Abstract
Objectives
The initial aim of this study was to use a systems biology approach to analyse a ciprofloxacin-selected multidrug-resistant (MDR) Salmonella enterica serotype Typhimurium, L664.
Methods
The whole genome sequence and transcriptome of L664 were analysed. Site-directed mutagenesis to recreate each mutation was carried out, followed by phenotypic characterization and mutation frequency analysis. As a mutation in the TCA cycle was detected we tested the controversial hypothesis regarding the bacterial response to bactericidal antibiotics, put forward by Kohanski et al. (Cell 2007; 130: 797–810 and Mol Cell 2010; 37: 311–20), that exposure of bacteria to agents such as ciprofloxacin produces reactive oxygen species (ROS), which transiently increase the mutation rate giving rise to MDR bacteria.
Results
L664 contained a mutation in ramR that conferred MDR. A mutation in tctA affected the TCA cycle and conferred the inability to grow on minimal agar. The virulence of L664 was not attenuated. Ciprofloxacin exposure produced ROS in L664 and SL1344 (tctA::aph), but it was reduced and occurred later. There were no significant differences in the rates of killing or mutations per generation to antibiotic resistance between the strains.
Conclusions
Whilst we confirm production of ROS in response to ciprofloxacin, we have no data to support the hypothesis that this leads to selection of MDR strains. Our results indicate that the mutations in tctA and glgA were random as they did not pre-exist in the parental strain, and that the mutation in tctA did not provide a survival advantage or disadvantage in the presence of antibiotic.
Keywords: TctA, RamR, efflux pumps, genome sequences
Introduction
Infections caused by multidrug-resistant (MDR) Gram-negative bacteria are a serious medical concern of the 21st century as there are few treatment options and few new drugs are being developed. For instance, antibiotic-resistant Salmonella enterica infections have been associated with increased risk of extra-intestinal infections, hospitalization and longer duration of illness, compared with infections due to susceptible isolates.1–4 Bacteria resistant to quinolone antibiotics (e.g. nalidixic acid) and fluoroquinolones (e.g. ciprofloxacin and norfloxacin) can be selected with single or several mutations in one or more chromosomal genes. Two types of mutant can be selected. Firstly, there are those with mutations that affect the interaction of the drug with the target topoisomerase proteins, DNA gyrase and DNA topoisomerase IV, encoded by gyrAB and parCE, respectively.5 Such mutants are only resistant to quinolones and fluoroquinolones. Secondly, MDR bacteria can be selected. Typically these contain mutations in the local repressor gene of a transcriptional activator, such as Escherichia coli marA6 or S. enterica serotype Typhimurium ramA,7–12 which is then overexpressed and in turn confers over-production of a resistance-nodulation-cell division (RND) type MDR efflux pump such as AcrAB-TolC.13
In the absence of a mutagen, mutation in bacteria has been classically considered to be a spontaneous random event, the consequences of which will only be detected under suitable selective conditions; for instance, an antibiotic-resistant sub-population of bacteria is only detected when the population is grown on antibiotic-containing agar. However, mutation rates can be affected by external influences, such as exposure to a mutagen. It has also been proposed that mutation occurs when an organism is exposed to a growth-limiting condition,14 and in 1997 we documented the occurrence of ciprofloxacin-resistant E. coli after prolonged antibiotic exposure.15 The phenomenon of so-called ‘late arising’, or ‘adaptive’ mutants, and the mechanisms by which such mutations occur, have been largely studied using the E. coli Lac system (Lac− to Lac+),16 and it has been suggested that under conditions of stress a sub-population of bacteria becomes transiently hypermutable,16,17 and that this is rpoS dependent.16 However, Koskiniemi et al.18 found that different levels of error-prone translesion polymerases and RpoS in Salmonella Typhimurium did not affect the mutation rate.
Antibiotic exposure is stressful to bacteria, and there have been several studies showing that exposure to a fluoroquinolone antibiotic gives rise to altered expression of hundreds of genes.19,20 Exposure to high concentrations of fluoroquinolone decreased expression of rpoS,20 whereas low concentrations decreased expression of sirA (which affects RpoS stability).21 Dwyer et al.19 also found three clusters of genes that responded to fluoroquinolone exposure that had not been described previously: (i) an iron uptake and utilization cluster; (ii) iron–sulphur cluster synthesis; and (iii) genes that respond to oxidative damage. They hypothesized that in response to fluoroquinolone exposure highly destructive hydroxyl radicals are produced. The same team21 also showed that after 30 min of exposure to a fluoroquinolone, a transient ≥5-fold increase in the ratio of NAD+ over NADH was seen, leading to their suggestion that there was a surge in NADH consumption upon antibiotic exposure. This induced a burst in superoxide generation via the respiratory chain and they postulated that this promoted destabilization of iron–sulphur clusters, stimulated a Fenton reaction and caused cell death. As NAD+ is reduced to NADH by tricarboxylic acid (TCA) cycle activity, it was proposed that loss of TCA cycle components would reduce the available pool of NADH, decrease superoxide generation and lead to increased survival in the presence of bactericidal antibiotics. Using E. coli mutants defective in components of the TCA cycle they found that loss of isocitrate dehydrogenase activity led to increased survival following norfloxacin treatment, whereas inactivation of genes that encode enzymes later in the TCA cycle had no effect. Most recently, Kohanski et al.22 have shown that there was a significant increase in mutation rate after exposure to a fluoroquinolone, and that there was significant correlation between the fold change in mutation rate and the peak signal when determining reactive oxygen species (ROS) formation. Thiourea, which mitigates the effects of hydroxyl radical damage, significantly reduced the mutation rate to near untreated levels. These data led Kaufmann and Hung23 to suggest that ROS transiently increase the bacterial mutation rate by generating DNA damage that is repaired in an error prone fashion. This led to the suggestion that bactericidal antibiotics, e.g. fluoroquinolones, behave as if they are mutagens. This could allow antibiotic-resistant bacteria to be selected at an elevated frequency whilst under pressure from antibiotics, and selection of resistant bacteria may occur in a few steps rather than many.
Our initial aims and objectives were to determine the genome and transcriptome of a ciprofloxacin-selected MDR Salmonella. However, following the discovery of the tctA mutation, its known role in the TCA cycle and subsequent links to survival during antibiotic exposure, we then used this mutant to explore the hypotheses of Kohanski et al.,21,22 which have been offered as a paradigm for the responses of bacteria to antibiotics in general. We hypothesized that exposure of Salmonella Typhimurium SL1344 to ciprofloxacin causes the generation of ROS, and that this transiently increases the mutation rate so that an MDR (L664) mutant containing mutations in three genes is selected. We further postulated that the mutation in ramR confers MDR, the mutation in tctA affects the TCA cycle (but allows survival of the mutant during elevated ROS levels) and mutation in glgA has no effect on antibiotic susceptibility or selection of resistant bacteria. Using mutants in which the mutations in L664 were introduced by site-directed mutagenesis, mutation experiments were carried out to test these hypotheses.
Materials and methods
Bacterial strains, mutant selection and determination of susceptibility to antibiotics, dyes, detergents and disinfectants
Mutant Salmonella Typhimurium SL134424 with decreased susceptibility to ciprofloxacin were selected as described previously.11,25 L664 ramR::aph mutants were created by P22 transduction from L1007.26 The SL1344 mutant lacking a functional tctA gene was constructed using the method of Datsenko and Wanner.27 The gene-inactivated mutants were complemented with their respective wild-type genes (amplified from SL1344 and cloned into pUC19). The site-directed substitution of glycine at position 109 of TctA with a serine was carried out with a commercial kit (QuikChange™ site-directed mutagenesis kit, Stratagene) (Table S1, available as Supplementary data at JAC Online). The MIC of each agent was determined by the standardized agar doubling-dilution method as described previously by the BSAC (http://www.bsac.org.uk).28
Total genome sequencing, analysis of sequence data and single nucleotide polymorphism (SNP) detection
Paired-end whole genome sequencing was performed on SL1344 and L664 at the University of Liverpool, using a 454 Life Sciences GS-FLX sequencer (Roche). The nucleotide sequence and gene predictions for Salmonella Typhimurium SL1344 NCTC 13347 were retrieved from the Sanger Centre web site (http://www.sanger.ac.uk/Projects/Salmonella/). Eight reads were obtained and the gene predictions were assigned a provisional functional annotation through homology searches: the best hit for BLASTP searches of each gene product prediction against the predicted Salmonella Typhimurium LT2 proteome. The read data were aligned against the annotated SL1344 sequence using xBASE-NG (http://www.ncbi.nlm.nih.gov/pubmed/17984072), which in turn uses the ‘runMapping’ component of Newbler (Roche). Newbler version 1.1.03.24 was used with default settings. High confidence differences as defined by Newbler were used, briefly: (i) at least three reads differing from the reference sequence; and (ii) at least one read aligned in the forward and reverse directions (see the manufacturers' instructions for details). These high-confidence differences were subsequently subjected to sorting and filtering by read depth and percentage coverage. The effects of individual nucleotide variations on predicted protein sequence were determined using xBASE-NG.SL1344 and L664 ramR and tctA were amplified by PCR, and the DNA was sequenced at the Functional Genomics Laboratory at the University of Birmingham.
RNA extraction and transcriptional analyses
Overnight cultures of Salmonella Typhimurium SL1344 and the test strain were grown in MOPS minimal medium (Teknova, USA) supplemented with histidine at 37°C, and the microarray experiments were carried out exactly as described by Webber et al.29 using the Pan-Salmonella Generation IV array generated at the Wellcome Trust Sanger Institute (Hinxton, UK). The Microarray dataset for L664 has been deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) with the experiment identifier E-MEXP-2696.
Growth kinetics and phenotype microarray (PM) system
The rate of growth in Luria–Bertani (LB) broth and MOPS minimal medium (Teknova, USA) supplemented with histidine for all strains, with and without the MIC of ciprofloxacin for each strain, were determined over 24 h at 37°C using FLUOstar OPTIMA (BMG Labtech, UK). All tests with the PM system were performed as described previously.30
Virulence assays, protein purification and western blotting of Sip proteins
Adhesion by, and intracellular survival within eukaryotic macrophages (RAW 264.7) by Salmonella Typhimurium SL1344 (and strains derived therefrom) were assessed exactly as described previously.29 A two-tailed Student's t-test was used to assess significance, using P < 0.05 as the cut-off. Caenorhabditis elegans survival assays were carried out exactly as described by Bailey et al.26 Protein purification and western blotting was performed exactly as described previously.29
Analysis of cell envelope components, and motility assays
Outer membranes were prepared by differential centrifugation, sonication and sarkosyl extraction, as described by Piddock et al.31 The ability of strains to migrate through (swimming) or across (swarming) semi-solid agar was determined exactly as described by Webber et al.29
Measurement of glycogen accumulation
The method described by Morán-Zorzano et al.,32 was followed. In brief, SL1344 and L664 were sub-cultured on LB agar supplemented with 50 mM glucose and incubated aerobically overnight at 37°C. Following overnight incubation, colonies were stained with iodine by overlaying the agar plates with 5 mL of iodine.
Measurement of production of ROS following antibiotic exposure
To establish whether ROS were produced following exposure to subinhibitory levels of ciprofloxacin, the method described by Kohanski et al.2 was followed, except that oxidation of the fluorescent reporter dye 3′-(p-hydroxyphenyl) fluorescein (HPF) was detected with a fluorescent plate reader (BMG FluoSTAR) and a fluorospectrophotometer (LS-30) with an excitation setting of 492 nm and an emission setting of 520 nm. Early logarithmic cultures (OD600 = 0.3) were exposed to 0.5× MIC of ciprofloxacin and 5 μM HPF simultaneously, then 100 μL of each culture was pipetted into black 96-well plates, the plates were placed in the FluoSTAR and a reading was taken every 3 min for 6 h. When using the LS-30 fluorospectrophotometer, 3 mL samples were taken every hour for 6 h, then washed three times in PBS prior to reading.
Results
MDR Salmonella Typhimurium are easily selected after exposure to ciprofloxacin
In this study we used ciprofloxacin as it is in widespread clinical use and is associated with the selection of MDR bacteria in vivo.33 Five separate, identical experiments were carried out to select mutants from SL1344 growing on agar containing ciprofloxacin. There was no significant difference (Kruskal–Wallis P = 0.406) between the frequencies of mutation to resistance or mutation rates, and MDR mutants were always selected (Table S2). L664 was one of the single colonies randomly selected in experiment 2 after 18 h of exposure of SL1344 on agar to double the MIC (0.06 mg/L) of ciprofloxacin. L664 was MDR with decreased susceptibility to a range of antibiotics of different classes, dyes, detergents and biocides (Table S3).11,25
L664 contains an SNP within ramR (SL0568) and tctA (SL2772)
Following whole genome sequencing of L664 and comparison with the genome of the parental SL1344 strain, four SNPs were found in L664 (Table S4). Two of the four SNPs conferred predicted amino acid substitutions in ORFs; these were in the genes ramR (SL0568) and tctA (SL2772). The mutations in L664 ramR and tctA were confirmed by PCR and DNA sequencing. Mutation in these genes in the SL1344 parental strain was never detected despite testing multiple stored cultures (freeze-dried, −20°C, beads and slopes). Inactivation of ramR in L664 and complementation with wild-type ramR conferred multidrug susceptibility as seen for the parental strain, SL1344. The ramR mutation resulted in a substitution, T50P, within a helix-turn-helix motif, so the mutation is likely to affect DNA binding at the operator region of ramA. The second SNP was found in tctA (SL2772). TctA is part of the tripartite system TctABC in Salmonella Typhimurium that transports tricarboxylates of the TCA cycle.34 The tricarboxylates citrate, isocitrate and cis-aconitate can be utilized by salmonellae as carbon and energy sources under aerobic and anaerobic conditions. A pre-requisite for metabolism is that the tricarboxylates are transported into the cells across the cytoplasmic membrane, and this is performed by the TctABC transport system. To establish the role, if any, of the SNP found in tctA, the observed mutation (G109S) was introduced into SL1344 by site-directed mutagenesis, to create L1207. The introduction of this mutation had no effect on antibiotic susceptibility (Table S3).
L664 also contains a 1 bp deletion in glgA (SL3502)
glgA encodes glycogen synthase. Morán-Zorzano et al.32 showed that mutants with a deletion of this gene lacked glycogen, so colonies of SL1344 and L664 growing on LB agar supplemented with 50 mM glucose were stained with iodine. SL1344 stained brown whereas L664 stained yellow, indicating that the mutant did not accumulate glycogen. To establish the role of GlgA, if any, in ciprofloxacin resistance, glgCAP was inactivated in SL1344 to give L1316. This mutation had no affect on susceptibility to antibiotics (Table S3).
The transcriptome of L664 revealed significant differential gene expression
Of the 386 genes (Table S5) for which expression was altered compared with the parental strain SL1344, 143 (37%) had increased expression and 243 (63%) had decreased expression. RT–PCR confirmed the expression pattern changes of 15 representative genes detected by microarray experiments, although, as found in other studies,26,29 the magnitude of the fold changes was different.
The expression of genes encoding components of drug efflux pumps was altered: expression of acrA and acrB was increased 2- and 5-fold, respectively, and expression of emrA, which encodes a major facilitator superfamily (MFS)-type transporter, was increased 1.5-fold. Expression of ompC, which encodes a porin, was decreased 5-fold (Table S4) and electrophoresis of outer membrane protein extracts by SDS–PAGE revealed the lack of OmpC in L664. The AraC-XylS family transcriptional activators MarA, SoxS and Rob, and local regulator AcrR, are proteins that influence expression of multidrug transporters (such as AcrB) and outer membrane proteins (such as OmpF and OmpC). However, no differential expression of marA, soxS or rob was observed in L664. Likewise, there was no change in the expression of envZ or ompR, the products of which also regulate the expression of ompF and ompC. However, compared with SL1344, expression of ramA, which encodes RamA and is also a member of the AraC-XylS family, was increased 8.7-fold in L664. Previous data26 suggest that porin genes are part of the regulon of RamA.
L664 has altered expression of numerous virulence genes, but is not attenuated
The ability of a bacterium, such as Salmonella, to infect the host is multifactorial and the expression of several virulence genes was different in L664 compared with SL1344. L664 had decreased expression (2–8-fold) of genes that encode proteins required for Salmonella to colonize and infect its host. These included 8 genes within salmonella pathogenicity island (SPI)-1 and 13 genes in SPI-2, as well as 5 genes in SPI-3, 4, 5 and 6 (Table S6). However, western blotting revealed that production of the secreted SPI-1 proteins SipA, SipB and SipC by L664 was similar to that by SL1344. It has been suggested that motility may be important in the virulence of Salmonella Typhimurium.35 Expression of flagella and chemotaxis genes was decreased in L664 (Table S6) suggesting that the MDR mutant was less motile than the susceptible parental strain, SL1344. The ability of L664 to migrate through or over semisolid agar was also evaluated. Compared with SL1344, L664 showed a significant (P < 0.000016) decrease in its ability to swim in 0.25% minimal agar. However, no significant difference was observed in its ability to swarm over 0.5% minimal agar. There was also no significant difference in the ability of L664 to adhere to, or survive in, mouse macrophage RAW 264.7 cells compared with SL1344 (Figure S1a). Furthermore, no attenuation of L664 was observed in the C. elegans infection model (Figure S1b). More detailed analysis of the expression of genes encoding proteins involved in virulence revealed that invF and hilA were not differentially expressed in L664. HilA is a member of the ToxR/OmpR family and is known to activate the sip operon, as well as to activate the transcriptional regulator InvF, which also induces the expression of the secreted proteins of the sip operon,36 which are important in invading host cells. InvF also has functions independent of HilA and is known to control expression of the effector protein, SopB.37 There was also no decrease in the expression of sopB. Furthermore, there was no decrease in the expression of the two-component regulator, BarA/SirA, which is known to control invasion.38 Taken together, despite changes in the level of transcription of some virulence genes, this was apparently insufficient to confer a distinct phenotype, and the ability of MDR L664 to infect the host was equal to that of its wild-type parental strain, SL1344.
Mutation in ramR confers a smaller effect on the transcriptome than inactivation of ramR
To determine which aspects of the L664 phenotype and subset of the transcriptome were due to mutation in ramR, the transcriptome of L664 was compared with that of a mutant in which ramR had been inactivated, L1007.26 Expression of the efflux pump genes acrA/B was similar in L1007 and in L664. However, compared with SL1344, expression of ramA was 25-fold higher in L1007, whereas L664 was ∼9-fold greater. These data suggest that the mutant RamR in L664 retained partial repression of ramA. There were other differences in the transcriptomes and phenotypes between L664 and L1007. For instance, L1007 had no differential expression of SPI genes, increased (not decreased) expression of flagellar and chemotaxis genes and had reduced virulence in various models of infection.
The transcriptome of L664 resembles that of E. coli exposed to the bactericidal fluoroquinolone, norfloxacin
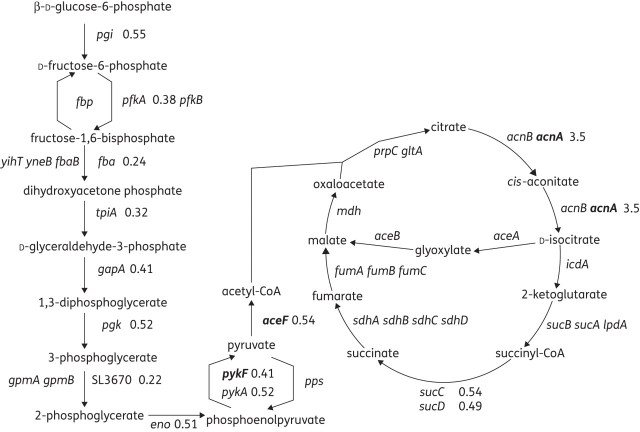
Transcriptional analysis of L664 showed a decrease in expression of 45 genes that encode proteins involved in cellular respiration and other energy-releasing pathways (Table S6). Pathway Tools39 analysis revealed that expression of 45 genes encoding enzymes involved in energy-producing pathways, such as glycolysis and the TCA cycle, was significantly different from that in SL1344 (Figure 1). The number of metabolic genes with differential expression was much lower in L1007 (ramR::aph), and where changes were seen they were opposite to those in L664. For instance, expression of glpBCF, frdABD, pckA and ilvM was increased in L1007 but decreased in L664. Kohanksi et al.21 also noted increased expression of nuo genes after fluoroquinolone treatment. These were also differentially expressed by L664; however, in the MDR mutant, expression of nuo genes was decreased.
Figure 1.
Pathway Tools output of gene expression data of L664 relative to SL1344. Values show the altered expression of 14 genes in the glycolysis and TCA cycle pathways. Where no value is shown, no change in expression was detected. Values in bold text indicate those where differential expression was verified by RT–PCR.
Due to the similarities between the transcriptome of L664 and data published by Kohanski et al.21 it led us to hypothesize that the response to ciprofloxacin had been ‘fixed’ by mutation. Therefore, to further characterize the involvement of the mutation observed in tctA of L664 (G109S), expression of four representative metabolic genes, acnA, aceF, accA and pykF, was determined for a mutant in which tctA had been inactivated (L1207), along with its complement (L1269) and another mutant in which the L664 tctA mutation had been introduced by site-directed mutagenesis (L1208; Table S3). Similar changes in expression of the four metabolic genes were seen in L664, L1207 and L1208 (Table 1). Biolog PM plates 1 and 2 were used to compare the ability of L664 with its parental strain, SL1344, to use 192 single carbon sources (Figure 2). SL1344 could utilize all; however, L664 was less able to utilize 65 (34%). L664 was not better able than SL1344 to utilize any of the tested carbon sources.
Table 1.
Generation time after growth in minimal medium and fold expression of accA, aceF, acnA and pykF observed in L664 and of tctA mutants compared with SL1344
| Fold changea relative to SL1344 |
|||||
|---|---|---|---|---|---|
| Strain | Generation time (min) | accA | aceF | acnA | pykF |
| SL1344 | 88.78 ± 6.3 | 1 | 1 | 1 | 1 |
| L1007 (SL1344 ramR::aph) | 93.43 ± 9.4 | (1) | (1) | (1) | (1.49) |
| L664 | 128.7 ± 8.5 | 1.45 | 0.67 | 2.43 | 0.65 |
| L1316 (SL1344 glgCAP::aph) | 89.26 ± 8.2 | ND | ND | ND | ND |
| L1207 (SL1344 tctA::aph) | 131.3 ± 6.2 | 1.63 | 0.57 | 2.05 | 0.59 |
| L1269 [SL1344 tctA::aph/pGEM-tctA(WT)] | 91.28 ± 5.3 | 1.01 | 0.99 | 0.97 | 1.18 |
| L1208 [SL1344 tctA::aph/pGEM-tctA(SDM G109S)] | 122.8 ± 8.6 | 1.55 | 0.55 | 1.95 | 0.64 |
WT, wild-type; SDM, site-directed mutagenesis; ND, not determined.
Bold text indicates statistically significant differences.
aData are from RT–PCR; values in brackets are from microarray data.
Figure 2.
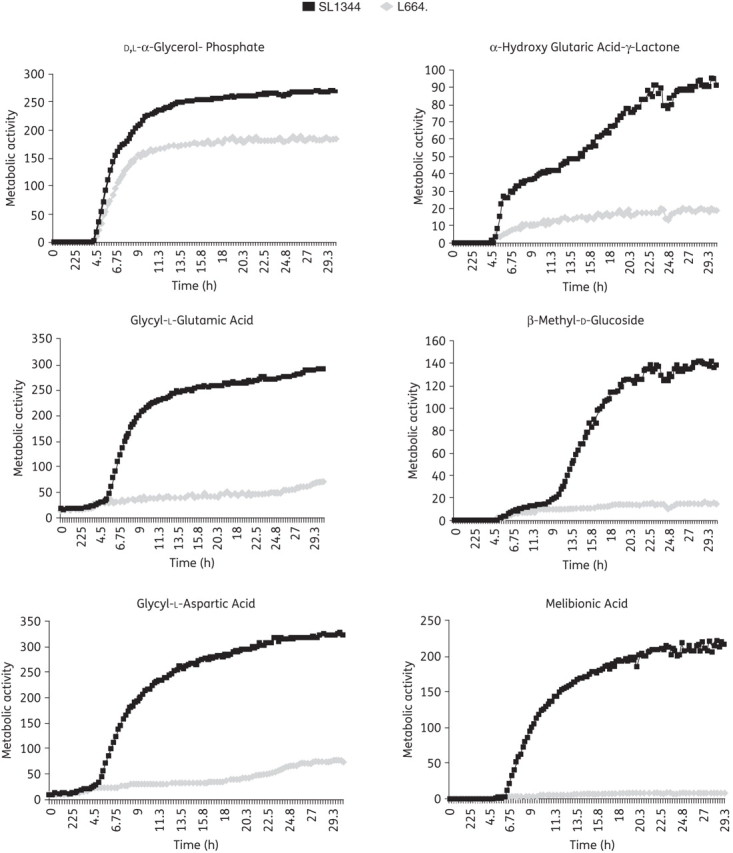
Selected carbon substrates that L664 poorly utilizes, as assayed on the Biolog PM.
The growth kinetics of SL1344, L664, L1316 (glgCAP::aph), L1207 (tctA::aph), L1269 (tctA::aph/pGEM-tctA) and L1208 [tctA::aph/pGEM-tctA(G109S)] were compared after growth of the bacteria in LB and minimal medium. No difference was observed in growth in liquid LB medium, or in time-to-grow or size of colonies on LB agar (Figure S2a). L664 and the mutants that contained a mutation in tctA (or had the gene inactivated) grew poorly in minimal liquid medium compared with those mutants that had wild-type tctA (Figure S2b). These mutants did not form colonies on minimal agar even after 72 h of incubation. Inactivation of ramR or glgCAP had no effect upon growth in minimal medium (Table 1). These data suggest that the growth defects of L664 are entirely due to the mutation in tctA and the consequent metabolic effects.
Is L664 hypermutable?
As L664 contained three mutations, the transcriptome of L664 was interrogated for expression of genes in which mutations or altered expression has been previously associated with conferring an altered mutable state; these comprised dinI, dinF, lexA, mutL, recA, rpoS, sulA, uvrB, uvrD, uvrA, umuD and umuC. Only lexA had altered expression: half that of SL1344. mutS was not on the microarray. Furthermore, the frequencies of mutation to resistance and mutations per generation in the presence of 100 mg/L rifampicin were not significantly different (frequency 5.6 × 10−9 versus 9.7 × 10−8, and 0.32 × 10−7 versus 0.48 × 10−7 mutations per generation for SL1344 and L664, respectively; P = 0.317). Taken together these data indicated that L664 was not hypermutable. Therefore, the mutations had arisen spontaneously and had become fixed in the population because they conferred an advantage (or no disadvantage) under the conditions used to select L664.
Does mutation in tctA or glgA confer a survival advantage in the presence of antibiotic?
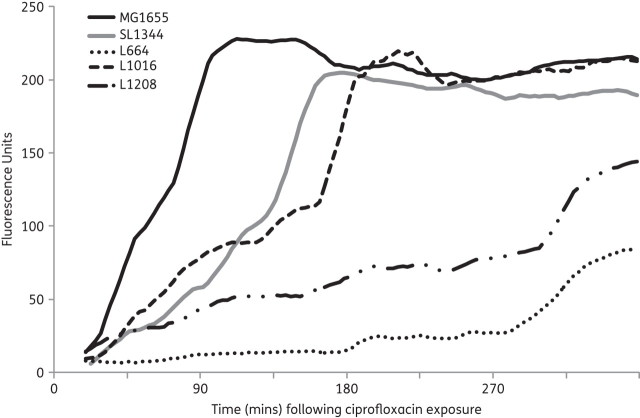
It has been suggested that: (i) mutants defective in components of the TCA cycle have increased survival during norfloxacin19,21,22 and nalidixic acid exposure;40,41 and (ii) exposure to a fluoroquinolone gives rise to ROS that transiently increase the rate of mutation. Exposure of SL1344, L664, L1016 and L1208 to subinhibitory levels of ciprofloxacin gave rise to the production of ROS when compared with E. coli MG1655 (Figure 3). However the onset of ROS production was delayed by approximately 3 h for tctA mutants compared with wild-type E. coli (MG1655) and Salmonella Typhimurium (SL1344). Based on these findings it was hypothesized that, compared with SL1344 and L1269 (tctA::aph/pGEM-tctA), L664 and L1208 [tctA::aph/pGEM-tctA(G109S)] would have increased survival to ciprofloxacin. However, at the MIC, 2× MIC, 5× MIC and 10× MIC of ciprofloxacin for each of SL1344, L664, L1208 and L1269 there was no difference in the rate of killing by the antibiotic over 18 h (Figure S3).
Figure 3.
Production of ROS (as indicated by fluorescence) in E. coli (MG1655), Salmonella Typhimurium (SL1344), L664, L1016 and L1208 following exposure to subinhibitory levels of ciprofloxacin (0.5× MIC).
It was also hypothesized that during antibiotic exposure there would be a greater transient increase in the rate of mutation for a tctA mutant than for SL1344. Furthermore, if mutation in tctA offered a survival advantage during antibiotic exposure, mutation in this gene would be found more frequently in mutants selected by antibiotic exposure. It was also postulated that under these conditions no revertants containing wild-type tctA would arise from the TctA mutant. To explore these hypotheses, SL1344, L664 and L1208 were exposed on two separate occasions to no antibiotic or to 0.5× MIC (for each strain) of ciprofloxacin, ampicillin or rifampicin. As described by Kohanski et al.,22 the strains were exposed to antibiotic for 4 h followed by a recovery period of 18 h in antibiotic-free medium. The mutation rates and frequencies of mutation to resistance were determined. There was no difference in the values for SL1344, L664 and L1208 exposed to ciprofloxacin, with or without prior exposure to ciprofloxacin or ampicillin (Table 2). Only the cultures exposed to ciprofloxacin gave rise to ampicillin-resistant colonies; all were MDR [resistant to chloramphenicol (8 mg/L), tetracycline (2 mg/L) and triclosan (0.25 mg/L)], suggesting they were ramR mutants. No rifampicin-resistant colonies were obtained irrespective of growth condition. Data for ciprofloxacin exposure, but not those for ampicillin exposure, were similar to those obtained by Kohanski et al.22 In this experiment, none of the mutants arising from SL1344 had a mutation in tctA. Additionally, tctA revertants were selected from L664 and L1208 after exposure to ciprofloxacin, but not after exposure to ampicillin. There were no statistically significant differences in the frequencies of mutation or mutations per generation for antibiotic-free cultures versus those exposed to ciprofloxacin, between SL1344, L664 and L1208. At this stage an additional experiment was added to those of Kohanksi et al.:22 the colonies obtained on the antibiotic-free agar were replica-plated onto antibiotic-containing LB and minimal agar. Except for SL1344 and the culture derived from the 4 h ampicillin exposure, both ciprofloxacin- and ampicillin-resistant colonies were obtained. In addition, rifampicin-resistant colonies were obtained from L664 in the cultures arising from the initial 4 h of exposure to ampicillin and ciprofloxacin, and from L1208 after 4 h of exposure to ciprofloxacin. Irrespective of whether exposed to antibiotic or not, all cultures of L664 and L1208 contained tctA revertants. When tested, the revertants all contained wild-type tctA, indicating a point mutation reversing the spontaneous mutation in L664 and the engineered site-directed mutation in L1208.
Table 2.
Frequency of mutation and mutations per generation of SL1344, L664, L1316 and L1208
| Mutation frequency (×10−9)/mutations per generation (×10−8) after plating onto agar containing: |
|||||
|---|---|---|---|---|---|
| Strain | 4 h of exposure to | CIP | AMP | RIF | reversion to wild-type tctA |
| After growth in the presence of antibiotic for 4 h and then 18 h recovery in antibiotic-free medium | |||||
| SL1344 | 1.9/0.075 | 0 | 0 | — | |
| AMP | 0 | 0 | 0 | — | |
| CIP | 3.3/0.22 | 4.4/0.27 | 0 | — | |
| L664 | 7.8/0.25 | 0 | 0 | 0 | |
| AMP | 9.2/0.28 | 0 | 0 | 0 | |
| CIP | 8.3/0.25 | 7.6/0.23 | 0 | 0.53/0.03 | |
| L1316 | 6.0/0.30 | — | — | — | |
| CIP | 2.5/0.19 | — | — | — | |
| L1208 | 9.9/0.25 | 0 | 0 | 0 | |
| AMP | 7.2/0.28 | 0 | 0 | 0 | |
| CIP | 5.9/0.22 | 8.9/0.3 | 0 | 0 | |
| After replica plating growth on antibiotic-free plates on to antibiotic-containing LB agar or minimal agar | |||||
| SL1344 | 1.6/0.06 | 0 | 0 | — | |
| AMP | 0 | 0 | 0 | — | |
| CIP | 3.3/0.22 | 5.2/0.31 | 0 | — | |
| L664 | 5.4/0.18 | 3.1/0.12 | 0 | 5.9/0.19 | |
| AMP | 6.5/0.21 | 0.77/0.04 | 0.46/0.03 | 4.5/0.15 | |
| CIP | 7.6/0.23 | 8.4/0.25 | 0.26/0.02 | 4.1/0.14 | |
| L1316 | 3.6/0.20 | — | — | — | |
| CIP | 2.5/0.19 | — | — | — | |
| L1208 | 7.9/0.22 | 0.47/0.027 | 0 | 6.1/0.19 | |
| AMP | 5.6/0.24 | 3.2/0.16 | 0 | 5.2/0.17 | |
| CIP | 5.1/0.20 | 8.4/0.29 | 0.27/0.03 | 3.2/0.12 | |
AMP, ampicillin; CIP, ciprofloxacin; RIF, rifampicin.
—, not tested.
Concentrations of antibiotics to which strains were exposed for 4 h in broth and then on agar plates following replica plating: SL1344, ampicillin (1 mg/L), ciprofloxacin (0.007 mg/L), rifampicin (100 mg/L);
L664, L1316 and L1208, ampicillin (2 mg/L), ciprofloxacin (0.06 mg/L), rifampicin (100 mg/L).
Inactivation of glgCAP had no effect on the frequency of mutation to antibiotic resistance (Table 2).
Discussion
Although, after ciprofloxacin exposure, the MDR mutant L664 was selected at a frequency suggesting a mutation in a single gene,25 whole genome sequencing of L664 revealed two SNPs: one in ramR, the second in tctA plus a 1 bp deletion in glgA. The mutation in ramR in L664 conferred increased expression of ramA, which resulted in MDR and organic solvent tolerance. L664 expressed less ramA than a ramR::aph mutant (L1007), and comparison of their transcriptomes suggested that the mutated RamR of L664 has retained some repressor activity. MDR and/or fluoroquinolone-resistant clinical isolates often contain multiple mutations.33 Marcusson et al.42 showed that in E. coli up to three mutations in a topoisomerase gene and marRO (homologous to ramR) conferred a fitness burden, but a fourth or fifth mutation in part ameliorated this effect. As virulence has been shown to be RamA-dependent,26 it is possible that this lower level of RamA, whilst sufficient to cause MDR, was insufficient to attenuate L664. There are several hypotheses to explain how, during antibiotic exposure, L664 was selected to contain three mutations. Firstly, one or both of the mutations was co-incidentally selected with the third, but these conferred no advantage or disadvantage to the mutant. Secondly, exposure to the antibiotic conferred conditions in which multiple mutations could occur more frequently than in antibiotic-free conditions. Thirdly, one of the mutations conferred antibiotic resistance, and another allowed survival of the mutant during antibiotic stress.
It has been suggested that antibiotic-resistant bacteria are less fit than their antibiotic-susceptible counterparts.43 Although counterintuitive, as the numbers of antibiotic-resistant bacteria isolated from infections in humans and animals continue to rise throughout the world, this has been explained not only by bacteria evolving to acquire mutations conferring antibiotic resistance, but also by various mechanisms that ameliorate the effects of such resistance.43,44 These include acquiring compensatory mutations. There have been several reports associating fitness costs in vivo with antibioticresistance mutations in Salmonella Typhimurium.43–47 Giraud et al.48 showed that highly ciprofloxacin-resistant mutants with one or more mutations in topoisomerase genes had longer generation times and were unable to colonize the gut of chickens. More recently, Wang et al.,49 Fabrega et al.50 and O'Regan et al.51 have suggested a link between decreased expression of Salmonella SPI-1 genes and fluoroquinolone resistance, and that such resistant strains were less fit. All mutants in these studies had one or more mutations in a topoisomerase gene plus a mutation in ramR, and were selected after multiple exposures to ciprofloxacin, or from a strain with pre-existing topoisomerase gene mutations. Therefore, the contribution of each mutation to the phenotype is difficult to define, especially as mutations affecting ramA expression confer pleiotropic cellular effects,26 as do those in topoisomerase genes (M. A. Webber and L. J. V. Piddock, unpublished data). We also previously showed that artificially engineered high-level expression of ramA in Salmonella Typhimurium conferred both MDR (due to enhanced efflux) and reduced fitness, and that this was associated with reduced virulencegene expression.26 However, inactivation of ramR did not give rise to high-level ramA expression and had no effect on virulence in vivo.26
Data arising in this study indicate that ciprofloxacin selected for the mutant containing the mutation in ramR, as this gave a selective advantage for growth in the presence of this antibiotic. Without the genome sequencing data it is unlikely that the mutations in tctA and glgA would have been detected. Therefore, the transcriptional and phenotypic changes may well have been incorrectly attributed to the mutation in ramR. Furthermore, the metabolic gene expression changes due to inactivation of ramR suggest that it is unlikely that the mutation in ramR can ameliorate the energy costs of the mutation in tctA. These data provide evidence that caution must be taken in the interpretation of transcriptome data for antibiotic-resistant clinical isolates and spontaneous mutants.
The mutations in glgA and tctA were unexpected. However, inactivation of glgCAP had no effect on antibiotic susceptibility or on selection of antibiotic-resistant bacteria, and so it is thought that this mutation was selected coincidentally with those in ramR and tctA. It was hypothesized that mutation in tctA affected the conformation of TctA and thus hindered the flow of tricarboxylates into the cell. The SNP found in tctA had no effect on antibiotic susceptibility, suggesting that TctABC did not transport antibiotics. The differential expression of genes encoding enzymes in energy-producing pathways, and altered growth kinetics in L664, were shown to be due to the mutation in tctA. The Biolog PM data confirmed the metabolic changes and revealed that L664 was unable to utilize several carbon sources. Eng et al.52 noted that carbon-source limitation in E. coli reduced the efficacy of killing by bactericidal drugs, including fluoroquinolones. Furthermore, Gruer et al.40 and Kohanski et al.21 found that E. coli icdA, acnA and acnB mutants had increased survival to nalidixic acid and fluoroquinolone treatment. However, despite the differential production of ROS due to antibiotic exposure, we found that the site-directed TctA mutant (L1208) was killed as rapidly as the parental strain (SL1344) by ciprofloxacin, and, in the absence of antibiotic in rich medium, had similar growth kinetics. L664 was also killed at the same rate as L1208 when exposed to the same multiple of the MIC. In addition, mutation in tctA did not give rise to a higher frequency of resistance or mutations per generation when L1208 was exposed to ciprofloxacin. Nonetheless, as found by Kohanksi et al.22 with E. coli and norfloxacin, exposure of SL1344, L664 and L1208 to ciprofloxacin gave rise to ampicillin-resistant colonies; these were all MDR. As approximately 50% of the spontaneous mutants selected after exposure of SL1344 to ciprofloxacin were MDR, and ampicillin resistance is typically part of this phenotype, this was not a surprise. No rifampicin-resistant mutants were obtained; this agent is not usually part of the MDR phenotype.8 Finally, spontaneous mutants with reversion to wild-type tctA were obtained with and without antibiotic pressure, and at similar frequencies.
While the hypothesis of Kohanski et al.21,22 that fluoroquinolones increase ROS, allowing selection of MDR bacteria, is both plausible and attractive (and we confirm production of ROS), we have no data to support the theory that this leads to selection of MDR strains. Our results indicate that the mutations in tctA and glgA were random as they did not pre-exist in the parental strain, and that although the mutation in tctA affected energy-producing pathways, it did not provide a survival advantage or disadvantage in the presence of an antibiotic, and so there was no pressure for selection. These data question not only the model proposed, but whether data obtained with E. coli offers a paradigm for the responses of other bacterial species to antibiotics.
Funding
This work was supported by a research grant (GO501415) from the UK MRC.
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We thank: Neil Hall and Margaret Hughes (University of Liverpool) for providing the whole genome sequencing service; Eirwen Morgan (Institute for Animal Health, UK) for carrying out the western blots; Jennifer Cottell for carrying out the C. elegans assays; and Sarah Coleman for the tissue culture data. We also thank Jeff Cole, Dan Andersson, Shea Fanning, Mark Webber, Martin Goldberg and Gerry Wright for helpful discussions and critical appraisal of this manuscript.
References
- 1.Angulo FJ, Nargund VN, Chiller TC. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B Infect Dis Vet Public Health. 2004;51:374–9. doi: 10.1111/j.1439-0450.2004.00789.x. doi:10.1111/j.1439-0450.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 2.Mølbak K, Baggesen DL, Aarestrup FM, et al. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N Engl J Med. 1999;341:1420–5. doi: 10.1056/NEJM199911043411902. doi:10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 3.Su LH, Wu TL, Chia JH, et al. Increasing ceftriaxone resistance in Salmonella isolates from a university hospital in Taiwan. J Antimicrob Chemother. 2005;55:846–52. doi: 10.1093/jac/dki116. doi:10.1093/jac/dki116. [DOI] [PubMed] [Google Scholar]
- 4.Varma JK, Greene KD, Ovitt J, et al. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg Infect Dis. 2005;11:943–6. doi: 10.3201/eid1106.041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett MJ, Jin YF, Ricci V, et al. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–6. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hächler H, Cohen SP, Levy SB. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–8. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abouzeed YM, Baucheron S, Cloeckaert A. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2008;52:2428–34. doi: 10.1128/AAC.00084-08. doi:10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey AM, Paulsen IT, Piddock LJ. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother. 2008;52:3604–11. doi: 10.1128/AAC.00661-08. doi:10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehrenberg C, Cloeckaert A, Klein G, et al. Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J Antimicrob Chemother. 2009;64:1175–80. doi: 10.1093/jac/dkp347. doi:10.1093/jac/dkp347. [DOI] [PubMed] [Google Scholar]
- 10.O'Regan E, Quinn T, Pagès JM, et al. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of RamA and other global regulators. Antimicrob Agents Chemother. 2009;53:1080–7. doi: 10.1128/AAC.01005-08. doi:10.1128/AAC.01005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricci V, Piddock LJ. Ciprofloxacin selects for multidrug resistance in Salmonella enterica serovar Typhimurium mediated by at least two different pathways. J Antimicrob Chemother. 2009;63:909–16. doi: 10.1093/jac/dkp054. doi:10.1093/jac/dkp054. [DOI] [PubMed] [Google Scholar]
- 12.Zheng J, Cui S, Meng J. Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J Antimicrob Chemother. 2009;63:95–102. doi: 10.1093/jac/dkn448. doi:10.1093/jac/dkn448. [DOI] [PubMed] [Google Scholar]
- 13.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–36. doi: 10.1038/nrmicro1464. doi:10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 14.Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riesenfeld C, Everett M, Piddock LJ, et al. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob Agents Chemother. 1997;41:2059–60. doi: 10.1128/aac.41.9.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez C, Hadany L, Ponder RG, et al. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet. 2008;4:e1000208. doi: 10.1371/journal.pgen.1000208. doi:10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torkelson J, Harris RS, Lombardo MJ, et al. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–11. doi: 10.1093/emboj/16.11.3303. doi:10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskiniemi S, Hughes D, Andersson DI. Effect of translesion DNA polymerases, endonucleases and RpoS on mutation rates in Salmonella typhimurium. Genetics. 2010;185:783–95. doi: 10.1534/genetics.110.116376. doi:10.1534/genetics.110.116376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dwyer DJ, Kohanski MA, Hayete B, et al. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw KJ, Miller N, Liu X, et al. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J Mol Microbiol Biotechnol. 2003;5:105–22. doi: 10.1159/000069981. doi:10.1159/000069981. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA, Dwyer DJ, Hayete B, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. doi:10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–20. doi: 10.1016/j.molcel.2010.01.003. doi:10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann BB, Hung DT. The fast track to multidrug resistance. Mol Cell. 2010;37:297–8. doi: 10.1016/j.molcel.2010.01.027. doi:10.1016/j.molcel.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Wray C, Sojka WJ. Experimental Salmonella typhimurium infection in calves. Res Vet Sci. 1978;25:139–43. [PubMed] [Google Scholar]
- 25.Ricci V, Tzakas P, Buckley A, et al. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob Agents Chemother. 2006;50:38–42. doi: 10.1128/AAC.50.1.38-42.2006. doi:10.1128/AAC.50.1.38-42.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey AM, Ivens A, Kingsley R, et al. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192:1607–16. doi: 10.1128/JB.01517-09. doi:10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. doi:10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews JM. BSAC standardized disc susceptibility testing method (version 7) J Antimicrob Chemother. 2008;62:256–78. doi: 10.1093/jac/dkn194. doi:10.1093/jac/dkn194. [DOI] [PubMed] [Google Scholar]
- 29.Webber MA, Bailey AM, Blair JM, et al. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol. 2009;191:4276–85. doi: 10.1128/JB.00363-09. doi:10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L, Lei XH, Bochner BR, et al. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol. 2003;185:4956–72. doi: 10.1128/JB.185.16.4956-4972.2003. doi:10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piddock LJ, Traynor EA, Wise R. A comparison of the mechanisms of decreased susceptibility of aztreonam-resistant and ceftazidime-resistant Enterobacteriaceae. J Antimicrob Chemother. 1990;26:749–62. doi: 10.1093/jac/26.6.749. doi:10.1093/jac/26.6.749. [DOI] [PubMed] [Google Scholar]
- 32.Morán-Zorzano MT, Alonso-Casajús N, Muñoz FJ, et al. Occurrence of more than one important source of ADPglucose linked to glycogen biosynthesis in Escherichia coli and Salmonella. FEBS Lett. 2007;581:4423–9. doi: 10.1016/j.febslet.2007.08.017. doi:10.1016/j.febslet.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Piddock LJ, White DG, Gensberg K, et al. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2000;44:3118–21. doi: 10.1128/aac.44.11.3118-3121.2000. doi:10.1128/AAC.44.11.3118-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widenhorn KA, Boos W, Somers JM, et al. Cloning and properties of the Salmonella typhimurium tricarboxylate transport operon in Escherichia coli. J Bacteriol. 1988;170:883–8. doi: 10.1128/jb.170.2.883-888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoramian-Falsafi T, Harayama S, Kutsukake K, et al. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb Pathog. 1990;9:47–53. doi: 10.1016/0882-4010(90)90039-s. doi:10.1016/0882-4010(90)90039-S. [DOI] [PubMed] [Google Scholar]
- 36.Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- 37.Eichelberg K, Galán JE. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect Immun. 1999;67:4099–105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altier C, Suyemoto M, Ruiz AI, et al. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:635–46. doi: 10.1046/j.1365-2958.2000.01734.x. doi:10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 39.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl 1):S225–32. doi: 10.1093/bioinformatics/18.suppl_1.s225. doi:10.1093/bioinformatics/18.suppl_1.S225. [DOI] [PubMed] [Google Scholar]
- 40.Gruer MJ, Bradbury AJ, Guest JR. Construction and properties of aconitase mutants of Escherichia coli. Microbiology. 1997;143:1837–46. doi: 10.1099/00221287-143-6-1837. doi:10.1099/00221287-143-6-1837. [DOI] [PubMed] [Google Scholar]
- 41.Helling RB, Kukora JS. Nalidixic acid-resistant mutants of Escherichia coli deficient in isocitrate dehydrogenase. J Bacteriol. 1971;105:1224–6. doi: 10.1128/jb.105.3.1224-1226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcusson LL, Frimodt-Moller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. doi:10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Björkman J, Nagaev I, Berg OG, et al. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–82. doi: 10.1126/science.287.5457.1479. doi:10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 44.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 45.Björkman J, Hughes D, Andersson DI. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:3949–53. doi: 10.1073/pnas.95.7.3949. doi:10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Björkman J, Samuelsson P, Andersson DI, et al. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol Microbiol. 1999;31:53–8. doi: 10.1046/j.1365-2958.1999.01142.x. doi:10.1046/j.1365-2958.1999.01142.x. [DOI] [PubMed] [Google Scholar]
- 47.Nagaev I, Björkman J, Andersson DI, et al. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol Microbiol. 2001;40:433–9. doi: 10.1046/j.1365-2958.2001.02389.x. doi:10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- 48.Giraud E, Cloeckaert A, Baucheron S, et al. Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J Med Microbiol. 2003;52:697–703. doi: 10.1099/jmm.0.05178-0. doi:10.1099/jmm.0.05178-0. [DOI] [PubMed] [Google Scholar]
- 49.Wang YP, Li L, Shen JZ, et al. Quinolone-resistance in Salmonella is associated with decreased mRNA expression of virulence genes invA and avrA, growth and intracellular invasion and survival. Vet Microbiol. 2009;133:328–34. doi: 10.1016/j.vetmic.2008.07.012. doi:10.1016/j.vetmic.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Fàbrega A, du Merle L, Le Bouguénec C, et al. Repression of invasion genes and decreased invasion in a high-level fluoroquinolone-resistant Salmonella typhimurium mutant. PLoS One. 2009;4:e8029. doi: 10.1371/journal.pone.0008029. doi:10.1371/journal.pone.0008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Regan E, Quinn T, Frye JG, et al. Fitness costs and stability of a high-level ciprofloxacin resistance phenotype in Salmonella enterica serotype Enteritidis: reduced infectivity associated with decreased expression of Salmonella pathogenicity island 1 genes. Antimicrob Agents Chemother. 2010;54:367–74. doi: 10.1128/AAC.00801-09. doi:10.1128/AAC.00801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eng RH, Padberg FT, Smith SM, et al. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother. 1991;35:1824–8. doi: 10.1128/aac.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.