Abstract
AIM: To profile the immunogenic proteins of Shigella flexneri (S. flexneri) expressed during human infection using a proteomic approach.
METHODS: Soluble and membrane protein extractions of S. flexneri 2457T were separated by two-dimensional gel electrophoresis (2-DE). Proteins were transferred to PVDF membrane and immunoblotted with sera from shigellosis patients. Reactive protein spots were matched to Coomassie stained gels run in parallel, cut out and trypsin digested. Matrix-assisted laser desorption/ionization time of flight-mass spectrometry (MALDI-TOF-MS) was used to determine the peptide mass fingerprints, which were searched in the MASCOT database to identify the protein.
RESULTS: A total of 8 immunoreactive proteins were successfully identified from the Coomassie stained gels in three repeats. Six of these proteins have not previously been reported as immunogenic in S. flexneri. These proteins could be potential candidates for vaccine or attenuation studies.
CONCLUSION: Soluble and membrane proteins of S. flexneri 2457T have been screened by 2-DE and immunoblotting with sera from shigellosis patients. Eight proteins are identified as immunogenic.
Keywords: Shigella flexneri, Immunogenetics, Vaccine antigen, Immunoblotting
INTRODUCTION
Shigellosis is a public health concern in developing countries, particularly for young children who make up 69% of all cases. The majority of shigellosis cases in the Asian, African and Central American regions are caused by Shigella flexneri (S. flexneri)[1]. Bacteria are transmitted via the fecal oral route and require as little as 100 organisms to cause the disease. S. flexneri penetrates the epithelial layer of the large intestine, invading and spreading throughout the intestinal epithelial cells. A number of proteins are known to be important for these invasion steps, particularly the products of the mxi-spa and ipa loci located on the virulence plasmid[2,3].
The immune response against S. flexneri appears to be predominantly a humoral response and it still remains unclear whether the host’s cellular immunity plays any protective role[4]. The humoral immunity appears to be protective against reinfection, generally against the homologous serotype[5-7]. This is because both the mucosal and systemic immune responses seem to be primarily directed against the lipopolysaccharide (LPS), the structure of which differs between the serotypes. Besides the LPS, S. flexneri proteins, which do not show serotype-specificity, have been identified as immunogenic during S. flexneri infection of higher primates. The invasion plasmid antigens (Ipa) are antigenic in monkey and human studies, and capable of inducing a protective immune response in mice and guinea pigs when delivered as a multiprotein complex[8-11]. Additionally, some major outer membrane proteins of 30-35 kDa are recognized by the mucosal immune response of Shigella patients and are protective in guinea pigs, mice and rabbits[8,12,13].
The escalation in antibiotic resistance in S. flexneri isolates is adding increased strain to the limited health services of developing countries. Consequently, the World Health Organization has prioritized the development of a safe and effective vaccine against S. flexneri[1]. However, despite promising results with attenuated live vaccine strains, the current vaccine candidates are either not sufficiently attenuated or immunogenic enough[14,15], suggesting that the identification of additional attenuation or protective antigens is warranted.
In order to identify such targets in S. flexneri, a profile of immunogenic proteins would be a useful tool. To date, a proteomic study of detectable immunoreactive proteins from natural Shigella infection of humans has not been reported. It is possible to identify antigens from two-dimensional gels using immunoproteomics, where proteins of a particular pathogen are separated by two-dimensional electrophoresis (2D-E) and blotted onto membranes and probed with patients’ sera. Subsequent matching and identification of any reactive protein spots generates an immunoproteome for the organism in question. This technique was initially used to visualize the antigens of Borrelia garinii and has since been used for a number of microorganisms including S. aureus, H pylori and C. albicans[16-19]. Similar studies have recently been performed with S. flexneri protein probed with sera from immunized mice or rabbits[20-22]. However, shigellosis is not a natural disease of mice or rabbits and it remains unclear what use immunoproteome data from laboratory animals will be in the study of human Shigella infection. Consequently the generation of an immunoproteome for S. flexneri with sera from human shigellosis patients would ultimately be a desirable tool for future vaccine and virulence research.
In this paper, we report the use of immunoproteomics to visualize and identify immunogenic S. flexneri 2457T serotype 2a soluble and membrane proteins, which are reactive to sera from S. flexneri infected patients.
MATERIALS AND METHODS
Bacterial cell culture
S. flexneri 2457T was grown in Luria Bertani broth overnight at 37°C in a shaking incubator. Overnight cultures were diluted 1:100 and shaken at 140 rpm until an OD600 of 0.5 was reached.
Isolation of soluble proteins
Cultures at OD600 0.5 were pelleted by centrifugation at 14 000 g, 4°C for 10 min. Pellets were washed in wash buffer (68 mmol/L NaCl, 2 mmol/L KCl, 1.5 mmol/L KH2PO4, 9 mmol/L NaH2PO4) and pelleted again. The pellet was resuspended in lysis buffer (8 mol/L urea, 4% w/v CHAPS, 1% w/v DTT, 0.8% 3/10 ampholytes, 35 mmol/L Trizma, 5 mmol/L EDTA, 1 mmol/L PMSF) and sonicated on ice at constant duty and an output of 8 for 10 × 10 s with 20 s breaks by a Branson 450 Sonifier (Branson Ultrasonics Corporation, Danbury, CT, USA). Unbroken cells and debris were removed by centrifugation at 100 000 × g for 1 h at 10°C. The supernatant was collected and stored at -80°C until required. The protein concentration of the remaining supernatant was determined by Bradford assay using BSA suspended in lysis buffer as the standard.
Isolation of outer membrane proteins
For the extraction of membrane proteins the method was modified from Molloy et al[23], with the following changes. Cultures at OD600 of 0.5 were collected by spinning at 1400 × g for 10 min. Harvested cells were resuspended in wash buffer (68 mmol/L NaCl, 2 mmol/L KCl, 1.5 mmol/L KH2PO4, 9 mmol/L NaH2PO4) and spun again. The pellet was resuspended in 50 mmol/L Tris-HCl pH 7.3 containing 0.2 g/L DNase I. The cells were disrupted by sonication as described above and unbroken cells and cell debris were removed by centrifugation at 6000 g for 10 min. The supernatant was collected and the pellet was resuspended in 50 mmol/L Tris-HCl pH 7.3 and submitted to sonication and centrifugation again. The supernatants were pooled, carbonate treated and collected as described by Molloy et al[23]. The collected membrane proteins were resuspended in 2% ASB-14, 1% DTT, 7 mol/L urea, 2 mol/L thiourea, 0.5% 3/10 ampholytes and stored at -80°C.
Two-dimensional electrophoresis
For the first dimension isoelectric focusing, 11 cm Immobiline Dry Strips with the pH gradient 4-7 were used (Amersham Pharmacia Biotech, Uppsala, Sweden). Strips were rehydrated for 16 h in 200 μL of rehydration solution (8 mol/L urea, 0.5% w/v CHAPS, 0.15% w/v DTT, 0.5% v/v Biolyte 3-10 carrier ampholytes and a trace of bromophenol blue). Strips were placed in a Multiphor II horizontal electrophoresis system (Amersham Pharmacia Biotech, Uppsala, Sweden) and 200 μL samples (about 1000 μg) of soluble and membrane protein preparations were cup loaded at the anode. Focussing was run at 20°C, 1 mA, 5 W at the following voltage gradients: 1 min 150 V, 20 min 150 V, 20 min 200 V, 2 h 300 V, 2 h on a linear gradient to 3500 V and 20 h at 3500 V. After isoelectric focussing, the strips were treated with equilibration solution I (40% v/v glycerol, 0.05 mol/L Tris-HCl pH 6.8, 6 mol/L urea, 2% w/v SDS and 2% w/v DTT) for 10 min and 10 min in equilibration solution II (40% v/v glycerol, 0.05 mol/L Tris-HCl pH 6.8, 6 mol/L urea, 2% w/v SDS and 2% w/v iodoacetamide and 0.005% bromophenol blue). Second dimension SDS-PAGE was performed on a horizontal Multiphor II electrophoresis system using precast Excel Gels with 12%-14% acrylamide gradient (Amersham Pharmacia Biotech, Uppsala, Sweden). Twin gels were loaded with a soluble protein and a membrane protein 11 cm Immobiline strip. Low molecular range markers (Amersham Pharmacia Biotech, Uppsala, Sweden) were loaded between the strips. Electrophoresis was performed at 200 V for 1 h 45 min and 600 V for 4-6 h, until the bromophenol blue front reached the edge of the gel. One of the twin gels was stained with CBB G250[24]. Approximately 150 spots were visualized for the soluble protein preparation and 25 for the membrane preparation.
Immunoblotting
The remaining gel was used for immunoblotting, where the proteins were transferred to PVDF membrane using the horizontal semi-dry electrophoretic Pharmacia LKB NovaBlot system (Amersham Pharmacia Biotech, Uppsala, Sweden) at a constant current of 0.8 mA/cm2. A reference map of proteins was produced by amido black staining of the membrane. The image was scanned before the membrane was destained[16].
The membrane was blocked with 5% skim milk powder/PBS/0.1% Tween-20 overnight at 4°C, washed three times with PBS/0.05% Tween-20 for 10 min each and incubated overnight at 4°C with 1:100 dilution of pooled sera from five S. flexneri patients (International Centre for Diarrhoeal Disease, Bangladesh) diluted in 1% skim milk powder/PBS/0.1% Tween-20. The serum samples were obtained by the International Centre for Diarrhoeal Disease, Bangladesh, from S. flexneri shigellosis patients during the acute stage of shigellosis, 3-6 d after the onset of diarrhoea. Sera were pooled due to the limited volume of patients’ sera available. The membrane was washed three times with PBS/0.05% Tween-20 for 10 min and incubated with 1:100 000 dilution of sheep anti-human Ig-HRP (Chemicon International, Temecula, CA, USA) in 1% skim milk powder/PBS/0.1% Tween-20 for 1.5 h, at 4°C. The membrane was washed with PBS/0.05% Tween three times and twice with PBS, before being incubated for 5 min with SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology Inc, Rockford, IL, USA) and developed by autoradiography. Membranes were stripped as per Jungblut and Bumann[25] and reprobed as above using sera from five healthy individuals obtained from the Australian Red Cross Blood Bank.
Peptide mass fingerprinting
Developed films, scanned pictures of the amido black stained membrane and the Coomassie stained twin gel were superimposed and compared using Adobe Photoshop® and transparencies prepared from the images. Protein spots from the Coomassie gel, which aligned to spots on the film, were excised from the gel. Some samples were sent for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis at the Australian Proteome Analysis Facility (APAF) at Macquarie University. Alternatively protein spots were digested with trypsin for 16 h at 37°C and the peptide mixture was mixed (1:1) with a saturated α-cyano-4-hydroxycinnamic acid solution in 30% acetonitrile-0.07% trifluoroacetic acid. One μL of mixture was spotted onto a sample template plate and mass spectra were obtained at the Research School of Biological Sciences (RSBS) Molecular Biology Facility (Australian National University) with an Omniflex mass spectrometer (Bruker Daltonics, Billerica, USA). Autolysing trypsin peptide 2211.1 Da was used as an internal calibration. Post-analysis data processing was done using XMASSTM software (Bruker Daltonics, Billerica, USA). Searches were performed against the MSDB database with the MASCOT peptide mass fingerprint search engine (Matrix Science)[26]. In all significant matches, the top match was a S. flexneri protein.
RESULTS
Soluble protein profile
Figure 1A shows the CBB G250 stained 2D-E gel of S. flexneri membrane and soluble protein preparations. This gel is a representative example of the three independent repeats of protein extraction and 2D electrophoresis performed for this study. Approximately 150 soluble protein spots were visualized from about 1 mg of soluble protein preparation applied to the CBB G250 stained gel. A twin gel was used for electroblotting onto PVDF membrane and membranes were transiently stained with amido black to aid in subsequent spot matching to the CBB gel. The membrane was probed with sera from S. flexneri patients and a map of immunoreactive proteins was visualized by chemiluminescence. The pattern of immunogenic proteins is shown on the film in panel B of Figure 1. This film is a representative of the three films from the immunoblot repeats. A total of 6 immunogenic proteins could be matched in each of the three repeats to a corresponding protein spot on the CBB G250 gel. These proteins were identified using peptide mass fingerprinting. The membranes from all three repeats were stripped and reprobed with sera from healthy volunteers. The resulting films had very little signal and displayed only 2 or 3 reactive spots (data not shown), none of which could be aligned to a visible S. flexneri protein spot on the CBB G250 gel and were not analysed by peptide mass fingerprinting.
Figure 1.
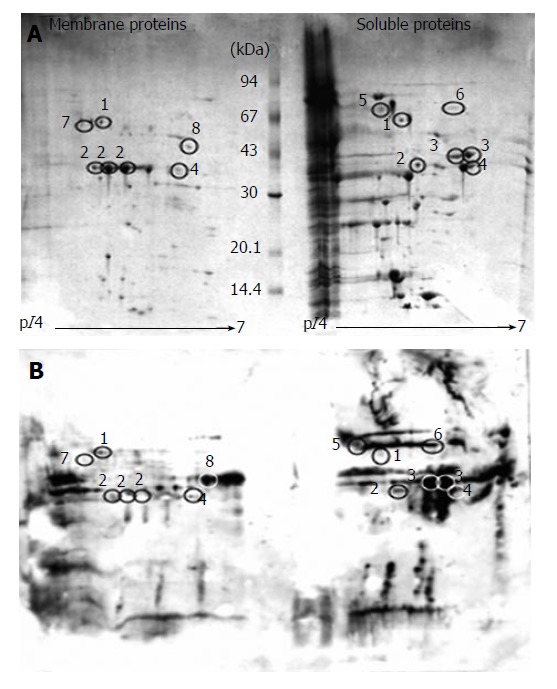
A: Coomassie blue stained 2D gel of S. flexneri membrane and soluble protein preparations; B: Immunoblot of S. flexneri membrane and soluble protein preparations. Circles indicate immunoreactive spots successfully matched and identified from the Coomassie gel (A). Numbers correspond to Table 1.
Outer membrane protein profile
The outer membrane protein extractions, stained by CBB G250, are shown on the representative gel in Figure 1A. Approximately 25 proteins were visualized for each of the three protein preparations. The film displaying the immunoreactive membrane proteins to S. flexneri patients’ sera is shown in panel B of Figure 1. It was possible to match five immunogenic spots on the film to the corresponding protein spots on the twin CBB G250 gel. Membranes, from each of the three repeats, were stripped and reprobed with control sera from healthy individuals. However, no immunoreactive spots were detected against the membrane protein preparations.
Identified immunoreactive proteins
Protein spots, which corresponded to an immunogenic spot from the immunoblot film were excised and submitted to in-gel tryptic digestion and MALDI-TOF. Of these protein spots, eight proteins were successfully identified by peptide mass matching. All eight proteins were shown as both the protein spot on the 2-DE gel and as the corresponding immunogenic spot on the matching film (Figure 1). Table 1 lists the identified proteins from the soluble and membrane preparations. Five of the eight proteins were detected in the membrane protein extraction: OmpA, AnsB, TolC, Ggt, TolB. However, OmpA, TolC and AnsB were also detected as immunogenic spots in the soluble protein extraction. Three proteins were identified exclusively as immunogenic soluble proteins: GroEL, Spa33 and IpaD. OmpA, TolC, IpaD and Spa33 were detected as immunogenic spots numerous times in the same gel, suggesting they are occurring as protein series, with each variation retaining its antigenic characteristics.
Table 1.
Proteins identified from the soluble and membrane protein immunoblots
| SpotNo. | Accession No. (GenBank) | Name | Description | Obs. pI/Mr1 | Exp. pI/Mr2 | Coverage (%) | n (peptides matched) |
| 1 | GI:30042642 | TolC | Outer membrane channel | 5.10/65.0 | 5.35/53.8 | 17 | 7 |
| 2 | GI:38000008 | OmpA | Outer membrane protein A (fragment) | 5.00-5.50/42.0 | 5.47/35.3 | 38 | 8 |
| 3 | GI:13310620 | IpaD | 37 K membrane antigen | 5.80/40.0 | 5.55/36.6 | 39 | 9 |
| 4 | GI:30042535 | AnsB | Periplasmic L-asparaginase | 6.10/38.0 | 5.95/36.8 | 22 | 6 |
| 5 | GI:17380384 | GroEL | 60 kDa chaperonin, protein Cpn60 | 4.80/68.0 | 4.85/57.3 | 18 | 7 |
| 6 | GI:32307045 | Spa33 | Surface presentation of proteins antigen | 5.80/68.0 | 5.67/33.4 | 23 | 4 |
| 7 | GI:30065275 | Ggt | Gamma-glutamyltranspeptidase | 5.00/65.0 | 5.55/61.8 | 15 | 4 |
| 8 | GI:30040341 | TolB | Involved in the tonB-independent uptake of group A colicins | 6.50/46.0 | 7.0/46.0 | 18 | 6 |
The observed pI and molecular weight of protein calculated from the protein spots position on the CBB G250 2D gel;
The predicted pI and molecular weight of the protein calculated from MSDB database.
DISCUSSION
A total of eight proteins were successfully identified in three separate 2D-E gel immunoblots with a pool of S. flexneri patients’ sera. Of the eight immunogenic proteins, only two are known to be immunoreactive for S. flexneri: IpaD and OmpA. IpaD is a secreted protein essential for entry into epithelial cells and has been reported to be antigenic in a number of S. flexneri studies[8,10,27-29]. However, it remains unclear whether anti-IpaD antibodies contribute to protective immunity in humans[2]. All four Ipa proteins have previously been reported to be immunogenic in natural infection, yet IpaA, IpaB and IpaC were not detected as antigens in this immunoproteome study[8]. A two-dimensional proteome map of S. flexneri 2457T generated by Liao et al[30] was also unsuccessful in detecting any of the Ipa proteins besides IpaD. Likewise, all three previous immunoproteomic studies for S. flexneri were unsuccessful in detecting any of the Ipa protein series as immunogenic[20-22]. It is possible that IpaA, IpaB and IpaC were not present in the 2-DE gels at sufficient protein concentrations for detection by the CBB G250 staining. The numbers of S. flexneri protein spots visualized on the CBB G250 gels were quite low, possibly due to insolubility problems during the protein extractions or low resolution of the pI 4-7 IEF strips used. A number of immunogenic spots detected on the immunoblot X-ray film could not be matched to a corresponding Coomassie stained spot, suggesting that some proteins were simply present at concentrations not detectable by CBB G250. It would be possible to improve the resolution of the 2D-E gels by using silver staining which increases the sensitivity of protein detection by an order of magnitude over CBB. Alternatively, fluorescent stains that are readily compatible with MALDI-TOF could increase the sensitivity of protein detection.
OmpA is a 35 kDa outer membrane protein and was identified a number of times during the immunoproteome analysis. OmpA is most probably a constituent of the major outer membrane proteins described as immunoreactive in Shigella patients and thus is known to be immunogenic[8].
The other six proteins have not previously been shown to be reactive to antibodies found in patients with a natural S. flexneri infection. However, the E. coli L-asparaginase II (AnsB) protein which shares 99.1% identity with S. flexneri AnsB, and the highly conserved chaperonin GroEL, in a number of bacterial species, have been reported as antigenic[31,32]. The remaining four proteins are potential candidates for Shigella-specific antigenic and immunization studies and could ultimately be useful in the development of new Shigella vaccines. However, being immunogenic does not necessarily mean that an antigen is capable of producing a protective immune response, with as little as 2.5% of antigens displaying protective properties[33]. Alternatively, because these antigenic proteins are clearly expressed during invasion of the host, as they are recognized by the host immune response, they may be important for pathogenesis. Any such proteins may be potential virulence factors and could be used in bacterial attenuation of live vaccine strains.
Membrane and surface associated proteins are often immunogenic due to their likelihood of being exposed to the host’s immune response. A number of the immunogenic proteins identified in this study are membrane or surface displayed proteins. Three outer membrane proteins were identified as immunogenic, OmpA, TolC and TolB. TolC plays a role in outer membrane permeability and has been used as an epitope carrier in Salmonella, while TolB is involved in the import of colicins[34,35]. Spa33 is a surface displayed protein involved in the regulation of the IpaB and IpaC secretion from the cell, an important virulence step during S. flexneri invasion[36]. Although Spa33 was identified as a significant match by MASCOT with peptide mass fingerprints produced from the corresponding protein spot, the observed molecular weight was inconsistent with the predicted mass. Spa33 was observed as a 68 kDa protein in Figure 1 but the predicted size was 33.4 kDa. This size discrepancy may be due to Spa33 interacting with itself as a dimer or possibly forming a complex with a protein which was not detected in the consequent peptide mass fingerprinting. There is no evidence in the literature for Spa33 forming homodimers although it is possible that Spa33 interacts with Spa32, which is predicted to be a protein of 35.6 kDa[36].
Often periplasmic proteins are immunogenic, particularly when they are found extracellularly where they are exposed to the host’s immune response. In this study, two metabolic periplasmic enzymes, Ggt and AnsB were identified; gamma glutamyltranspeptidase (Ggt) is a periplasmic enzyme, which is essential for the utilization of gamma-glutamyl peptide as an amino acid source and L-asparaginase II (AnsB) is a high affinity periplasmic enzyme, induced by anaerobiosis[37,38]. Both AnsB and Ggt were identified from the membrane extraction of S. flexneri proteins. It is possible that the membrane preparation contains some contaminating soluble proteins. The protein sequence of Ggt contains a signal peptide and has 99.7% aa identity to E. coli K12 Ggt which also contains a signal peptide and localizes to the periplasm[39]. However, the mammalian form of Ggt is linked to the plasma membrane and Neisseria meningitidis Ggt associates with the inner membrane, suggesting that the presence of S. flexneri Ggt in the membrane fraction may not simply be inefficient separation of membrane proteins from soluble proteins but perhaps due to a membrane association of the protein.
Control immunoblots were performed with each of the three membranes, using sera from five healthy individuals. It can be assumed that these five individuals have previously been exposed to commensal gut microflora, including E. coli of which Shigella is a clone[40]. Consequently, the control sera should contain antibodies reactive to E. coli and other commensal bacterial proteins, which may potentially recognize homologous Shigella proteins[41]. Thus, the control immunoblots could be used as a baseline, where any Shigella proteins recognized by the control sera were most likely reacting with antibodies raised against the commensal gut microflora. Consequently, immunogenic proteins, which were only detected in the Shigella immunoblots and not in control blots could be considered as part of a Shigella-specific immune response. The control immunoblots did not generate any spots that aligned to the eight S. flexneri protein spots from the S. flexneri patients, suggesting that the eight antigenic proteins identified reacted to anti-Shigella specific antibodies in the Shigella patients’ sera.
This is the first S. flexneri proteomic study of the immunoreactive proteins expressed during natural Shigella infection of humans. A similar study has been performed using immunized mouse sera in 2-DE gel immunoblots of S. flexneri outer membrane protein and soluble protein preparations. That study identified 13 seroreactive proteins, none of which correspond with our identified proteins[20]. Another group has visualized immunoproteomes for S. flexneri protein probed with sera from immunized rabbits[20-22]. Only two proteins, OmpA and TolC identified as immunoreactive in the present study were also detected in that work. As S. flexneri does not naturally infect or cause shigellosis-like symptoms in mice or rabbits and the experimental animals were not immunized mucosally, it remains unclear how relevant these seroreactive proteins are to the natural human anti-S. flexneri immune response.
In this study, eight proteins with reactivity to sera from patients with S. flexneri infection were identified using immunoproteomics. Six of these identified proteins have not previously been reported as immunogenic in S. flexneri natural infection. These immunoreactive proteins could be novel candidates for vaccine development. Additionally, such proteins have great potential for roles in virulence, as it seems likely they are expressed by the bacteria during the infection of the host, making them prospective attenuation targets for live vaccine construction.
ACKNOWLEDGMENTS
We would like to thank Dr. Ulrike Mathesius for her assistance with the 2D electrophoresis work, the Australian Red Cross Blood Bank for providing the control sera samples and Julie Christie of RSBS-MBF for technical advice and assistance with the MALDI-TOF.
Footnotes
S- Editor Wang J L- Editor Zhu LH E- Editor Bi L
References
- 1.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 2.Ménard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol. 1988;170:2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Way SS, Borczuk AC, Goldberg MB. Thymic independence of adaptive immunity to the intracellular pathogen Shigella flexneri serotype 2a. Infect Immun. 1999;67:3970–3979. doi: 10.1128/iai.67.8.3970-3979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mel DM, Arsić BL, Nikolić BD, Radovanić ML. Studies on vaccination against bacillary dysentery. 4. Oral immunization with live monotypic and combined vaccines. Bull World Health Organ. 1968;39:375–380. [PMC free article] [PubMed] [Google Scholar]
- 6.Jennison AV, Verma NK. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev. 2004;28:43–58. doi: 10.1016/j.femsre.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 7.DuPont HL, Hornick RB, Snyder MJ, Libonati JP, Formal SB, Gangarosa EJ. Immunity in shigellosis. II. Protection induced by oral live vaccine or primary infection. J Infect Dis. 1972;125:12–16. doi: 10.1093/infdis/125.1.12. [DOI] [PubMed] [Google Scholar]
- 8.Oaks EV, Hale TL, Formal SB. Serum immune response to Shigella protein antigens in rhesus monkeys and humans infected with Shigella spp. Infect Immun. 1986;53:57–63. doi: 10.1128/iai.53.1.57-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminski RW, Turbyfill KR, Oaks EV. Mucosal adjuvant properties of the Shigella invasin complex. Infect Immun. 2006;74:2856–2866. doi: 10.1128/IAI.74.5.2856-2866.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale TL, Oaks EV, Formal SB. Identification and antigenic characterization of virulence-associated, plasmid-coded proteins of Shigella spp. and enteroinvasive Escherichia coli. Infect Immun. 1985;50:620–629. doi: 10.1128/iai.50.3.620-629.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turbyfill KR, Hartman AB, Oaks EV. Isolation and characterization of a Shigella flexneri invasin complex subunit vaccine. Infect Immun. 2000;68:6624–6632. doi: 10.1128/iai.68.12.6624-6632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamus G, Mulczyk M, Witkowska D, Romanowska E. Protection against keratoconjunctivitis shigellosa induced by immunization with outer membrane proteins of Shigella spp. Infect Immun. 1980;30:321–324. doi: 10.1128/iai.30.2.321-324.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witkowska D, Mulczyk M, Adamus G, Romanowska E. Studies on virulence of Shigella flexneri. Protective effect of outer membrane proteins. Arch Immunol Ther Exp (Warsz) 1985;33:625–628. [PubMed] [Google Scholar]
- 14.Kotloff KL, Noriega FR, Samandari T, Sztein MB, Losonsky GA, Nataro JP, Picking WD, Barry EM, Levine MM. Shigella flexneri 2a strain CVD 1207, with specific deletions in virG, sen, set, and guaBA, is highly attenuated in humans. Infect Immun. 2000;68:1034–1039. doi: 10.1128/iai.68.3.1034-1039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coster TS, Hoge CW, VanDeVerg LL, Hartman AB, Oaks EV, Venkatesan MM, Cohen D, Robin G, Fontaine-Thompson A, Sansonetti PJ, et al. Vaccination against shigellosis with attenuated Shigella flexneri 2a strain SC602. Infect Immun. 1999;67:3437–3443. doi: 10.1128/iai.67.7.3437-3443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jungblut PR, Grabher G, Stoffler G. Comprehensive detection of immunorelevant Borrelia garinii antigens by two-dimensional electrophoresis. Electrophoresis. 1999;20:3611–3622. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3611::AID-ELPS3611>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Vytvytska O, Nagy E, Bluggel M, Meyer HE, Kurzbauer R, Huber LA, Klade CS. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics. 2002;2:580–590. doi: 10.1002/1615-9861(200205)2:5<580::AID-PROT580>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Pitarch A, Diez-Orejas R, Molero G, Pardo M, Sanchez M, Gil C, Nombela C. Analysis of the serologic response to systemic Candida albicans infection in a murine model. Proteomics. 2001;1:550–559. doi: 10.1002/1615-9861(200104)1:4<550::AID-PROT550>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 19.Krah A, Miehlke S, Pleissner KP, Zimny-Arndt U, Kirsch C, Lehn N, Meyer TF, Jungblut PR, Aebischer T. Identification of candidate antigens for serologic detection of Helicobacter pylori-infected patients with gastric carcinoma. Int J Cancer. 2004;108:456–463. doi: 10.1002/ijc.11557. [DOI] [PubMed] [Google Scholar]
- 20.Peng X, Ye X, Wang S. Identification of novel immunogenic proteins of Shigella flexneri 2a by proteomic methodologies. Vaccine. 2004;22:2750–2756. doi: 10.1016/j.vaccine.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Ying T, Wang H, Li M, Wang J, Wang J, Shi Z, Feng E, Liu X, Su G, Wei K, et al. Immunoproteomics of outer membrane proteins and extracellular proteins of Shigella flexneri 2a 2457T. Proteomics. 2005;5:4777–4793. doi: 10.1002/pmic.200401326. [DOI] [PubMed] [Google Scholar]
- 22.Ying TY, Wang JJ, Wang HL, Feng EL, Wei KH, Huang LY, Huang PT, Huang CF. Immunoproteomics of membrane proteins of Shigella flexneri 2a 2457T. World J Gastroenterol. 2005;11:6880–6883. doi: 10.3748/wjg.v11.i43.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- 24.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 25.Jungblut PR, Bumann D. Immunoproteome of Helicobacter pylori. Methods Enzymol. 2002;358:307–316. doi: 10.1016/s0076-6879(02)58097-6. [DOI] [PubMed] [Google Scholar]
- 26.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Van de Verg LL, Herrington DA, Boslego J, Lindberg AA, Levine MM. Age-specific prevaleance of serum antibodies to the invasion plasmid and lipopolysaccharide antigens of Shigella species in Chilean and North American populations. J Infect Dis. 1992;166:158–161. doi: 10.1093/infdis/166.1.158. [DOI] [PubMed] [Google Scholar]
- 28.Oaks EV, Picking WD, Picking WL. Antibody response of monkeys to invasion plasmid antigen D after infection with Shigella spp. Clin Diagn Lab Immunol. 1996;3:242–245. doi: 10.1128/cdli.3.2.242-245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A, Rong ZC, Ekwall E, Forsum U, Lindberg AA. Serum antibody responses against shigella lipopolysaccharides and invasion plasmid-coded antigens in shigella-infected Swedish patients. Scand J Infect Dis. 1993;25:569–577. doi: 10.3109/00365549309008545. [DOI] [PubMed] [Google Scholar]
- 30.Liao X, Ying T, Wang H, Wang J, Shi Z, Feng E, Wei K, Wang Y, Zhang X, Huang L, et al. A two-dimensional proteome map of Shigella flexneri. Electrophoresis. 2003;24:2864–2882. doi: 10.1002/elps.200305519. [DOI] [PubMed] [Google Scholar]
- 31.Zügel U, Kaufmann SH. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung NK, Chau IY, Coccia PF. Antibody response to Escherichia coli L-asparaginase. Prognostic significance and clinical utility of antibody measurement. Am J Pediatr Hematol Oncol. 1986;8:99–104. [PubMed] [Google Scholar]
- 33.Bumann D, Jungblut PR, Meyer TF. Helicobacter pylori vaccine development based on combined subproteome analysis. Proteomics. 2004;4:2843–2848. doi: 10.1002/pmic.200400909. [DOI] [PubMed] [Google Scholar]
- 34.Spreng S, Dietrich G, Goebel W, Gentschev I. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing protective listerial epitopes within a surface-exposed loop of the TolC-protein. Vaccine. 2003;21:746–752. doi: 10.1016/s0264-410x(02)00594-7. [DOI] [PubMed] [Google Scholar]
- 35.Lazzaroni JC, Dubuisson JF, Vianney A. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie. 2002;84:391–397. doi: 10.1016/s0300-9084(02)01419-0. [DOI] [PubMed] [Google Scholar]
- 36.Schuch R, Maurelli AT. Spa33, a cell surface-associated subunit of the Mxi-Spa type III secretory pathway of Shigella flexneri, regulates Ipa protein traffic. Infect Immun. 2001;69:2180–2189. doi: 10.1128/IAI.69.4.2180-2189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harms E, Wehner A, Jennings MP, Pugh KJ, Beacham IR, Rohm KH. Construction of expression systems for Escherichia coli asparaginase II and two-step purification of the recombinant enzyme from periplasmic extracts. Protein Expr Purif. 1991;2:144–150. doi: 10.1016/1046-5928(91)90063-o. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki H, Kumagai H, Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: formation and localization. J Bacteriol. 1986;168:1332–1335. doi: 10.1128/jb.168.3.1332-1335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percival RS, Marsh PD, Challacombe SJ. Serum antibodies to commensal oral and gut bacteria vary with age. FEMS Immunol Med Microbiol. 1996;15:35–42. doi: 10.1111/j.1574-695X.1996.tb00356.x. [DOI] [PubMed] [Google Scholar]


