Abstract
Macroautophagy (autophagy hereafter) recycles intracellular components to sustain mitochondrial metabolism that promotes the growth, stress tolerance and malignancy of lung cancers, suggesting that autophagy inhibition may have antitumor activity. To assess the functional significance of autophagy in both normal and tumor tissue, we conditionally deleted the essential autophagy gene, autophagy-related-7, Atg7, throughout adult mice. Here we report that systemic ATG7 ablation caused susceptibility to infection and neurodegeneration that limited survival to 2–3 months. Moreover, upon fasting, autophagy-deficient mice suffered fatal hypoglycemia. Prior autophagy ablation did not alter the efficiency of non-small-cell lung cancer (NSCLC) initiation by activation of oncogenic KrasG12D and deletion of the Trp53 tumor suppressor. Acute autophagy ablation in mice with pre-existing NSCLC, however, blocked tumor growth, promoted tumor cell death, and generated more benign disease (oncocytomas). This anti-tumor activity occurred prior to destruction of normal tissues, suggesting that, acute autophagy inhibition may be therapeutically beneficial in cancer.
Keywords: autophagy, fasting, hypoglycemia, cancer, oncocytoma
Introduction
Autophagy is a process that captures cytoplasmic proteins and organelles in vesicles called autophagosomes, which then fuse with lysosomes and are degraded. Autophagy functions at a low basal level to remove damaged cellular components to maintain protein and organelle quality, thereby preventing the gradual toxic accumulation of intracellular waste material (1). Autophagy is dramatically upregulated by starvation where the catabolism and recycling of cellular components sustains energy homeostasis essential for cells and newborn mice to survive without nutrients (2–4). These functions of autophagy are conserved from yeast through mammals and are controlled by the autophagy related genes (Atgs).
Mice constitutively deficient in Atg5 or Atg7 are born developmentally normal but fail to survive the neonatal starvation period due to metabolic insufficiency, illustrating the importance of autophagy to supply metabolic substrates to bridge gaps in nutrient availability (2, 3). Neuronal-specific deficiency in Atg5 or Atg7 results in the accumulation of autophagy substrates such as aggregated and ubiquitinated proteins and damaged organelles, motor and behavioral defects, neurodegeneration, and lethality between 1–6 months after birth (5, 6). These findings suggest that autophagy is critical for preventing the toxic accumulation of damaged proteins and organelles in post-mitotic tissues, although there is a potential additional contribution of autophagy to brain energy metabolism. Mosaic or liver-specific deletion of Atg5 or liver-specific deficiency in Atg7 also causes accumulation of autophagy substrates, steatosis and eventual hepatoma development, suggesting that autophagy prevents liver damage and limits liver cancer initiation (2, 7, 8). Additional tissue-specific knockout studies underscore the importance of autophagy in tissue homeostasis, metabolism, and stem cell maintenance (1).
Autophagy has a context-dependent role in cancer (9). It is upregulated and required for the survival of tumor cells in hypoxic tumor regions (9). Oncogenic Ras transformation upregulates basal autophagy required for maintenance of mitochondrial metabolism and progression of tumorigenesis (10–12). Moreover, studies knocking out essential autophagy genes in genetically engineered mouse models (GEMMs) for cancer have demonstrated a pro-tumorigenic role for autophagy (13). Deletion of Atg7 in KrasG12D-and BrafV600E-NSCLC in adult mice causes tumors to accumulate defective mitochondria and become metabolically impaired. Without ATG7, tumor cell proliferation is suppressed and tumors progress to benign oncocytomas rather than adenomas and carcinomas, extending the mice's lifespan (14–16). In some but not all contexts autophagy promotes tumorigenesis by mitigating p53 activity (14–19). Although these studies point to a pro-tumorigenic role for autophagy, they involved Atg gene deletion specifically in tumor cells, and thus do not demonstrate that autophagy deficiency is selectively detrimental to tumor tissue. Furthermore, as Atg gene deletion occurred concurrently with activation of oncogenic mutations that initiate tumorigenesis, these prior studies do not model acute, systemic autophagy ablation as would occur during autophagy inhibition for cancer therapy.
To address the tumor selectivity of autophagy ablation in cancer, we engineered the mice to conditionally (Tamoxifen [TAM])-inducible) and systemically delete Atg7 throughout adult mice. We found that adult mice with Atg7 acute whole-body deletion (Atg7Δ/Δ mice) manifested systemically blocked autophagy without extensive organ damage at 5 weeks post-deletion, although a small fraction of mice succumbed to Streptococcus infection. In contrast, by 6 to 12 weeks post deletion, extensive liver and muscle damage were evident and neurodegeneration limited survival to 2–3 months. Atg7Δ/Δ mice suffered lethality upon fasting, where mice displayed extreme muscle wasting and died of hypoglycemia, indicating that autophagy is required for glucose homeostasis. This reveals a new role for autophagy in the management of systemic energy balance. In the setting of cancer, 5 weeks of acute autophagy ablation converted established lung adenocarcinomas to oncocytomas and blocked mTOR and MAP kinase signaling, and cell proliferation and survival. Compared to tumor-specific autophagy deficiency produced concurrently with tumor initiation, acute systemic autophagy ablation was more destructive to tumors. Thus, established tumors have greater autophagy dependency than both newly developing tumors and most normal tissues. This suggests that, with proper control of the extent and/or timing of autophagy inhibition, there is likely to be a therapeutic window to suppress tumorigenesis while mostly sparing normal tissue.
Results
Acute Atg7 deletion in adult mice produces systemic autophagy defect
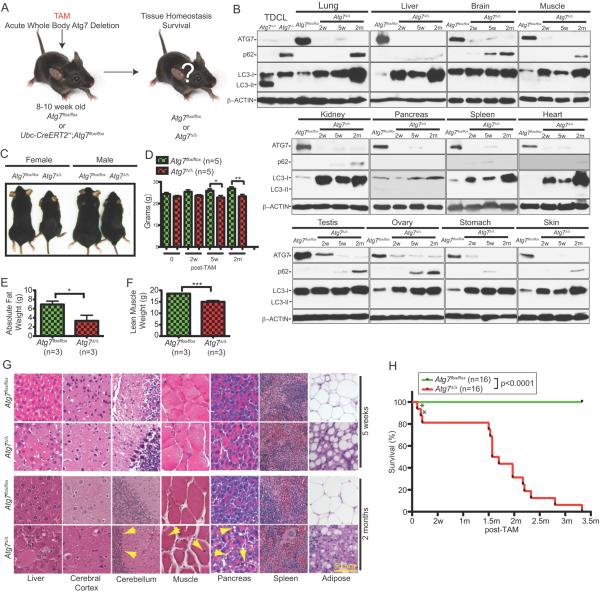
Adult mice were engineered with floxed alleles of Atg7 (2) and a transgene expressing the TAM-regulated Cre-recombinase fusion protein under the control of the ubiquitously expressed ubiquitin C (Ubc) promoter (20). 8–10 week old adult mice retain intact Atg7 (wild type mice or mice with Atg7 floxed alleles with or without the Ubc-CreERT2 allele) and express ATG7 protein (Supplementary Fig. S1A), but when provided TAM, the Cre is activated only in mice with Atg7 floxed and Ubc-CreERT2 alleles, deleting the Lox-P sites and Atg7, producing a near complete and sustained loss of ATG7 protein (Atg7Δ/Δ mice) in all tissues examined by 2 weeks following the last day of 5-day course of TAM administration (Fig 1A and B and Supplementary Fig. S1B). Atg7 deletion throughout mouse tissues was also confirmed by PCR (data not shown).
Figure 1. Conditional whole-body deletion of Atg7 abrogates autophagy and impairs long-term survival.
A. Experimental design for generation of Atg7Δ/Δ mice. Atg7flox/flox or Ubc-CreERT2/+;Atg7flox/flox mice were treated with TAM at 8–10 week of age by intraperitoneal injection and analyzed at times thereafter.
B. Western blotting for ATG7, p62 and LC3 at the indicated times (w: weeks, m: months) of the indicated tissues from TAM-treated Atg7flox/flox or AtgΔ/Δ adult mice. Atg7flox/flox control tissues are from a 5 weeks post-TAM-treated mouse. Tumor derived cell lines (TDCL) from Atg7-intact and -deficient tumors (16) were used as controls for ATG7, LC3 and p62 protein expression. β-ACTIN serves a protein loading control.
C. Representative pictures of 2 months post-TAM-treated Atg7flox/flox or Atg7Δ/Δ female and male mice (n=3 for each genotype).
D. Body weight difference of TAM-treated Atg7flox/flox or Atg7Δ/Δ adult mice at the indicated times (w: weeks, m: months). Error bar represents SEM, *p<0.05, **p<0.01 (two-way ANOVA with Bonferroni post-test).
E. Body fat composition as determined by EchoMRI of 10 days post-TAM-treated Atg7flox/flox or Atg7Δ/Δ adult mice. Error bar represents SEM, *p<0.05 ((two-way ANOVA with Bonferroni post-test).
F. Lean muscle mass as determined by EchoMRI of 10 days post-TAM-treated Atg7flox/flox or Atg7Δ/Δ adult mice. Error bar represents SEM, ***p<0.001 (two-way ANOVA with Bonferroni post-test).
G. Representative liver, cerebral cortex, cerebellum, muscle, pancreas, spleen and adipose histology (Hematoxylin and Eosin stained; H&E) of TAM-treated Atg7flox/flox or Atg7Δ/Δ adult mice at indicated times. Arrowheads in cerebellum point to purkinje cells, arrowheads in muscle point to centrally nucleated myofibers, arrowheads in pancreas point to intra-acinar vacuolization.
H. Kaplan-Meier survival curve of TAM-treated Atg7flox/flox or Atg7Δ/Δ adult mice (w: weeks, m: months). Asterisks denote mice that died of infection. p<0.0001 (log-rank Mantel-Cox test).
The loss of ATG7 correlated with accumulation of the autophagy substrate p62 and the unprocessed form of microtubule-associated protein 1A/1B-light chain 3 (LC3-I) and the absence of the cleaved, lipidated and active (autophagosome associated) form LC3-II (Fig. 1B and Supplementary Fig. S1). These findings are consistent with efficient, conditional ablation of ATG7 expression and loss of autophagy in adult mouse tissues, allowing the assessment of the role of autophagy in adult mice for the first time.
ATG7 deficiency causes depletion of white adipose tissue (WAT) and damage to liver and muscle
In comparison to Atg7 wild type mice, Atg7Δ/Δ mice at 2 months post-TAM were smaller in size and had reduced body weight (Fig. 1C and D), despite similar feeding behavior and energy expenditure. Liver weight was increased in Atg7Δ/Δ mice (Supplementary Fig. S1C), consistent with steatosis that results also from liver-specific Atg7 or Atg5 deletion (2, 7, 8, 21). Echo Magnetic Resonance Imaging (EchoMRI) analysis of body composition showed less absolute fat and lean body mass in Atg7Δ/Δ compared to control mice at 2 months post-TAM (Fig. 1E and F) explaining the decreased body size of Atg7Δ/Δ mice. Histological examination of tissues from the Atg7Δ/Δ mice revealed limited tissue damage at 5 weeks post-TAM that included the beginning of liver abnormalities, splenic enlargement, testicular degeneration and depletion of the lipid content of WAT (Fig. 1G and Supplementary Fig. S1D and S1I). Spleens of Atg7Δ/Δ mice were enlarged with accumulation of megakaryocytes (Fig. 1G) although bone marrow appeared normal (data not shown). We observed testicular degeneration in Atg7Δ/Δ mice, which was partially due to Cre toxicity; testes were smaller in Atg7Δ/Δ mice with elevated p62 aggregates mainly in Leydig cells (Fig. 1B and Supplementary Fig. S1I). Autophagy is essential for adipocyte differentiation and formation of WAT (22, 23). Here, acute autophagy ablation rapidly depleted lipid stores and reduced established WAT by converting WAT to brown adipose tissue (BAT) or by blocking BAT to WAT transdifferentiation. In contrast, marked liver, brain, muscle and pancreas damage and further depletion of WAT were apparent in Atg7Δ/Δ mice at 2 months post-TAM (Fig. 1G). Thus, systemic Atg7 deficiency for 5 weeks produces limited toxicities but this is not the case at 2–3 months.
Tissue damage resulting from acute, systemic Atg7 deficiency in adult mice for 2 months included increased liver enlargement where hepatocytes accumulated p62 and LC3-I aggregates, mitochondria (as indicated by the mitochondrial marker TOM20) and lipid droplets (Oil red O staining) (Fig. 1B and G and Supplementary Fig. S1E–G). These mice also had decreased number of large pyramidal neurons and Purkinje cells (Fig. 1G). At the same time, we observed degenerative changes in muscle with centrally nucleated, small myofibers (Fig. 1G). These findings are consistent with a time-dependent destructive effect to normal tissues upon loss of autophagy as reported with tissue-specific knockout of Atg5 or Atg7 (5, 6, 21–26). We also observed intra-acinar vacuolization in the pancreas (Fig. 1G). Consistent with the role of autophagy in lipid metabolism, kidney and lung tissues accumulated lipids (Supplementary Fig. S1G). Other tissues were not visibly affected by acute Atg7 ablation. Interestingly, constitutive autophagy deficiency impairs lung function in neonates (27), but as this did not occur in Atg7Δ/Δ adult mice, this suggests that developing tissues can more reliant on autophagy than adult ones.
ATG7 extends adult lifespan by suppressing susceptibility to infection and neurodegeneration
A cohort of wild type and Atg7Δ/Δ mice was followed to assess the consequence to overall survival. Lifespan of Atg7Δ/Δ mice was shortened to 2–3 months, demonstrating that retention of Atg7 for functional autophagy is required for the viability of adult mice (Fig. 1H). Death of Atg7Δ/Δ mice resulted from two distinct causes. Several mice (12%) succumbed to Streptococcus infection shortly after deletion (Fig. 1H, asterisks, and Supplementary Fig. S1G), consistent with the finding that autophagy is required for eliminating intracellular Streptococcus in cells in vitro (28). The vast majority of the Atg7Δ/Δ mice, however, died between 2–3 months due to neurodegeneration (Fig. 1H), similar to findings with neuronal-specific deficiency in Atg5 or Atg7 (5, 6). With conditional whole-body deletion of Atg7 we observed progressive motor and behavioral deficits such as tremors and ataxic walking and limb-clasping reflexes when held by their tails whereas control animals tend to extend their limbs (Supplementary Movie S1). Atg7Δ/Δ mice also displayed very severely impaired balance and bradykinesia (abnormally slow movements) shortly before death, collectively indicating that death results from neurological defects. Thus the predominant toxicity of autophagy deficiency is neurodegeneration.
Autophagy is required for adult mice to survive fasting
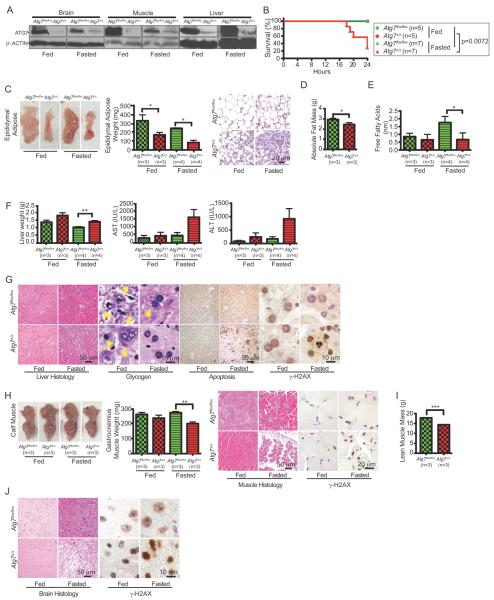
The above analysis focused on unstressed mice. A major function of autophagy conserved from yeast to mammals is stress survival. This is evident in the requirement of autophagy to survive the stress of the neonatal starvation period (2, 3). Similarly, autophagy is robustly induced in adult mouse tissues in response to starvation (4). To test if autophagy is functionally important for adult mice to survive fasting, Atg7 was deleted in 8–10 week old mice by TAM administration (mice with Atg7 floxed alleles plus the Ubc-CreERT2 allele; Atg7Δ/Δ) or left intact (mice with Atg7 floxed alleles without the Ubc-CreERT2 allele; Atg7flox/flox). 10 days following the last day of a 5-day schedule of TAM administration when ATG7 expression was extinguished (Fig. 2A), mice were subjected to fasting (free access to water without food) for 24 hours. We chose this short-term autophagy inactivation to establish the acute and direct requirement for autophagy. All fed mice (Atg7 intact or Atg7Δ/Δ) and all fasted mice with Atg7 intact survived for 24 hours as expected (Fig. 2B). In contrast, the majority of Atg7Δ/Δ mice failed to survive fasting for 24 hours (Fig. 2B). Thus, similar to neonates, adult mice also require autophagy to tolerate starvation. Serum insulin and leptin levels were downregulated by fasting independent of autophagy status (Supplementary Fig. S2), suggesting that the hormonal response to starvation was normal and thus not the cause of the fasting lethality of ATG7-deficient mice.
Figure 2. Autophagy is required for adult mice to survive fasting.
A. ATG7 Western blot of tissue lysates from Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated, fed and fasted. β-ACTIN serves a protein loading control.
B. Kaplan-Meier survival curve of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted; p=0.0072 (log-rank Mantel-Cox test).
C. Representative male pictures (left), tissue weight (middle) and histology (H&E) (right) of epididymal adipose tissues of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted (n=4, for each). Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
D. Body fat composition as determined by EchoMRI of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated. Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
E. Quantification of serum FFA levels of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
F. Liver weight (left) and serum level of liver enzymes AST (middle) and ALT (right) from Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. Error bar represents SEM, **p<0.01 (two-way ANOVA with Bonferroni post-test).
G. Representative liver histology (H&E) and liver glycogen levels (PAS staining) of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. Arrowheads point to glycogen. IHC staining for Active Caspase-3 shows apoptosis induction and IHC staining for γ-H2AX shows DNA damage response activation in ATG7-deficient liver tissue when fasted.
H. Representative gross image of calf muscle (left) and weight of gastrocnemius muscle and representative muscle histology (H&E) from Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. IHC staining for γ-H2AX shows DNA damage response activation in ATG7-deficient muscle tissue when fasted. Error bar represents SEM, **p<0.01 (two-way ANOVA with Bonferroni post-test).
I. Lean muscle mass as determined by EchoMRI of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated. Error bar represents SEM, ***p<0.001 (two-way ANOVA with Bonferroni post-test).
J. Representative brain histology (H&E) of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. IHC staining for γ-H2AX shows DNA damage response activation in ATG7-deficient brain tissue when fasted.
Autophagy maintains fat stores and the mobilization of lipids during fasting
The physiology and metabolic requirements of neonates are distinct from that of adult mice, raising the question as to whether the role of autophagy in supporting survival during fasting also differed. Neonates require autophagy to maintain serum amino acid but not glucose levels (3). To address the mechanism by which autophagy contributes to the survival of adult mice during fasting, tissues and serum were examined during fed and fasted conditions in the presence and absence of ATG7.
There was a striking depletion of WAT, apparent with short-term Atg7 deletion under fed conditions (Fig. 2C). Histological examination revealed marked reduction of epididymal adipose mass due to a reduction in the size of fat droplets and accumulation of mitochondria within adipocytes in Atg7Δ/Δ mice in the short-term (Fig. 2C and Supplementary Fig. S1F). Thus, the presence of ATG7 favors WAT whereas ATG7 deficiency favors BAT. To assess absolute total body fat mass, live Atg7 intact or Atg7Δ/Δ mice were subjected to EchoMRI. Fed Atg7Δ/Δ mice displayed reduced absolute fat mass compared to mice with Atg7 intact, indicating that acute autophagy deficiency causes depletion of fat stores (Fig. 2D). ATG7 deficiency promotes adipose differentiation favoring BAT formation, possibly due to a decreased capacity for lipid storage and increased lipid metabolism through β-oxidation. This suggests that the reduction in dedicated lipid stores may compromise survival during fasting. Indeed, fat mass was more severely depleted in fasted Atg7Δ/Δ mice than in those with Atg7 intact (Fig. 2C). Mobilization of free fatty acids (FFAs) in serum during fasting was also defective without ATG7, revealing a metabolic deficiency that can contribute to lethality (Fig. 2E).
Glycogen stores are depleted in fasted ATG7-deficent mice
Glycogen store mobilization, particularly in liver, provides an important source of glucose in fasted conditions that contributes to serum glucose homeostasis. Fasting reduced liver size to a greater extent in Atg7Δ/Δ mice (17% with Atg7 intact, compared to a 29% in Atg7Δ/Δ mice), and these mice had higher serum levels of the liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT) when fasted, indicative of some liver damage. Indeed, these mice had increased apoptosis coincident with DNA damage response activation (γ-H2AX positive hepatocytes) although healthy liver tissue was still present (Fig. 2F and G). Importantly, in contrast to fasted Atg7-intact mice, fasted Atg7Δ/Δ mice had complete depletion of liver glycogen stores (Fig. 2G). Thus, autophagy is critical to prevent depletion of glycogen stores during fasting. Since acute deficiency in Atg7 produced accelerated depletion of lipid and glycogen stores, this indicates a systemic requirement for autophagy that may contribute to their failure to tolerate fasting.
ATG7 prevents muscle wasting during fasting
Degradation of muscle protein sustains homeostasis during fasting (29). Gastrocnemius muscle mass was slightly reduced in Atg7Δ/Δ mice with short-term deletion under fed condition in comparison to that from Atg7 intact mice (Fig. 2H), as was a reduction in total muscle mass revealed by EchoMRI (Fig. 2I). Fasting, however, induced severe muscle wasting with DNA damage response activation (γ-H2AX positive myofibers) in Atg7Δ/Δ compared to Atg7-intact mice (Fig. 2H). Thus, without autophagy, fasting induces severe muscle wasting. Ultimately, metabolic failure can lead to brain damage and death. In contrast to fed and fasted mice with Atg7 intact, fasted Atg7Δ/Δ mice showed loss of motor control indicative of neurologic impairment, which was consistent with the occurrence of extensive brain damage (Fig. 2J).
Fasted Atg7-deficient adult mice die from hypoglycemia
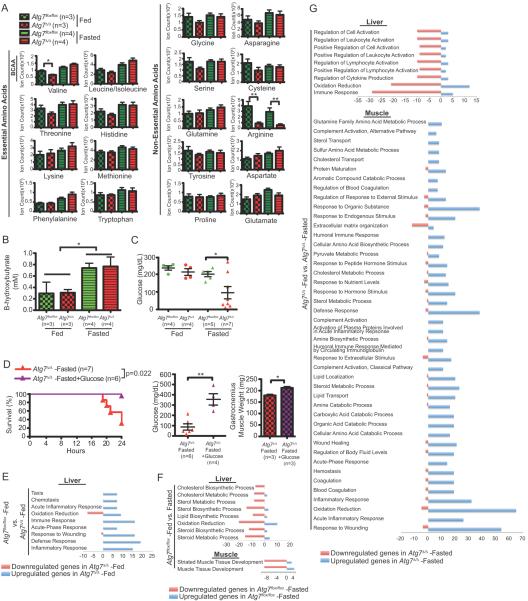
To address the mechanism by which autophagy sustains mouse survival during fasting, we first examined levels of serum amino acids by Liquid Chromatography-Mass Spectrometry (LC-MS) in fed and fasting conditions, in Atg7 intact and Atg7Δ/Δ mice. In contrast to Atg7-deficient neonates undergoing neonatal starvation, adult Atg7Δ/Δ mice with short-term Atg7 deletion sustained serum amino acid levels with the exception of arginine (Fig. 3A). Liver amino acids were also similar in fed and fasted mice (data not shown) suggesting that this was not responsible for the survival defect.. Serum β-hydroxybutyrate levels were elevated in fasting in both Atg7 intact and Atg7Δ/Δ mice, suggesting that serum amino acids were sufficient to induce ketogenesis (Fig. 3B). Extensive muscle wasting in fasted Atg7Δ/Δ mice may sustain serum amino acids and ketogenesis, partly compensating for depleted fat and glycogen stores.
Figure 3. Autophagy sustains glucose homeostasis required to survive fasting.
A. Quantification of serum amino acid levels in Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. BCAA: branched chain amino acid. Error bar represents SEM, *p<0.05, **p<0.01 (two-way ANOVA with Bonferroni post-test).
B. Quantification of serum β-hydroxybutyrate levels in Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
C. Quantification of blood glucose levels of Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated fed and fasted. Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
D. Kaplan-Meier survival curve (left), serum glucose levels (middle) and weight of gastrocnemius muscle (right) from Atg7Δ/Δ adult mice 10 days post-TAM-treated, fasted, without or with glucose supplemented p=0.0022 (log-rank Mantel-Cox test). Error bar represents SEM, *p<0.05, **p<0.01 (two-way ANOVA with Bonferroni post-test).
E–G. Analysis of differentially expressed genes from liver and muscle tissue in Atg7flox/flox or Atg7Δ/Δ adult mice 10 days post-TAM-treated, fed and fasted showing upregulated (blue) or downregulated (pink) genes for significant biological processes as determined by GO term DAVID Analysis. Number scale represents the number of up- (positive number) and down- (negative number) regulated genes in the indicated biological processes.
We next examined serum glucose levels and found that the majority of fasted Atg7Δ/Δ mice were severely hypoglycemic consistent with loss of viability (Fig. 3C). Indeed, glucose supplementation was sufficient to rescue serum glucose levels, muscle wasting and survival of fasted Atg7Δ/Δ mice (Fig. 3D). Thus, autophagy in adult mice is required to maintain fasting serum glucose levels, preventing hypoglycemia and death, revealing different metabolic roles for autophagy in the systemic metabolism of neonatal and adult mice.
Atg7 deletion alters gene expression responses to fasting
Functional analysis of differentially expressed genes revealed that molecular functions related to inflammatory response were highly enriched in livers from Atg7Δ/Δ mice (Fig. 3E and Supplementary Fig. S3A and Supplementary Tables S1 and S3). This suggests that liver function is altered by Atg7 deficiency under fed conditions, although this was not the case for muscle, consistent with normal histology (Fig. 1G). Fasting Atg7-intact mice repressed gene expression for lipid metabolic processes and insulin signaling and elevated those for fatty acid oxidation in liver (Fig. 3F and Supplementary Tables S1 and S3) but induced few changes in muscle gene expression (Fig. 3F, and Supplementary Fig. S3B and Supplementary Tables S2 and S4). Fasting Atg7Δ/Δ mice reduced signatures for liver immune response-related genes; however, there was not a significant change in lipid metabolism-related gene expression as opposed to Atg7-intact mice (Fig. 3G and Supplementary Fig. S3A and Supplementary Tables S1 and S3). Gluconeogenesis predominantly occurs in the liver, producing glucose that supports metabolism in the brain and other tissues. Genes associated with gluconeogenesis were induced by fasting in liver in Atg7Δ/Δ mice (e.g. Ppargc1a, Atf4, Arntl, Pdk4, Got1, Lepr) (Supplementary Fig. S3A and C and Supplementary Table S3) suggesting that it may be defective because of insufficient substrates. For example, depletion of FFAs may cause more use of amino acids for ketogenesis instead of gluconeogenesis. Depletion of WAT would limit the availability of glycerol for use as a substrate for gluconeogenesis. Fasting caused activation of catabolic processes for autophagy and atrophy (e.g. Ulk1, Bnip3, Foxo1, Fbxo32, Mt1 and Mt2) (Fig. 3G, Supplementary Fig. S3B, D–E, Supplementary Tables S2 and S4). The muscle atrophy signature in the fasted Atg7Δ/Δ mice is consistent with extensive muscle wasting.
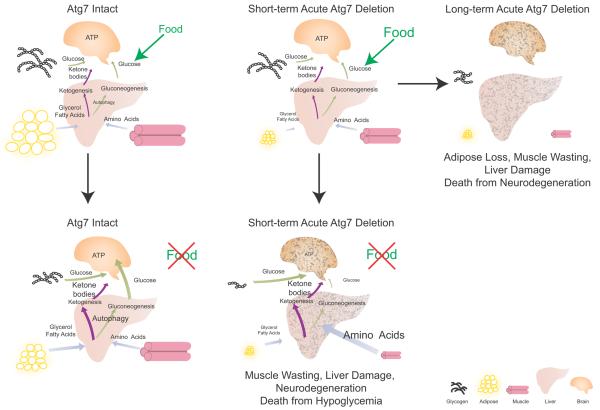
In summary, healthy adult mice have lipid and glycogen stores that are mobilized during fasting, reducing the demand for muscle catabolism to supply amino acids for gluconeogenesis. Liver and muscle autophagy may also supply substrates for gluconeogenesis. Lipid and glycogen stores in Atg7Δ/Δ mice were depleted even under fed conditions, and fasting accelerates glycogen elimination and caused excessive muscle wasting (Fig. 4 and Supplementary Fig. S4). While serum amino acid levels and ketogenesis were sustained in Atg7Δ/Δ mice, they died nonetheless from hypoglycemia. Therefore, this suggests that adult mice require autophagy to provide substrates for glucose homeostasis to survive fasting.
Figure 4. Mechanism by which autophagy supports survival of adult mice during fasting.
See text for explanation.
ATG7 deficiency does not alter lung tumor initiation
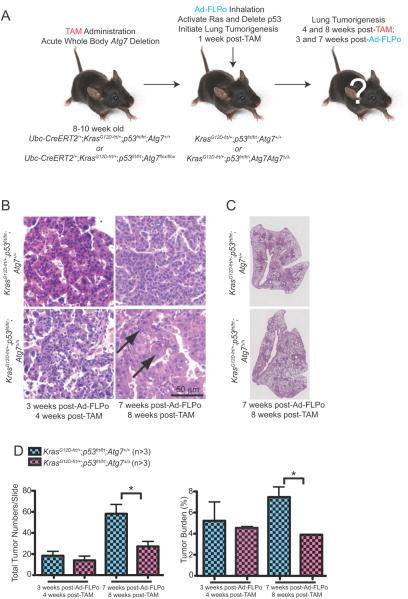
To test if autophagy influences tumor initiation, mice were engineered with Frt alleles of both KrasG12D and Trp53 (p53) without and with the conditional Floxed system for generating Atg7Δ/Δ mice. Mice can then have Atg7 ablated by TAM administration and deletion of the Lox-P sites in the Atg7 alleles by Cre recombinase followed later by initiation of lung tumorigenesis by intranasal administration of adenovirus expressing Flp recombinase (Ad-FLPo) and simultaneous activation of KRAS and deletion of p53 by excision of Frt sites (Fig. 5A).
Figure 5. Acute, systemic autophagy ablation does not alter the efficiency of tumor initiation.
A. Experimental design to induce conditional whole-body Atg7 deletion prior to tumor induction. Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7+/+ and Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7flox/flox mice were treated with TAM at 8–10 weeks of age by intraperitoneal injection to delete Atg7 throughout the mouse, then these mice were infected with Ad-FLPo at 1 week post-TAM to activate RAS and delete p53 and were analyzed at times thereafter.
B. Representative lung tumor histology (H&E) at 3 weeks post-Ad-FLPo, 4 weeks post-TAM (left) and 7 weeks post-Ad-FLPo, 8 weeks post-TAM (right). Arrows point to oncocytes.
C. Representative lung lobes at 7 weeks post-Ad-FLPo and 8 weeks post-TAM in the indicated genotypes.
D. Quantification of tumor numbers (left) and tumor burden (right) at the indicated times. Error bar represents SEM, *p<0.05 (two-way ANOVA with Bonferroni post-test).
When Atg7 was deleted and lung tumorigenesis was initiated by RAS activation and p53 deletion, the number and size of tumors was monitored 4 and 8 weeks post-TAM (3 and 7 weeks post Ad-FLPo). There was no difference in tumor frequency or burden 3 weeks post Ad-FLPo regardless of Atg7 status, indicating that the functional status of autophagy does not alter the ability of KrasG12D activation and p53 deficiency to induce formation of lung tumors (Fig. 5B–D). At 7 weeks post FLPo, however, there were fewer tumors and a reduction in tumor burden in Atg7Δ/Δ mice (Fig. 5D and Supplementary Fig. S5), coincident with emergence of oncocytomas rather than adenocarcinomas (Fig. 5B). This suggests that loss of autophagy does not affect the ability of activated RAS and p53 deficiency to initiate tumorigenesis but rather it diminishes tumor growth over time.
Acute ATG7 ablation alters tumor fate and compromises maintenance
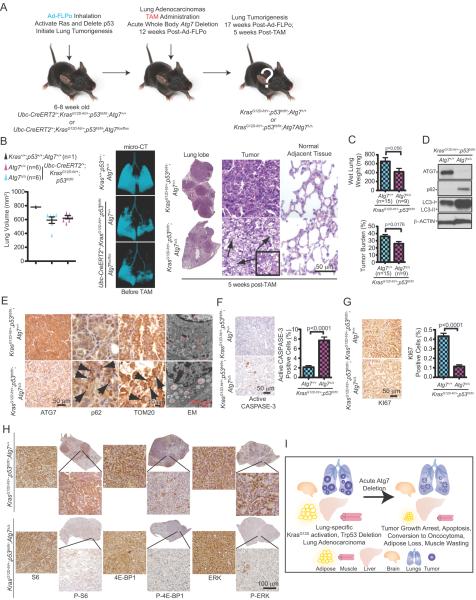
To perform the converse experiment, lung tumors were generated, and at 12 weeks post-Ad-FLPo administration, TAM administration produced acute autophagy deficiency both in established tumors and normal tissues (Fig. 6A). Tumorigenesis was then assessed 5 weeks post-TAM (17 weeks post-Ad-FlpO) at which point there was little effect of autophagy ablation to normal tissues (Fig. 1G). Lung tumor burden was similar among all mice at the time of TAM administration as determined by micro-computed tomography (micro-CT) (Fig. 6B). At 17 weeks post-FLPo and 5 weeks post-TAM, as expected, lung tumor burden, with KRAS activation and p53 deficiency produced large adenocarcinomas, encompassing most of the airspace in the lobes of the lungs with intact ATG7 (Fig. 6B and Supplementary Fig. S6A). In contrast, in Atg7Δ/Δ mice at 17 weeks post FLPo and 5 weeks post-TAM, tumors were strikingly smaller, had poorly-defined margins, and were composed of oncocytes and dead tumor cells (Fig. 6B and Supplementary Fig. S6A). There was no detectable difference in normal adjacent lung tissue in tumor-bearing Atg7Δ/Δ mice (Fig 6B). The difference in tumor burden in Atg7Δ/Δ mice with 5 weeks of Atg7 deletion was apparent by examination of wet lung weight and by determining tumor burden in scanned lung sections with tumor area reduced by one third (Fig. 6C and Supplementary Fig. S6A).
Figure 6. Acute, systemic Atg7 deficiency compromises tumorigenesis.
A. Experimental design to induce lung tumors prior to conditional whole-body Atg7 deletion. Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7+/+ and Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7flox/flox mice were infected with Ad-FLPo at 6–8 weeks of age to activate RAS and delete p53, then these mice were treated with TAM at 12 weeks post-Ad-FLPo by intraperitoneal injection to create Atg7Δ/Δ mice. Tumor progression was analyzed at various times thereafter.
B. Representative micro-CT 3-dimensional reconstruction and quantification of lung volume showing healthy air space before TAM treatment and equivalent tumor burden at the time of TAM treatment of mice with the indicated genotypes. Kras+/+;p53+/+;Atg7+/+ mice were used as a control for normal lung airspace. Tumor histology and normal adjacent lung tissue (H&E images) were analyzed at 5 weeks post-TAM (last three panels). Arrows point to dead cells.
C. Quantification of wet lung weight (top) and tumor burden (bottom). Error bar represents SEM, p values are calculated using two-way ANOVA with Bonferroni post-test.
D. Representative western blotting for ATG7, p62, S6, P-S6 and LC3 of Atg7+/+ or AtgΔ/Δ tumor tissues. β-ACTIN serves a protein loading control.
E. Representative IHC for ATG7, p62, TOM20 and EM images of tumor tissue. Arrows in second panel point to p62 aggregates and arrows in the third panel point to TOM20 accumulation in tumors from ATG7-deficient mice. N: nucleus, M: mitochondria, Ly: Lysosome, AP: Autophagosome, L: Lipid.
F. Representative IHC images for active CASPASE-3 (left) with quantification (right). Error bar represents SEM, p values are calculated using two-way ANOVA with Bonferroni post-test.
G. Representative IHC images for KI67 (left) with quantification (right). Error bar represents SEM, p values are calculated using two-way ANOVA with Bonferroni post-test.
H. Representative IHC images for S6, P-S6, 4E-BP1, P-4E-BP1, ERK and P-ERK.
I. A model for destructive effect of autophagy on established tumors.
Loss of ATG7 protein expression and autophagy ablation in tumors in the Atg7Δ/Δ mice were confirmed by Western blotting and immunohistochemistry (IHC). These tumors specifically accumulated p62 and LC3 aggregates, and abnormal mitochondria as detected by Tom20 IHC and electron microscopy (EM) (Fig. 6D–E and Supplementary Fig. S6B). Tumors in Atg7Δ/Δ mice also displayed lipid accumulation (Fig. S6C) consistent with defective mitochondrial fatty acid oxidation as reported previously with tumor-specific Atg7 deficiency (16). The impaired tumorigenesis in Atg7Δ/Δ mice was attributed to increased apoptosis (active CASPASE-3) (Fig. 6F) and ablation of proliferation, mTOR and MAP kinase signaling (KI67, loss of phosphorylated S6 [P-S6], 4E-BP1 [P-4E-BP1] and ERK [P-ERK] IHC) (Fig. 6G–H and Supplementary Fig. S6D). Serum glucose levels were similar in wild type and Atg7Δ/Δ tumor-bearing mice, indicating that hypoglycemia was not a factor contributing to defective tumor growth and survival (data not shown). While tumor-specific deletion of Atg7 concurrently with tumor initiation by KRAS activation and p53 deletion also reduces tumor burden, generates oncocytomas, suppresses proliferation and induces tumor cell death progressively beyond 6 weeks post deletion (16), the kinetics and magnitude of these events are greatly accelerated by acute ablation of Atg7 throughout the mouse (Fig. 6I).
Discussion
Acute autophagy ablation using conditional whole-body deletion of Atg7 in adult mice demonstrated the essential, systemic role of autophagy in host defense against Streptococcus infection and in preventing neurodegeneration that together limit lifespan to no more than 3 months. This contrasts constitutive autophagy-deficient mice where neonates fail to survive much more than 24 hours after birth even when force-fed (2, 3), suggesting that autophagy is more critical in newborn than adult mice.
Loss of autophagy in adult mice rapidly decreases the mass of WAT suggesting that autophagy deficiency creates systemic metabolic deficiency even under fed conditions. Autophagy deficiency may necessitate the catabolism of dedicated fat stores in WAT by cytosolic lipases or may cause browning of adipose tissue that increases β-oxidation and lipid consumption, which is consistent with mitochondrial accumulation (30). This defective adipose homeostasis and mobilization of FFAs in starved ATG7-deficient mice can contribute to systemic metabolic impairment.
One property common to adult and neonatal mice is the requirement for autophagy to survive starvation, although the underlying mechanisms appear distinct. In contrast to wild type adult mice, we found that those with acute autophagy ablation are intolerant to fasting and die of hypoglycemia. As WAT is depleted in autophagy-deficient mice, fasting results in accelerated depletion of glycogen stores and excessive muscle catabolism in comparison to wild type mice. The autophagy-deficient adult mice sustain serum amino acid levels and ketogenesis, but not FFA and glucose levels, indicating that gluconeogenesis in the liver is insufficient for survival. Moreover, despite excessive muscle wasting and maintenance of serum amino acid levels, this is apparently insufficient for gluconeogenesis. This suggests that liver autophagy and catabolism of lipid stores may be critical either directly or indirectly for gluconeogenesis and glucose homeostasis during fasting. The fasted autophagy-deficient mice display rapid liver, muscle and brain damage demonstrating the essential requirement for systemic autophagy to survive fasting. Thus, autophagy is required by both neonates and adults to survive starvation but for different reasons. Neonates need autophagy to generate amino acids to sustain metabolic homeostasis, in contrast to adult mice that need autophagy to prevent fatal hypoglycemia. Fasted adult mice deficient for autophagy have depleted lipid stores and compensate for metabolic impairment by extensive muscle wasting, which is ultimately insufficient for viability. Liver-specific deficiency in autophagy is not fatal to fasted mice (2, 31), suggesting that autophagy in other tissues, most likely including WAT (which is lost in the absence of autophagy), is essential for survival and glucose homeostasis.
RAS-driven cancers often upregulate autophagy and are dependent on autophagy for survival although the point at which autophagy functions during tumorigenesis was not known (13). To address this, we ablated Atg7 and then initiated tumorigenesis by activating RAS and deleting p53 in the lung. Autophagy appeared dispensable for tumor initiation in this setting, although over time the suppressed proliferation and emergence of oncocytomas as seen previously in the lung tumor-specific Atg7 knockouts were apparent. This suggests that functional autophagy status does not alter the efficiency by which KrasG12D activation and p53 loss initiate lung tumor formation, but rather it is instead critical for tumor maintenance.
Acute knockdown of essential autophagy genes in many RAS mutant human cancer cell lines can rapidly suppress growth and survival, whereas knockout of essential autophagy genes in GEMMs at the time of cancer initiation suppresses tumor growth gradually over time (10, 11, 16, 18, 19). This indicates that acute autophagy ablation may be more detrimental to tumorigenesis than constitutive ablation. To test this hypothesis, Atg7 was acutely ablated in mice with established KrasG12D-activated and p53-deficient NSCLC. Five weeks of systemic autophagy deficiency was dramatically destructive to these pre-existing NSCLCs, which underwent loss of mTOR and MAP kinase signaling, proliferative arrest, apoptosis and conversion of adenocarcinomas to oncocytomas. Branched chain amino acids and arginine are mTOR inducers and there were low levels of serum valine and arginine in response to autophagy inhibition, which may contribute to loss of mTOR signaling along with defective mitochondrial metabolism (Fig. 3A). Acute autophagy ablation more severely compromises tumorigenesis in comparison to tumors that evolve with deficiency in essential autophagy genes. Established tumors may be more autophagy-dependent, systemic autophagy deficiency may be more destructive to tumors than tumor-specific deficiency, or tumors evolving without autophagy may have some capacity to adapt, diminishing the impact of autophagy loss. In support of systemic autophagy deficiency being more detrimental to tumorigenesis, it also causes the elimination of WAT, perhaps creating a systemic environment metabolically unsupportive of tumor growth.
The key to determining if autophagy inhibition will be potentially effective to diminish tumorigenesis is establishing if tumors are more susceptible to acute autophagy inactivation than normal tissues, therefore providing a therapeutic window. Our data demonstrate that complete systemic autophagy inhibition for prolonged durations is not likely to be tolerable from a safety perspective. That said, the most serious observed effects on normal health were bacterial infection and neurodegeneration. Infection can presumably be managed through prophylactic antibiotics, and central nervous system toxicities should be considered in the design of pharmacological agents. Note that, genetic ablation as we have done here is complete and irreversible inhibition of autophagy, not likely achievable with small molecule inhibitors and as such this represents an extreme situation. It would, however, be interesting to compare genetic and pharmacological inhibition of autophagy (with chloroquine, for example) to help establish how effective autophagy inhibition needs to be to impair tumor growth. To what extent the detrimental effects of acute autophagy ablation are reversible in normal and tumor tissue also remains to be addressed. Finally, it will be interesting to test systemic deletion of other autophagy-related genes in this model to resolve any potential autophagy-independent functions associated with loss of specific ATG genes. With these issues addressed, our observation that 5 weeks of acute Atg7 ablation was sufficient to shut off mTOR and MAP kinase signaling and cause widespread tumor proliferative arrest and death while largely sparing normal tissues raises great hopes for antitumor efficacy.
Methods
Mice
All animal care and treatments were carried out in compliance with Institutional Animal Care and Use Committee (IACUC) guidelines. Ubc-CreERT2 mice (20) (Jackson Laboratory, Bar Harbor, ME) and Atg7flox/flox mice (2) (provided by Dr. M. Komatsu, Tokyo Metropolitan Institute of Medical Science) were cross-bred to generate Ubc-CreERT2/+;Atg7flox/flox mice. Details of the TAM treatment to induce Atg7 deletion can be found in the Supplementary Methods.
To assess the consequence of acute Atg7 deletion in response to fasting, 10 days following the last day of 5-day TAM administration, mice were fasted for 24 hours with free access to water but no food. At 20–24 hours post fasting, plasma was collected for hormonal and, biochemical analysis and tissues were harvested for histological examination. For glucose supplementation, mice were provided drinking water containing 10% glucose throughout fasting.
To assess the consequence of acute ATG7 deletion to tumorigenesis, compound Ubc-CreERT2;Atg7 mice were bred to FSF-KrasG12D mice (32) (Jackson Laboratory, Bar Harbor, ME) and Trp53frt/frt mice (33) (provided by Dr. L. Johnson, Genentech) to generate Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7+/+ and Ubc-CreERT2/+;KrasG12D-frt/+;p53frt/frt;Atg7flox/flox mice. To activate RAS and delete p53 in the lung and produce tumors, mice were infected intranasally with recombinant, replication-deficient adenovirus expressing Flp recombinase (Ad-FlpO, University of Iowa Adenoviral Core, Iowa City, IA) at 1.2 × 108 pfu/mouse at 6–8 weeks of age as described previously (16).
Gene expression analysis
Liver and muscle tissues were snap-frozen for RNA extraction. Extracted RNAs were processed, labeled and hybridized to the Affymetrix GeneChip Mouse Genome 430A 2.0 array. The raw Affymetrix CEL files for liver and muscle samples were analyzed with Agilent GeneSpring GX11 software. See Supplementary Methods for details of gene expression analysis.
Serum assays
Blood glucose was measured with a True2Go glucose meter (Nipro Diagnostics). Serum insulin and leptin levels were determined with ultra sensitive mouse insulin (Crystal Chem Inc., 90080) and leptin (Crystal Chem Inc., 90030) ELISA kits. Serum FFAs and β-hydroxybutyrate were measured using commercial kits from Cayman Chemical (700310 and 700190, respectively). Serum AST and ALT levels were determined using Olympus AU2700 (Idexx Bioresearch, West Sacramento, CA).
Metabolomic analysis by LC-MS
Samples were analyzed by LC-MS which involved reversed-phase ion-pairing chromatography coupled by negative mode electrospray ionization to a stand-alone orbitrap mass spectrometer (Thermo Scientific). See Supplementary Methods for details.
Histology
See Supplementary Methods for details about tissue fixation and antibodies used.
Western blotting
Tissues and tumor samples were ground in liquid nitrogen and lysed in Tris lysis buffer (1 M Tris HCl, 1 M NaCl, 0.1 M EDTA, 10% NP40), and probed with antibodies against ATG7 (Sigma, A2856), LC3 (Novus Biologicals, NB600-1384), p62 (10) and β-ACTIN (Sigma, A1978).
Other detailed experimental procedures are described in the Supplementary Methods.
Supplementary Material
Statement of Significance.
We systemically ablated cellular self-cannibalization by autophagy in adult mice and determined that it is dispensable for short-term survival, but required to prevent fatal hypoglycemia and cachexia during fasting, delineating a new role for autophagy in metabolism. Importantly, acute, systemic autophagy ablation was selectively destructive to established tumors compared to normal tissues, thereby providing the preclinical evidence that strategies to inhibit autophagy may be therapeutically advantageous for Ras-driven cancers.
Acknowledgements
We thank Dr. M. Komatsu for the Atg7flox/flox mice, Dr. L. Johnson and Exelexis for Trp53frt/frt mice. We also thank Dr. J. Storch for access to the EchoMRI, N. Campbell for micro-CT analysis, W. Chen for histology quantification, R. Patel for EM and CINJ Shared Resources and White laboratory members for helpful comments. This research was supported by the Histopathology and Imaging, Transgenic/Knockout Mouse and Preclinical Imaging Shared Resources and a pilot grant from the Rutgers Cancer Institute of New Jersey (P30 CA072720). This work was supported by NIH grants R37 CA53370 and R01 CA130893 to E. W., and RC1 CA147961 and R01 CA163591 to J. D. R. and E. W. and the Val Skinner Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 7.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicological sciences : an official journal of the Society of Toxicology. 2012;127:438–50. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–9. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth and fate of KRAS (G12D) -driven lung tumors. Autophagy. 2013;9 doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-drive lung tumors. Cancer discovery. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nature communications. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 20.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell stem cell. 2007;1:113–26. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 22.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–5. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10:45–56. doi: 10.4161/auto.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 29.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. The FEBS journal. 2013;280:4294–314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. International journal of cell biology. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–36. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer research. 2011;71:4040–7. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nature biotechnology. 2010;28:585–93. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.