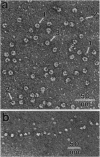
Abstract
The trp RNA-binding attenuation protein of Bacillus subtilis, TRAP, regulates both transcription and translation by binding to specific transcript sequences. The optimal transcript sequences required for TRAP binding were determined by measuring complex formation between purified TRAP protein and synthetic RNAs. RNAs were tested that contained repeats of different trinucleotide sequences, with differing spacing between the repeats. A transcript containing GAG repeats separated by two-nucleotide spacers was bound most tightly. In addition, transmission electron microscopy was used to examine the structure of TRAP and the TRAP-transcript complex. TRAP was observed to be a toroid-shaped oligomer when free or when bound to either a natural or a synthetic RNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antson A. A., Brzozowski A. M., Dodson E. J., Dauter Z., Wilson K. S., Kurecki T., Otridge J., Gollnick P. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J Mol Biol. 1994 Nov 18;244(1):1–5. doi: 10.1006/jmbi.1994.1698. [DOI] [PubMed] [Google Scholar]
- Antson A. A., Otridge J., Brzozowski A. M., Dodson E. J., Dodson G. G., Wilson K. S., Smith T. M., Yang M., Kurecki T., Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995 Apr 20;374(6524):693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Gollnick P., Yanofsky C. The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis. J Bacteriol. 1992 Apr;174(7):2059–2064. doi: 10.1128/jb.174.7.2059-2064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Stults J. T., Shire S. J., Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994 Jun 17;269(24):16597–16604. [PubMed] [Google Scholar]
- Babitzke P., Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear D. G., Hicks P. S., Escudero K. W., Andrews C. L., McSwiggen J. A., von Hippel P. H. Interactions of Escherichia coli transcription termination factor rho with RNA. II. Electron microscopy and nuclease protection experiments. J Mol Biol. 1988 Feb 20;199(4):623–635. doi: 10.1016/0022-2836(88)90306-3. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987 Mar 27;48(6):945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Kuroda M. I., Henner D., Yanofsky C. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol. 1988 Jul;170(7):3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M. I., Shimotsu H., Henner D. J., Yanofsky C. Regulatory elements common to the Bacillus pumilus and Bacillus subtilis trp operons. J Bacteriol. 1986 Sep;167(3):792–798. doi: 10.1128/jb.167.3.792-798.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Brutlag D. L. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1986 Feb;83(3):696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen J. A., Bear D. G., von Hippel P. H. Interactions of Escherichia coli transcription termination factor rho with RNA. I. Binding stoichiometries and free energies. J Mol Biol. 1988 Feb 20;199(4):609–622. doi: 10.1016/0022-2836(88)90305-1. [DOI] [PubMed] [Google Scholar]
- Otridge J., Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Kuroda M. I., Yanofsky C., Henner D. J. Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon. J Bacteriol. 1986 May;166(2):461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slock J., Stahly D. P., Han C. Y., Six E. W., Crawford I. P. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J Bacteriol. 1990 Dec;172(12):7211–7226. doi: 10.1128/jb.172.12.7211-7226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]