP, 0.25 mM GTP, 10 mM glutamine and a 2-μL aliquot of CTPS. The reaction was incubated without glutamine for 5 min. Then the reaction was initiated by addition of glutamine, and the formation of CTP was monitored at 291 nm at 25 or 37°C until it reached saturation after 12-15 min. Units of activity were calculated using equation (1), where ε = 1338 M−1 cm−1 at 291nm and K = 1.
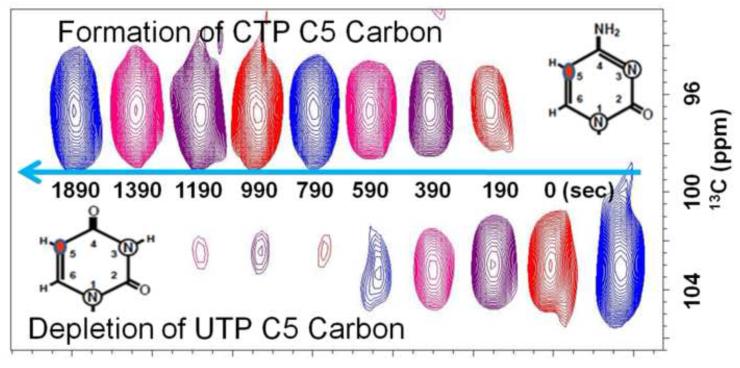
As an independent method for cross-validating the spectrophotometric method, 1H-13C heteronuclear single quantum correlation (HSQC) NMR experiment was performed on a 600 MHz Bruker Avance III spectrometer at 37°C. The NMR assay is based on the appearance of 13C –labeled CTP and disappearance of 13C –labeled UTP as monitored through the C5 aromatic carbons of UTP and CTP. The UTP C5 carbon resonates in a spectral region (~103 ppm) distinct from CTP’s C5 (~97 ppm) carbon. This separation makes it very straightforward to monitor the formation of CTP from UTP using two-dimensional NMR. Unfortunately, 1D NMR fails because the H5 proton resonances of both UTP and CTP overlap with their ribose H1′ proton resonances. The carbon resonances remove this degeneracy. Under our reaction conditions, CTP is made quantitatively in 30 min from UTP. The assay mixture (250μL) contained 90% D2O, 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM 13C-15N-UTP (Sigma-Aldrich), 1 mM ATP, 0.25 mM GTP, 10 mM glutamine and a 0.5-μL aliquot of CTPS. The reaction was incubated without glutamine for 5 min. The reaction was initiated by the addition of glutamine. The formation of CTP and disappearance of UTP were monitored at 37°C until the reaction reached saturation after 30 min. The results were fit to Equation (2) were Yo is the y-intercept, Ysat is the y-value at saturation, R is the first-order rate constant and t is the time elapsed. Data processing and analysis were performed with TopSpin and Prism 5 softwares.
| Equation(2) |
Results
Labeled nucleotides are valuable for use in in vitro transcription of labeled RNA for structural and dynamics studies, yet a number of enzymes of the pentose phosphate pathway needed to make these nucleotides cost-effectively are currently not commercially available. We have therefore focused on elaborating optimal conditions of facile protein production to help in making these nucleotides for NMR studies of RNA.
Cloning and subcloning of genes encoding for RK, APRT, UPRT and XGPRT
The RK gene was subcloned from the genomic DNA into pGEMT vectors and transformed into DH5α E. coli cell line for selection of positive colonies, and subcloned into the pET15b vector, encoding an N-terminal His6 tag. The new construct was transformed into E. coli BL21(AI) cell line for protein overexpression. Sequencing, preparative scale protein expression and diagnostic SDS-PAGE confirmed the successful cloning and expression of the RK construct.
Similarly, the genes for APRT, UPRT and XGPRT, all initially in a pKK223-3 vector, were subcloned into a pCR4-TOPO plasmid and the subsequent selection, propagation, isolations and subcloning were identical to those performed for the RK gene. The new constructs were again inserted into a pET15b vector and the resulting genes were expressed efficiently in BL21(AI) E. coli. Sequencing, preparative scale protein expression and diagnostic SDS-PAGE confirmed the successful cloning and expression of the gene constructs. Similarly, for the clones of PRPPS and CTPS, the respective vectors were transformed into E. coli strain BL21(AI) for protein overexpression.
Expression and purification of proteins
Subcloning and transforming all the genes into identical expression hosts facilitated the expression and purification of these six enzymes, using a simplified and generalized protocol by a one-step NiNTA affinity chromatography. With this protocol, the overexpression and purification of all six enzymes was performed in two days.
The overexpression of the proteins produced reproducible milligram amounts of protein. Test expressions were performed with a combination of different concentrations of L-(+)-arabinose, lactose and IPTG to optimize conditions for overexpression. Induction of one-liter cultures for 2 h under 0.05% L-(+)-arabinose and immediately after for 3 h under 1mM IPTG produced the greatest amount of protein per liter culture, with an average of 8 g of wet cell pellet for all constructs. Nevertheless, the amount of final purified protein ranged from 1 to 24 mg per liter culture and each protein construct gave reproducible quantities.
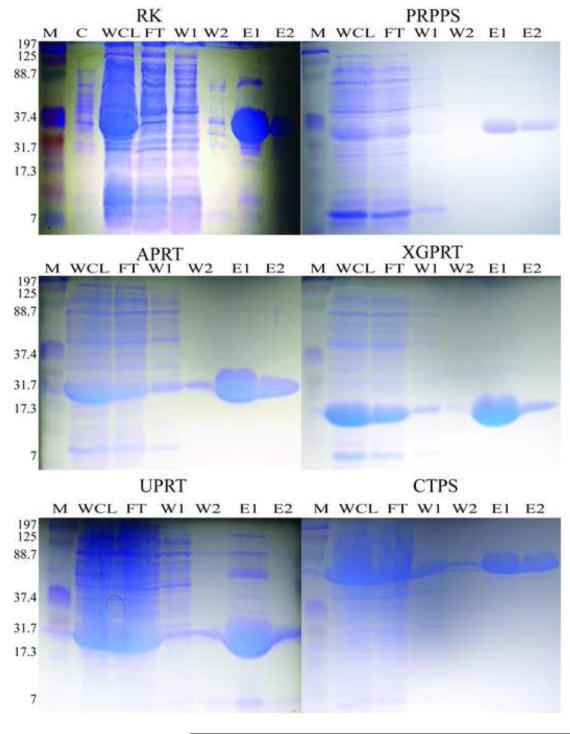
For the His-tag purification, it was found that purification was most efficient for all six proteins using 50 and 400 mM imidazole in the wash and elution buffers, respectively, as judged by SDS-PAGE (Fig. 2). The percent recovery of protein from the crude supernatant of the whole cell lysate ranged from 0.5 to 5% considering that total protein in the crude lysate ranged from 200 to 580 mg. The strain overexpressing PRPPS yielded the lowest amount of protein, whereas the strain overexpressing APRT gave the highest (Table 1). This low yield of PRPPS could be due to downregulation of protein expression, since PRPP is known to be a potent signaling molecule and intracellular concentrations above normal would be detrimental. The SDS-PAGE analysis indicated that all six proteins have a purity of >90% (Fig. 2). This was also observed in the activity assays, as activities remained unaffected or were even higher than previously reported.
Figure 2.
Representative SDS-PAGEs from each of the six enzyme purifications. (M) Marker (kDa) (C) Uninduced control (WCL) Supernatant of the whole cell lysate (FT) Nickel column flow-through (W1, W2) Column wash 1 and 2 (E1, E2) Fractions of imidazole eluted protein 1 and 2. The apparent molecular weights are 37, 34, 19, 15, 19 and 67 KDa for RK, PRPPS, APRT, XGPRT, UPRT and CTPS, respectively.
Table 1.
| Purification Steps |
Enzyme | Total Protein /mg |
Yield /% |
Total Activity /U |
Specific Activity /Umg−1 |
Purification /fold |
|---|---|---|---|---|---|---|
| Crude Supernatant |
RK | 427 ± 20c | 100 | 15 400 ± 520 | 36.1 ± 1.4 | 1 |
| PRPPS | 197 ± 13 | 100 | 2250 ± 154 | 11.4 ± 0.8 | 1 | |
| APRT | 454 ± 32 | 100 | 9620 ± 415 | 21.2 ± 1.2 | 1 | |
| XGPRT | 568 ± 53 | 100 | 3740 ± 176 | 6.6 ± 0.5 | 1 | |
| UPRT | 581 ± 60 | 100 | 15 600 ± 652 | 26.9 ± 2.1 | 1 | |
| CTPS | 427 ± 43 | 100 | 31 400 ± 1281 | 73.5 ± 5.7 | 1 | |
| Ni-NTA Column |
RK | 21.8 ± 2.2 | 5.1 | 1530 ± 62 | 70.2 ± 5.3 | 1.9 |
| PRPPS | 1.0 ± 0.1 | 0.5 | 21.7 ± 0.9 | 21.7 ± 1.7 | 1.9 | |
| APRT | 22.8 ± 2.1 | 5.0 | 483 ± 32 | 21.2 ± 1.7 | 1.0 | |
| XGPRT | 24.4 ± 2.5 | 4.3 | 3120 ± 128 | 128 ± 10 | 20 | |
| UPRT | 14.8 ± 2.0 | 2.5 | 2120 ± 70 | 143 ± 14 | 5.3 | |
| CTPS | 20.2 ± 2.5 | 4.7 | 2280 ± 131 | 113 ± 11 | 1.5 |
The starting material was 40ml of crude E. coli supernatant.
One unit is defined as the conversion of 1 μmol of substrate/min at 25°C or 37C for UPRT and CTPS.
All results except enzyme activity are averages from 3 separate isolation procedures. The enzyme activity data are averages from 3 analyses for one of the isolations.
Uncertainties are expressed as standard deviation.
Enzymatic activity of purified His-tagged proteins
Direct and indirect continuous spectrophotometric assays were performed to check the activity of all six enzymes at the two stages of purification. XGPRT, UPRT and CTPS were assayed directly by monitoring the appearance of the nucleotide product at their respective wavelength. RK, PRPPS and APRT were assayed indirectly by coupling their activities with NADH oxidation (Figures 3 and 4). Initial rates and maximal activity were obtained from every reaction, and the purified RK, UPRT, XGPRT and CTPS yielded robust total activities of 1500 to 2300 U per liter of bacterial culture, whereas PRPPS yielded 22 U per liter and APRT 484 U per liter (Table 1). In addition, the following specific activities for each enzyme were obtained: 70 U/mg for RK (75 U/mg in [45]), 22 U/mg for PRPPS (25 U/mg in [46]), and 21 U/mg for APRT (14 U/mg in [47]). Moreover, the specific activities for XGPRT, UPRT and CTPS were higher than previously reported, 128, 144 and 113 U/mg, respectively (95 U/mg in [48]; 7 U/mg in [34]; and 8 U/mg in [49]). Purification folds calculated for these enzymes ranged from 1- to ~ 20-fold: APRT had a similar specific activity and negligible purification fold and XGPRT attained almost a 20-fold purification (Table 1). Given that the specific activity of CTPS was unusually high with respect to recent work [49], a 13C NMR assay was conducted as an independent validation method. The NMR method afforded a direct means of simultaneously monitoring the depletion of UTP and the accumulation of CTP using the distinct chemical shifts of uracil’s C5 atom (at 103 ppm) and cytosine’s C5 atom (at 97 ppm). The resulting total activity of CTPS was 2640 U per liter, which yielded a specific activity of 131U/mg, an activity comparable to that found using the spectrophotometric assay at 37°C. This experiment conducted in 90% H2O gave a similar specific activity of 169 U/mg. The values obtained in this work are consistently higher than previously reported.
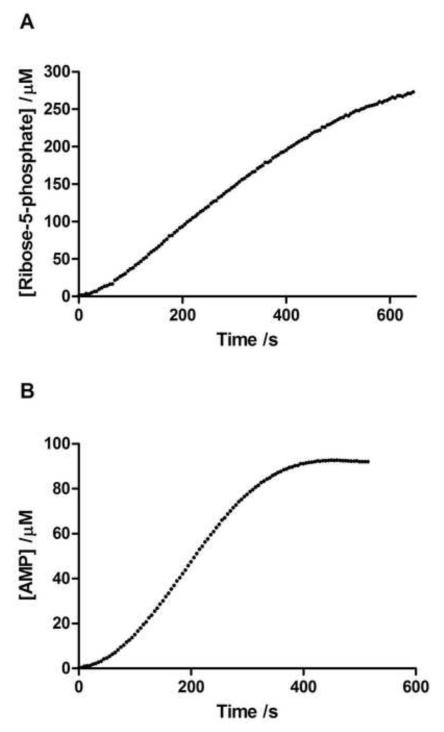
Figure 4.
Representative progress curves obtained from the activity assays. (a) RK progress curve assayed with [RK] =0.67nM, [Ribose]=5mM, [ATP]=3mM. (b) APRT progress curve assayed with [APRT]=2.30nM, [PRPP]=1.5mM, [adenine-HCl]=1.5mM.
Discussion
The synthesis of nucleotides using the pentose phosphate pathway requires 17-18 enzymes (17 for systems using creatine phosphokinase for ATP regeneration) when starting from glucose or 12-13 when starting from ribose. When starting with glucose as the carbon source, 6 out of the 18 enzymes required for nucleotide synthesis are not commercially available and must be produced from overexpressing E. coli strains. Of these 6, only APRT was reported to have a significant activity of 350 U. The other five are only moderately overexpressed with activities of 28-40 U, making the enzyme preparation labor intensive [17,24,26]. Similarly, when starting with ribose as the carbon source, half of the enzymes required for nucleotide synthesis are not commercially available and other than ribokinase, most of the enzymes are only moderately active. Thus, our goal was to find cheaper avenues to make labeled nucleotides using constructs that highly overexpress several of the commercially unavailable enzymes. In this study, six His6-tagged recombinant proteins from the pentose phosphate pathway were, therefore, successfully expressed in E. coli and purified to homogeneity, yielding a single protein band of the expected molecular weight on SDS-PAGE for each protein (Fig. 2). In the current study, the yields of pure recombinant protein, up to 25 mg per liter of crude E. coli supernatant (Table 1), are higher than that obtained previously for purification of recombinant UPRT, APRT, XGPRT [20]. A number of steps were taken to improve the yield. Previous studies used vectors which contain additional features not present in the pET15b vector used in the current study. The pET15b vector contains only a His6-tag and a thrombin cleavage site which facilitates straightforward purification and optional cleavage of the His6-tag, should this be deemed to interfere with function; we did not find it necessary for any of the constructs as activities were similar or higher than previously reported. Finally, while the current study used affinity chromatography for the end stage, previous studies used streptomycin sulfate to precipitate the nucleic acids, ammonium sulfate to precipitate the proteins followed by a DEAE chromatographic step and a final ammonium sulfate precipitation steps [24,26]. Affinity chromatography is known to typically produce a higher protein yield and higher protein purity, compared to ammonium sulfate precipitations and gel filtration [29]. Use of uniform expression hosts and single step affinity purification means that all six enzymes can be overexpressed and purified in two days, saving time and making the process less laborious.
In summary, His6-tagged recombinant proteins have been purified to homogeneity, and found to possess comparable or superior activity to previously reported preparations. The method permits the production of substantial amounts of recombinant enzymes required for conducting enzymatic synthesis of nucleotides for biophysical studies such as NMR spectroscopy, Raman spectroscopy and Mass spectrometry.
Figure 5.
Activity assay for cytidine triphosphate synthetase monitored by 1H-13C HSQC NMR. The assay mixture (250μl) contained 0.64nM CTPS, 50mM Tris-HCl (pH 8.0), 10mM MgCl2, 10mM glutamine, 1mM 15N2-13C9-UTP, 1mM ATP, 0.25mM GTP and 90% D2O. The reaction was initiated by addition of glutamine and monitored over the course of 30 min. The arrow depicts the progression of the reaction.
Acknowledgement
This work was supported in part by NIH grant R01 GM77326 (TKD), Henry C. Welcome Fellowship (TKD), Nano-Biotechnology Award (TKD), and UMD Travel Award (PKA). We thank Dr. Sheng Li, Graduate school of the Chinese Academy of Sciences, Shanghai for the plasmid used for expression of full length human PRPP synthetase, Dr. Stephen L. Bearne, Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, Nova Scotia for the expression vector for CTP synthetase, and Dr. James Williamson, The Scripps Research Institute for the UPRT, XGPRT, and APRT clones.
Abbreviations used
- NMR
nuclear magnetic resonance
- LB
Luria-Bertani
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- Xgal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
- PEG
polyethylene glycol
- BSA
Bovine Serum Albumin
- PEP
phosphoenol pyruvate
- NAD+
nicotinamide adenine dinucleotide
- MK
myokinase
- PK
pyruvate kinase
- LDH
lactate dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: LJ Alvarado, none; PK Arthur, none; TK Dayie, none
References
- [1].Toor N, Keating KS, Pyle AM. Structural insights into RNA splicing. Curr. Opin. Struct. Biol. 2009;19:260–266. doi: 10.1016/j.sbi.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- [3].Dayie KT, Padgett RA. A glimpse into the active site of a group II intron and maybe the spliceosome, too. RNA. 2008;14:1697–1703. doi: 10.1261/rna.1154408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr. Opin. Chem. Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell. Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- [6].Wahl MC, Will CL, Luhrman R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- [7].Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- [8].Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Serganov A. Determination of riboswitch structures: Light at the end of the tunnel? RNA Biol. 2010;7:98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- [10].Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- [11].Weigand JE, Suess B. Aptamers and riboswitches: perspectives in biotechnology. Appl. Microbiol. Biotechnol. 2009;85:229–236. doi: 10.1007/s00253-009-2194-2. [DOI] [PubMed] [Google Scholar]
- [12].Mattick JS. A new paradigm for developmental biology. J. Exp. Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- [13].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- [14].Rodor J, Letelier I, Holuigue L, Echeverria M. Nucleolar RNPs: from genes to functional snoRNAs in plants. Biochem. Soc. Trans. 2010;38:672–676. doi: 10.1042/BST0380672. [DOI] [PubMed] [Google Scholar]
- [15].Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- [16].Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- [17].Tolbert TJ, Williamson JR. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. J. Am. Chem. Soc. 1996;118:7929–7940. [Google Scholar]
- [18].Simon ES, Grabowski S, Whitesides GM. Preparation of phosphoenolpyruvate from D-(−)-3-phosphoglyceric acid for use in regeneration of ATP. J. Am. Chem. Soc. 1989;111:8920–8921. [Google Scholar]
- [19].Rising KA, Schramm VL. Enzymic synthesis of NAD+ with the specific incorporation of atomic labels. J. Am. Chem. Soc. 1994;116:6531–6536. [Google Scholar]
- [20].Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- [21].Hall KB. Uses of 13C- and 15N-labeled RNA in NMR of RNA-protein complexes. Methods Enzymol. 1995;261:542–559. doi: 10.1016/s0076-6879(95)61024-3. [DOI] [PubMed] [Google Scholar]
- [22].Pardi A. Multidimensional heteronuclear NMR experiments for structure determination of isotopically labeled RNA. Methods Enzymol. 1995;261:350–380. doi: 10.1016/s0076-6879(95)61017-0. [DOI] [PubMed] [Google Scholar]
- [23].Puglisi JD, Wyatt JR. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–350. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- [24].Tolbert TJ, Williamson JR. Preparation of specifically deuterated and 13C-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997;119:12100–12108. [Google Scholar]
- [25].Dayie KT, Tolbert TJ, Williamson JR. 3D C(CC)H TOCSY experiment for assigning protons and carbons in uniformly 13C- and selectively 2H-labeled RNA. J. Magn. Reson. 1998;130:97–101. doi: 10.1006/jmre.1997.1286. [DOI] [PubMed] [Google Scholar]
- [26].Scott LG, Tolbert TJ, Williamson JR. Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol. 2000;317:18–38. doi: 10.1016/s0076-6879(00)17004-1. [DOI] [PubMed] [Google Scholar]
- [27].Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]; J. Mol. Biol. 2006;360:742. Erratum in: [Google Scholar]
- [28].Vallurupalli P, Scott L, Hennig M, Williamson JR, Kay LE. New RNA labeling methods offer dramatic sensitivity enhancements in 2H NMR relaxation spectra. J. Am. Chem. Soc. 2006;128:9346–9347. doi: 10.1021/ja0632512. [DOI] [PubMed] [Google Scholar]
- [29].Dayie KT. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008;9:1214–1240. doi: 10.3390/ijms9071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Park J, van Koeverden P, Singh B, Gupta RS. Identification and characterization of human ribokinase and comparison of its properties with E. coli ribokinase and human adenosine kinase. FEBS Letters. 2007;581:3211–3216. doi: 10.1016/j.febslet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- [31].Li S, Lu Y, Peng B, Ding J. Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J. 2007;401:39–47. doi: 10.1042/BJ20061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sinha SC, Smith JL. The PRT protein family. Curr. Opinion Struct. Biol. 2001;11:733–739. doi: 10.1016/s0959-440x(01)00274-3. [DOI] [PubMed] [Google Scholar]
- [33].Sin IL, Finch LR. Adenine phosphoribosyltransferase in Mycoplasma mycoides and Escherichia coli. J. Bacteriol. 1972;112:439–444. doi: 10.1128/jb.112.1.439-444.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lundegaard C, Jensen KF. Kinetic mechanism of uracil phosphoribosyltransferase from Escherichia coli and catalytic importance of the conserved proline in the PRPP binding site. Biochemistry. 1999;38:3327–3334. doi: 10.1021/bi982279q. [DOI] [PubMed] [Google Scholar]
- [35].Vos S, de Jersey J, Martin JL. Crystal structure of Escherichia coli xanthine phosphoribosyltransferase. Biochemistry. 1997;36:4125–4134. doi: 10.1021/bi962640d. [DOI] [PubMed] [Google Scholar]
- [36].Lunn FA, MacDonnell JE, Bearne SL. Structural requirements for the activation of Escherichia coli CTP synthase by the allosteric effector GTP are stringent, but requirements for inhibition are lax. J. Biol. Chem. 2008;283:2010–2020. doi: 10.1074/jbc.M707803200. [DOI] [PubMed] [Google Scholar]
- [37].Andersson CE, Mowbray SL. Activation of ribokinase by monovalent cations. J. Mol. Biol. 2002;315:409–419. doi: 10.1006/jmbi.2001.5248. [DOI] [PubMed] [Google Scholar]
- [38].Wood T. Spectrophotometric assay for D-ribose-5-phosphate ketol-isomerase and for D-ribulose-5-phosphate 3-epimerase. Anal. Biochem. 1970;33:297–306. doi: 10.1016/0003-2697(70)90300-3. [DOI] [PubMed] [Google Scholar]
- [39].Gross A, Abril O, Lewis JM, Geresh S, Whitesides GM. Practical synthesis of 5-phospho-D-ribosyl-alpha-1-pyrophosphate (PRPP): enzymatic routes from ribose 5-phosphate or ribose. J. Am. Chem. Soc. 1983;105:7428–7435. [Google Scholar]
- [40].Parkin DW, Leung HB, Schramm VL. Synthesis of nucleotides with specific radiolabels in ribose. Primary 14C and secondary 3H kinetic isotope effects on acid-catalyzed glycosidic bond hydrolysis of AMP, dAMP, and inosine. J. Biol. Chem. 1984;259:9411–9417. [PubMed] [Google Scholar]
- [41].Hove-Jensen B, Maigaard M. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J. Bacteriol. 1993;175:5628–5635. doi: 10.1128/jb.175.17.5628-5635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Keough DT, McConachie LA, Gordon RB, de Jersey J, Emmerson BT. Human hypoxanthine-guanine phosphoribosyltransferase. Development of a spectrophotometric assay and its use in detection and characterization of mutant forms. Clin. Chim. Acta. 1987;163:301–308. doi: 10.1016/0009-8981(87)90248-8. [DOI] [PubMed] [Google Scholar]
- [43].Guddat LW, Vos S, Martin JL, Keough DT, de Jersey J. Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase. J. Protein Sci. 2002;11:1626–1638. doi: 10.1110/ps.0201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Anderson PM. CTP synthetase from Escherichia coli: an improved purification procedure and characterization of hysteretic and enzyme concentration effects on kinetic properties. Biochemistry. 1983;22:3285–3292. doi: 10.1021/bi00282a038. [DOI] [PubMed] [Google Scholar]
- [45].Chuvikovsky DV, Esipov RS, Skoblov YS, Chupova LA, Muravyova TI, Miroshnikov AI, Lapinjoki S, Mikhailopulo IA. Ribokinase from E. coli: Expression, purification, and substrate specificity. Bioorg. Med. Chem. 2006;14:6327–6332. doi: 10.1016/j.bmc.2006.05.057. [DOI] [PubMed] [Google Scholar]
- [46].Nosal JM, Switzer RL, Becker MA. Overexpression, purification, and characterization of recombinant human 5-phoshoribosyl-1-pyrophosphate synthetase isozymes I and II. J. Biol. Chem. 1993;268:10168–10175. [PubMed] [Google Scholar]
- [47].Hochstadt J. Adenine phosphoribosyltransferase from Escherichia coli. Methods Enzymol. 1978;51:558–567. doi: 10.1016/s0076-6879(78)51078-1. [DOI] [PubMed] [Google Scholar]
- [48].Krenitsky TA, Neil SM, Miller RL. Guanine and xanthine phosphoribosyltransferase activities of Lactobacillus casei and Escherichia coli. J. Biol. Chem. 1970;245:2605–2611. [PubMed] [Google Scholar]
- [49].Bearne SL, Hekmat O, MacDonnell JE. Inhibition of Escherichia coli CTP synthase by glutamate γ-semialdehyde and the role of the allosteric effector GTP in glutamine hydrolysis. Biochem. J. 2001;356:223–232. doi: 10.1042/0264-6021:3560223. [DOI] [PMC free article] [PubMed] [Google Scholar]