Abstract
Aim:
Expression of liver low-density lipoprotein receptor (LDLR), a determinant regulator in cholesterol homeostasis, is tightly controlled at multiple levels. The aim of this study was to examine whether proteasome inhibition could affect LDLR expression and LDL uptake in liver cells in vitro.
Methods:
HepG2 cells were examined. Real-time PCR and Western blot analysis were used to determine the mRNA and protein levels, respectively. DiI-LDL uptake assay was used to quantify the LDLR function. Luciferase assay system was used to detect the activity of proprotein convertase subtilisin/kexin type 9 (PCSK9, a major protein mediating LDLR degradation) promoter. Specific siRNAs were used to verify the involvement of PCSK9.
Results:
Treatment of HepG2 cells with the specific proteasome inhibitor MG132 (0.03–3 μmol/L) dose-dependently increased LDLR mRNA and protein levels, as well as LDL uptake. Short-term treatment with MG132 (0.3 μmol/L, up to 8 h) significantly increased both LDLR mRNA and protein levels in HepG2 cells, which was blocked by the specific PKC inhibitors GF 109203X, Gö 6983 or staurosporine. In contrast, a longer treatment with MG132 (0.3 μmol/L, 24 h) did not change LDLR mRNA, but markedly increased LDLR protein by reducing PCSK9-mediated lysosome LDLR degradation. Furthermore, MG132 time-dependently suppressed PCSK9 expression in the HepG2 cells through a SREBP-1c related pathway. Combined treatment with MG132 (0.3 μmol/L) and pravastatin (5 μmol/L) strongly promoted LDLR expression and LDL uptake in HepG2 cells, and blocked the upregulation of PCSK9 caused by pravastatin alone.
Conclusion:
Inhibition of proteasome by MG132 in HepG2 cells plays dual roles in LDLR and PCSK9 expression, and exerts a beneficial effect on cholesterol homeostasis.
Keywords: cholesterol homeostasis, LDLR, MG132, proteasome inhibition, PKC, PCSK9, pravastatin, HepG2 cells
Introduction
Liver low-density lipoprotein receptor (LDLR) plays a central role in the maintenance of human plasma cholesterol levels by mediating LDL clearance from the bloodstream. Loss-of-function mutations in the LDLR gene have been identified as the major cause of familial hypercholesterolemia (FH), which is characterized by elevated levels of plasma LDL-cholesterol (LDL-c) and an increased risk of cardiovascular disease1. The enhancement of LDLR activity is therefore an effective method of manipulating cholesterol homeostasis2.
LDLR expression is coordinately regulated in cells at multiple levels. Sterol-responsive element-binding proteins (SREBPs), especially SREBP-2, predominantly regulate the LDLR transcription in a cholesterol-negative feedback pathway1. Post-translational regulation is another important approach to modulate LDLR expression. Proprotein convertase subtilisin/kexin type 9 (PCSK9), a newly identified protein, is a critical regulator for LDLR. PCSK9 binds directly to the epidermal growth factor repeat A(EGF-A) of LDLR and blocks LDLR recycling by promoting its degradation in lysosome3. Individuals with PCSK9 loss-of-function mutations display reduced LDL-c levels and risk of coronary heart disease4. In earlier clinical trials, monoclonal antibodies against PCSK9 achieved a substantial reduction in the LDL-c levels of FH and non-FH patients5. Therefore, PCSK9 is the most promising target for an LDL-C reducing treatment since the discovery of the HMG-CoA inhibitor-statins.
Statins, the most widely used lipid-lowering drugs, increase LDLR expression by enhancing SREBP-2 function. However, therapies based on SREBP-2 activation also induce the expression of PCSK9, another SREBP-2 target gene, which reduces LDLR expression by promoting its degradation6. A recent clinical trial revealed that patients, including those with familial hypercholesterolemia, who were treated with a combination of statins and PCSK9 antibody achieved target LDL-c concentrations more effectively than those treated with statins alone7. Thus, the development of PCSK9 inhibitors for statin supplementation and the identification of novel dual modulators for LDLR and PCSK9 may be promising methods to lower LDL-c.
Proteasome inhibition is reported to confer cardioprotective effects through multiple mechanisms. MG132, a specific proteasome inhibitor, reduces the degradation of IκB (inhibitory κB), GRK2 (G-protein-receptor kinase 2), ARC (apoptosis repressor with caspase recruitment domain), and HSP (heat shock proteins), all of which exert beneficial effects on acute myocardial ischemia8. Moderate proteasome inhibition by MG132 results in the upregulation of anti-oxidative enzymes, and it protects rat myocytes against H2O2-mediated oxidative stress9. Moreover, proteasome inhibition may benefit atherosclerosis through the improvement of reverse cholesterol transport (RCT) in vivo and in vitro by preventing the degradation of ABCA1 and ABCG1 in macrophages10. Accordingly, we speculated that proteasome inhibition plays a role in maintaining cholesterol homeostasis, which contributes to its cardioprotective effects.
In the current study, we employed MG132 to evaluate the potential effects of proteasome inhibition on LDL-c clearance via detection of the LDLR expression and function in HepG2 cells. Our data showed that MG132 upregulates LDLR transcription and function in a dose- and time-dependent manner. Moreover, MG132 appears to augment LDLR transcription through a PKC-related pathway. Unexpectedly, MG132 suppressed PCSK9 expression, sequentially decreased LDLR degradation and promoted LDL uptake in HepG2 cells. These results clearly support a role of MG132 as a dual modulator of LDLR and PCSK9 expression. In addition, a combination of MG132 and pravastatin enhanced the LDLR expression while suppressing the increase in PCSK9 stimulated by pravastatin alone.
Materials and methods
Cell lines and reagents
HepG2 (ATCC® HB-8065) and HEK293 cells (ATCC® CRL-1573™) were maintained in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Gibco Invitrogen China Limited, Shanghai, China). The cells were treated with various compounds in the presence of 2% LPDS, unless otherwise indicated. Stable transfectants of PCSK9-FLAG-HEK293 were obtained by G418 (1 mg/mL) selection for 2 weeks.
The primers for reverse transcription and real-time PCR were synthesized by Sangon Inc (Shanghai, China). Other PCR-related reagents were obtained from Bio-Rad Inc (California, USA). The si-RNAs were synthesized by Genepharma Inc (Shanghai, China). The luciferase reporter assay system was obtained from Promega Inc (Wisconsin, USA). Antibodies for the LDLR and PCSK9 were purchased from Abcam (Cambridge, United Kingdom), SREBP-2 from Proteintech (Chicago, USA), and ERK, pERK, ubiquitin, and GAPDH from Cell Signaling Technologies (Massachusetts, USA). MG132 was acquired from MP Biomedicals Co, Ltd (California, USA). Gö 6983 and staurosporine were purchased from Santa Cruz Biotechnology, Inc (Texas, USA). Actinomycin D, β-ACTIN, and all other reagents were obtained from Sigma Aldrich (Saint Louis, USA), unless otherwise specified.
Real-time PCR and Western blot analysis
The total RNA was isolated (Trizol, Invitrogen Life technologies, California, USA) and reverse-transcribed to cDNA (Promega). The mRNA expression of LDLR, PCSK9, SREBP-1c, FAS, SCD, and SREBP-2 was assessed by real-time PCR using specific primers and the SYBR green PCR Master Mix (Bio-Rad, USA). The primer sequences are listed in Table 1.
Table 1. Sequences of primers used to quantify expression of genes implicated in LDLR and PCSK9 regulation.
| Gene name (Human) | Accession No | Primer (5′–3′) | Product length (bp) |
|---|---|---|---|
| GAPDH | NM_002046.3 | AAGAAGGTGGTGAAGCAGG AGGTGGAGGAGTGGGTGTCG | 109 |
| PCSK9 | NM_174936.3 | CCAAGCCTCTTCTTACTTCACC GCATCGTTCTGCCATCACT | 111 |
| LDLR | NM_000527.4 | CTGAAATCGCCGTGTTACTG GCCAATCCCTTGTGACATCT | 92 |
| SREBP-1c | NM_001005291.2 | GCAGGGGTAGGGCCAACGGCCT CATGTCTTCGAAAGTGCAATCC | 80 |
| FAS | NM_004104.4 | TGTGGACATGGTCACGGAC GGCATCAAACCTAGACAGGTC | 100 |
| SCD | NM_005063.4 | AGTCTGGCTTTGCTCTATTAAGAT CTTCCCTTAGATCCTTTTCTACTG | 192 |
| SREBP-2 | NM_004599.3 | CTCTGACCAGCACCCACACT CACACCATTTACCAGCCATAAG | 118 |
Protein (30 μg) from the HepG2 cell lysates or 40 μL of medium (without FBS) was separated using SDS-PAGE and transferred to a PVDF membrane (Bio-Rad). The membranes were incubated with antibodies specific for LDLR (1:5000), PCSK9 (1:2500), SREBP-2 (1:500), ERK and pERK (1:1000), ubiquitin (1:1000), β-ACTIN (1:2000), and GAPDH (1:2000), respectively, at 4 °C overnight, followed by incubation with the corresponding secondary antibodies. The bands were visualized with an enhanced ECL system (Thermo Fisher, Massachusetts, USA).
Lipoprotein isolation and DiI-LDL preparation
Human LDL and lipoprotein-deficient serum (LPDS) were separated from the pooled plasma of normal cholesterolemic volunteers by ultracentrifugation and were extensively dialyzed against dialysis buffer and PBS. The LDL was labeled with the fluorescent probe-DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, Biotium, California, USA) as previously described, with some modifications11. Briefly, DiI in DMSO (15 mg/mL) was added to the LDL-LPDS mixture (v/v, 1:2) to a final concentration of 300 mg DiI/mg LDL protein. The DiI-LDL-LPDS solution was incubated overnight at 37 °C. The DiI-labeled LDL was isolated by ultracentrifugation, dialyzed against a dialysis buffer, sterilized using a 0.45 μm filter (Millipore, Massachusetts, USA) and stored at 4 °C. All of the procedures conformed to the declaration of Helsinki and were approved by Ethics Committee of Shanghai Xuhui Central Hospital.
DiI-LDL uptake assay
The LDLR activity of the HepG2 cells was determined using a previously described method, with minor modifications11. Briefly, HepG2 cells grown on 24-well plates were incubated in DMEM with 2% LPDS and 20 μg/mL DiI-LDL for 4 h at 37 °C in the dark. After incubation, the cells were rinsed twice with PBS containing 0.4% albumin and were washed three times with PBS. For the fluorescence microscopy, the cells were fixed with 4% formalin, and the nuclei were subsequently stained with Hoechst dye. For the fluorescence quantification, 400 μL of isopropanol was added into each well of a 24-well plate followed by a 20-min incubation at room temperature under constant shaking, and 200 μL aliquots were used for the analysis with a SpectraMax M2e Microplate Reader (Molecular Devices, 520–570 nm). NaOH (0.5 mol/L) was used to lyse the remaining cells for 30 min, and aliquots of 10 μL were used for the protein determination (Coomassie Brilliant Blue G-250, Bio-Rad, USA).
Proteasome activity assay
The HepG2 cells were seeded in 6-well plates, treated with MG132 for the recommended time, and trypsinized. The chymotrypsin activity was detected using the protocol of the Proteasome-Glo™ Assay Systems (G8621, Promega).
LDLR mRNA stability detection
HepG2 cells were treated with MG132 or a vehicle for 6 h, 5 mg/L actinomycin D was added to all of the wells, and cells were harvested for 30, 60, or 120 min. The LDLR mRNA levels were assessed with a real-time PCR and plotted as a percentage of the remaining LDLR mRNA. The decay curves were plotted versus the time.
RNA interference with small anti-PCSK9 RNA
HepG2 cells seeded in the 6-well plate were transiently transfected with PCSK9-specific siRNAs (40 nmol/L) using Lipofectamine™ 2000 (Invitrogen) in serum-free DMEM medium according to the kit's instructions. This medium with siRNA-PCSK9 was maintained for 6 h, and it was then replaced by a new medium (high glucose-DMEM with 2% LPDS) for 24 h before being treated with the drugs.
si-PCSK9: GGUCUGGAAUGCAAAGUCAdTdT;
Negative control: UUCUCCGAACGUGUCACGUdTdT
Promoter analysis
The luciferase activity was assessed using the luciferase assay system according to the manufacturer's instructions (Promega, USA), and β-galactosidase expression (pCH110) was used for the normalization. HepG2 cells were transfected with pCH110 and the indicated luciferase promoter construct plasmids for PCSK9 (D4, D4-SRE, and D4-HNF). The D4-HNF mutant plasmid was constructed as described previously12.
PCSK9 activity assay
HEK293 cells with or without the stable transfection of PCSK9-Flag constructs were cultured in DMEM supplemented with 1% FBS for 24 h to obtain the conditioned medium. The conditioned medium was used to detect PCSK9 activity for reducing the LDLR level in HepG2 cells. Pre-seeded HepG2 cells were washed twice with PBS and cultured for 6 h at 37 °C in conditioned medium with or without 30 min of exposure to MG132. Following the culture, the cells were either measured for DiI-LDL uptake ability or lysed in RIPA buffer for a Western blot analysis.
Statistical analysis
The continuous variables are presented as mean±SEM. Student's t-test was used to compare the differences between the two groups, and one-way ANOVA followed by Dunnett's Multiple Comparison Test was used to compare the differences for more than two groups. P<0.05 was considered statistically significant.
Results
MG132 upregulates LDLR mRNA and protein levels in HepG2 cells
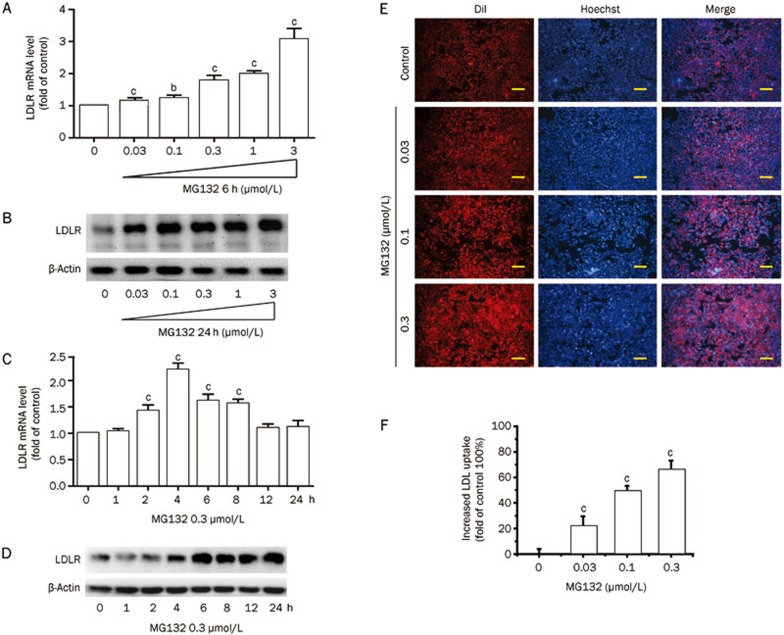
MG132 promoted a remarkable increase in both the LDLR mRNA and protein levels (Figure 1A–1D). LDLR mRNA was upregulated transiently and the levels peaked at 4 h (2.2-fold that of the control, P<0.01) and then declined to background levels after 12 h (MG132 0.3 μmol/L). However, the LDLR protein level continued to show a robust elevation throughout the 4 to 24 h treatment (Figure 1D), indicating the involvement of a post-translation pathway.
Figure 1.
Effects of MG132 on LDLR expression in and LDL uptake by HepG2 cells. (A) Real-time quantification of the LDLR mRNA level and (B) Western blot analysis of LDLR protein in HepG2 cells treated with MG132 (0.03, 0.1, 0.3, 1, and 3 μmol/L) for 6 h. Time-course of the LDLR mRNA (C) and protein (D) levels in HepG2 cells exposed to MG132 (0.3 μmol/L) for 24 h. DiI-LDL uptake was assessed in cells treated with the indicated concentrations of MG132 for 24 h; (E) Representative images of cells associated with DiI-LDL (Red) and Hoechst-stained nuclei (Blue). Scale bar 50 μm; (F) The normalized fluorescence of isopropanol-extracted DiI (520 and 570 nm). Data are presented as the mean±SEM of at least three independent experiments. bP<0.05, cP<0.01 vs the vehicle-treated groups.
MG132 promotes LDL uptake in HepG2 cells
We further investigated whether LDL uptake was increased by MG132. As expected, MG132 (0, 0.03, 0.1, 0.3 μmol/L, 24 h) increased LDL uptake in HepG2 cells in a dose-dependent manner, with a 70% (P<0.01) increase at the highest dose (0.3 μmol/L) compared with the untreated cells (Figure 1E, 1F).
We additionally tested the effects of MG132 on primary mouse hepatocytes (PMH). MG132 consistently increased the LDLR expression and DiI-LDL uptake in the PMH from C57BL/6 mice (Supplementary Figure 1A, 1B). However, upregulation of the DiI-LDL uptake was not observed in the PMH from ldlr−/− mice (Supplementary Figure 1C, 1D), indicating that this increased uptake of LDL was mediated by LDLR but not by other receptors.
Low dose of MG132 modestly inhibited proteasome activity in HepG2 cells
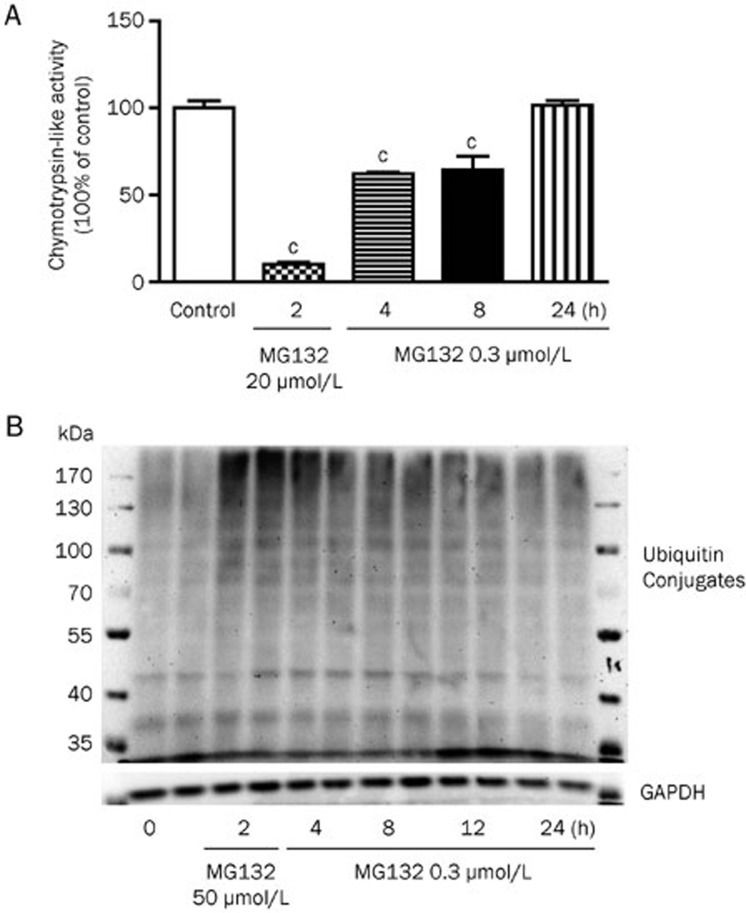
To determine the proteasome inhibition ability of MG132 under currently used doses, we examined the chymotrypsin-like proteasome activity in the HepG2 cells treated with MG132. The proteasome activity was greatly inhibited by a high concentration of MG132 (20 μmol/L), but it was only partially inhibited by 37.8%±1.5% and 35.6%±13.4% after 4 and 8 h of treatment with 0.3 μmol/L MG132, respectively, and the activity returned to normal levels after 24 h of incubation (Figure 2A). In addition, a modest increase in the ubiquitinated proteins was observed after 4 and 8 h of treatment, but no significant increase was observed at 24 h, which was comparable to the results obtained from the chymotrypsin-like proteasome activity (Figure 2B). Proteasome inhibition is reported to induce an Nrf-1 protein-dependent compensatory increase in the proteasome subunit gene expression and to restore proteasome function13. MG132 consistently induced an increase in the expression of specific proteasome subunit genes (Supplementary Figure 2), indicating that the restoration of proteasome function at 24 h may be caused by the compensatory expression of proteasome subunit genes.
Figure 2.
Proteasome activity. (A) Chymotrypsin-like protease activities of HepG2 cells treated with vehicle, 20 μmol/L MG132 (positive control) or 0.3 μmol/L MG132 for indicated times. (B) Western blot analysis of ubiquitinated proteins in HepG2 cells. The data are representatives of three independent experiments. cP<0.01 vs control.
MG132 upregulates LDLR mRNA through a PKC-dependent pathway
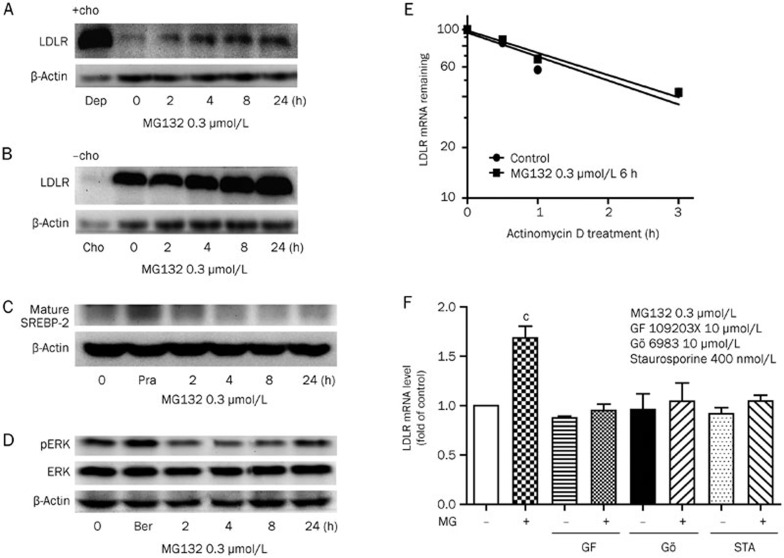
Next, we explored the mechanisms involved in MG132-mediated LDLR mRNA enhancement in HepG2 cells. High cellular cholesterol levels inhibit SREBP-2 maturation, thus decreasing LDLR transcription, and vice versa. To determine whether these processes are related to LDLR transcription upregulation by MG132, high levels of sterols (10 mg/L cholesterol and 2 mg/L 25-OH cholesterol) were added to the culture media. The suppressive sterol levels almost abolished the LDLR expression but did not block the effect of MG132 (0.3 μmol/L) in increasing the LDLR expression (Figure 3A). Moreover, depleting the cellular cholesterol levels by adding 10 μmol/L pravastatin and 100 μmol/L mevalonlactone in the medium also had no effect on the induction of LDLR by MG132 (Figure 3B). We also detected the mature form of SREBP-2 by immunoblotting (Figure 3C), and no significant increase was observed, although SREBP is reported to be degraded by the proteasome14. This lack of a significant increase may be attributed to the low doses of MG132 used in this experiment, leading to only partial proteasome inhibition, while the majority of activity was preserved, as noted above in Figure 2. Our data confirmed that the MG132-mediated augmentation of the LDLR mRNA level is independent of the SREBPs.
Figure 3.
MG132 modulates LDLR mRNA and protein levels through a PKC-dependent pathway. Western blot analysis of LDLR expression in HepG2 cells incubated with MG132 (0.3 μmol/L) under cholesterol addition (A, +cho, 2 mg/L 25-OH cholesterol and 10 mg/L cholesterol) or cholesterol depletion (B, -cho, 10 μmol/L pravastatin and 100 μmol/L mevalonlactone) conditions for the indicated times, (dep-, control of cholesterol depletion group; cho-, control of cholesterol addition group; great increase of LDLR expression in the dep group and decrease in the cho group indicating increased or decreased SREBP-2 activity). Western blot analysis of (C) mature SREBP-2 and (D) phosphorylated ERK (Pra-, pravastatin 5 μmol/L; Ber-, berberine 15 μg/L as positive controls, respectively). (E) LDLR mRNA was measured via real-time PCR, and the decay curves were plotted versus the time (MG132 0.3 μmol/L, 6 h). (F) Real-time PCR analysis of LDLR transcription in HepG2 cells incubated with GF 109203X (10 μmol/L), Gö 6983 (10 μmol/L) and staurosporine (400 nmol/L) in the presence or absence of MG132 0.3 μmol/L for 6 h, respectively. All of the values are presented as the mean±SEM of three or more independent experiments. bP<0.05, cP<0.01 vs the control groups.
The LDLR mRNA has a short half-life due to the ARE site on its 3′-UTR, and phosphorylation of the extracellular signal-regulated kinase (ERK) is reported to stabilize it1,15. To determine whether these processes are involved, we examined the LDLR mRNA decay rates and ERK phosphorylation levels in the presence or absence of MG132. LDLR mRNA stability remained unchanged after incubation with MG132 (6 h) compared with the control group, while the phosphorylation of ERK was slightly decreased initially (0–12 h) and restored after the 24 h treatment (Figure 3D, 3E). Accordingly, we conclude that MG132-induced LDLR transcription is not mediated by mRNA stabilization.
In experiments using inhibitors of different pathways, including NF-κB, ERK, PI3K/Akt, JAK, p38, JNK, and PKC (data not shown), the specific inhibitors of protein kinase C (PKC)-GF 109203X (10 μmol/L), Gö 6983 (10 μmol/L), and staurosporine (400 nmol/L) completely abolished the LDLR transcription increase induced by the short-term treatment with MG132 (0–6 h) (Figure 3F), strongly indicating the involvement of the PKC pathway.
Inhibition of PCSK9 expression promotes LDLR protein levels during long-term MG132 treatment
LDLR mRNA expression in cells treated with MG132 was comparable to that of the vehicle group after 12–24 h (Figure 1C), but the protein level remained higher. These findings suggested the involvement of a post-translation regulatory pathway. We further investigated whether PCSK9, a primary modulator of LDLR degradation, contributes to the increased LDLR protein level.
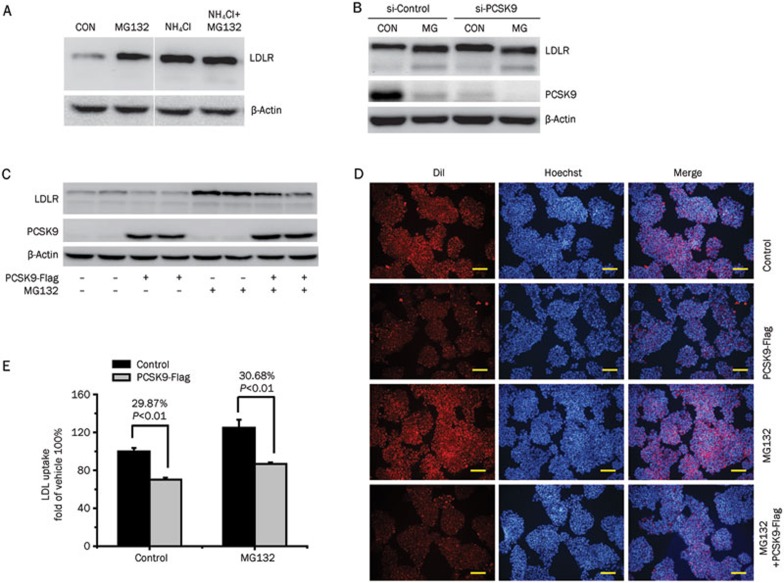
LDLR is synthesized in a premature form by ribosomes bound to the endoplasmic reticulum (ER) and then translocated into the ER lumen and subsequently processed to its mature form in the Golgi apparatus. Mature LDLR is guided to the cell surface, where it functions as an LDL-clearing mediator. After LDL binding and endocytosis, LDLR releases its ligand in endosomes, followed by recycling to the cell surface or degradation in the lysosome16. PCSK9 affects LDLR recycling by directly binding to LDLR and increasing its degradation in the lysosome in a pH-dependent manner3. NH4Cl, a lysosome pH disruptor, abolished MG132-induced LDLR protein augmentation at 24 h, suggesting that MG132 influences LDLR degradation in the lysosome (Figure 4A). Moreover, the disruption of endogenous PCSK9 expression by the targeted siRNA abrogated the elevation of the mature LDLR by MG132 at 24 h (Figure 4B). Our data support the involvement of endogenous PCSK9 in stabilizing the LDLR protein during long-term MG132 treatment.
Figure 4.
PCSK9 is involved in MG132-mediated LDLR protein upregulation. (A) Western blot analysis of LDLR expression in HepG2 cells treated with MG132 (0.3 μmol/L, 24 h) in the presence or absence of NH4Cl (a lysosome pH desruptor). (B) Western blot analysis of the LDLR expression in HepG2 cells transfected with siRNA-PCSK9 and then treated with MG132 (0.3 μmol/L, 24 h). (C) Western blot analysis of the LDLR protein level following the PCSK9-Flag addition (conditioned medium, 6 h). DiI-LDL uptake of the HepG2 cells with or without the exogenous addition of PCSK9 using the conditioned medium (6 h) of HEK293 cells transfected with PCSK9-Flag constructs. (D) Representative fluorescence microscopy images of cell-associated DiI-LDL (Red) and Hoechst-stained nuclei (Blue). Scale bar 50 μm. (E) Fluorescence of isopropanol-extracted DiI (520–570 nm, normalized to the cell protein). The data are the representative results of three independent experiments. Mean±SEM.
To further determine whether MG132 directly inhibits PCSK9 function, HepG2 cells were incubated with conditioned medium from 293T cells stably expressing PCSK9-Flag protein for 6 h, followed by a DiI-LDL uptake assay and Western blot analysis. The LDLR level was reduced upon the addition of conditioned medium with recombinant PCSK9-Flag (Figure 4C), in turn, leading to a ∼30 % decrease in LDL uptake (Figure 4D, 4E). The PCSK9 activity was not affected by MG132 pre-incubation, as is evident from decreased LDLR protein and LDL uptake. Based on these findings, we concluded that MG132 is not a direct inhibitor of PCSK9 function.
MG132 decreases PCSK9 mRNA and protein expression in HepG2 cells
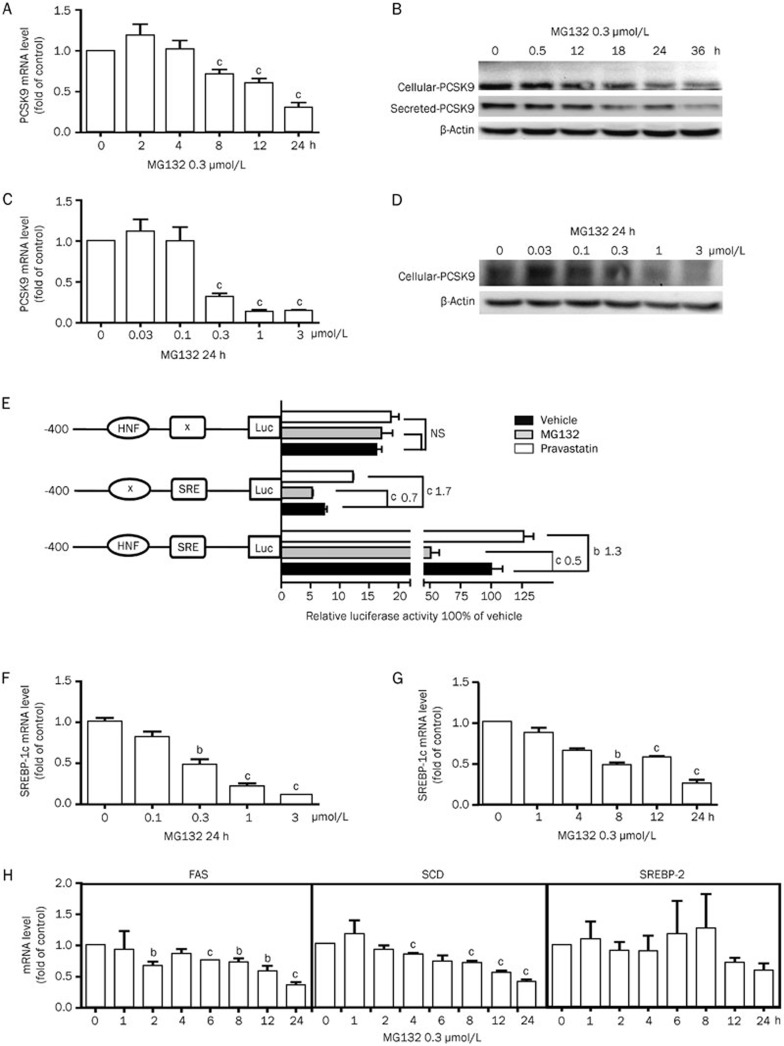
MG132 suppressed PCSK9 mRNA levels in a time- and dose-dependent manner (Figure 5A and 5C). The PCSK9 mRNA level was reduced by 70% compared with the control after 24 h treatment with 0.3 μmol/L MG132. The cellular and secreted PCSK9 protein levels were additionally diminished in a time- and dose-dependent manner in the presence of MG132 (Figures 5B, 5D).
Figure 5.
MG132 downregulates PCSK9 expression in HepG2 cells. (A, C) Real-time PCR and (B, D) Western blot analyses of the PCSK9 expression under MG132 treatment for the indicated time and dose. (E) Luciferase activities of the HepG2 cells transfected with PCSK9 promoter constructs containing wild-type, HNF mutant or SRE mutant sequences treated with MG132 (0.3 μmol/L, 24 h, normalized to β-galactosidase activity). Real-time PCR quantification of SREBP-1c (F, G), FAS, SCD, and SREBP-2 (H) mRNA levels. The data are representatives of three independent experiments. bP<0.05, cP<0.01.
The inhibition of the PCSK9 promoter activity was not altered by the HNF site mutation (Figure 5E). Therefore, the regulation of PCSK9 transcription by MG132 is not mediated by hepatic nuclear factor-1 alpha (HNF 1α). However, mutation of the SRE sequence on the PCSK9 promoter fully reversed the downregulation of PCSK9 transcription by MG132. As PCSK9 is a target gene of SREBP-2, we examined whether cellular cholesterol conditions affect the PCSK9 mRNA level modulated by MG132. However, neither the increase nor the depletion of cellular cholesterol affected the PCSK9 transcription reduction (data not shown), confirming the non-involvement of SREBP-2. We then detected the mRNA level of SREBP-1c, another member of the SREBPs that is reported to induce PCSK9 transcription, which is regulated primarily via transcription control rather than sterol-dependent cleavage17. MG132 suppressed the mRNA level of SREBP-1c in a dose- and time-dependent manner, similar to that of PCSK9 (Figure 5F, 5G). The mRNA levels of two other target genes of SERBP-1c, fatty acid synthase (FAS) and stearoyl CoA desaturase (SCD), were additionally decreased by MG132 (Figure 5H). Examination of the SREBP-2 mRNA expression revealed a decreasing tendency, but conventional levels of significance were not attained.
Furthermore, we employed a bundle of inhibitors separately targeting the NF-κB, p38, PI3K/Akt, ERK, JAK, JNK, and PKC pathways to identify the related signaling mechanisms (data not shown). Downregulation of the PCSK9 mRNA by MG132 appeared to be partially related to the PKC pathway (Supplementary Figure 3A, 3B) but independent of the other signaling pathways examined. The precise mechanisms underlying PCSK9 modulation by MG132 require further study.
We additionally explored whether MG132 interferes with PCSK9 synthesis and secretion using HEK293T cells over-expression of PCSK9. However, no differences were observed in the abundance of the cellular and secreted forms of PCSK9 or LDLR between the MG132-treated and untreated cells (Supplementary Figure 3C). Accordingly, we conclude that MG132 does not affect the general PCSK9 synthesis and secretion processes.
A combination of MG132 and pravastatin further enhances LDLR protein expression and function while suppressing PCSK9 upregulation by pravastatin alone in HepG2 cells
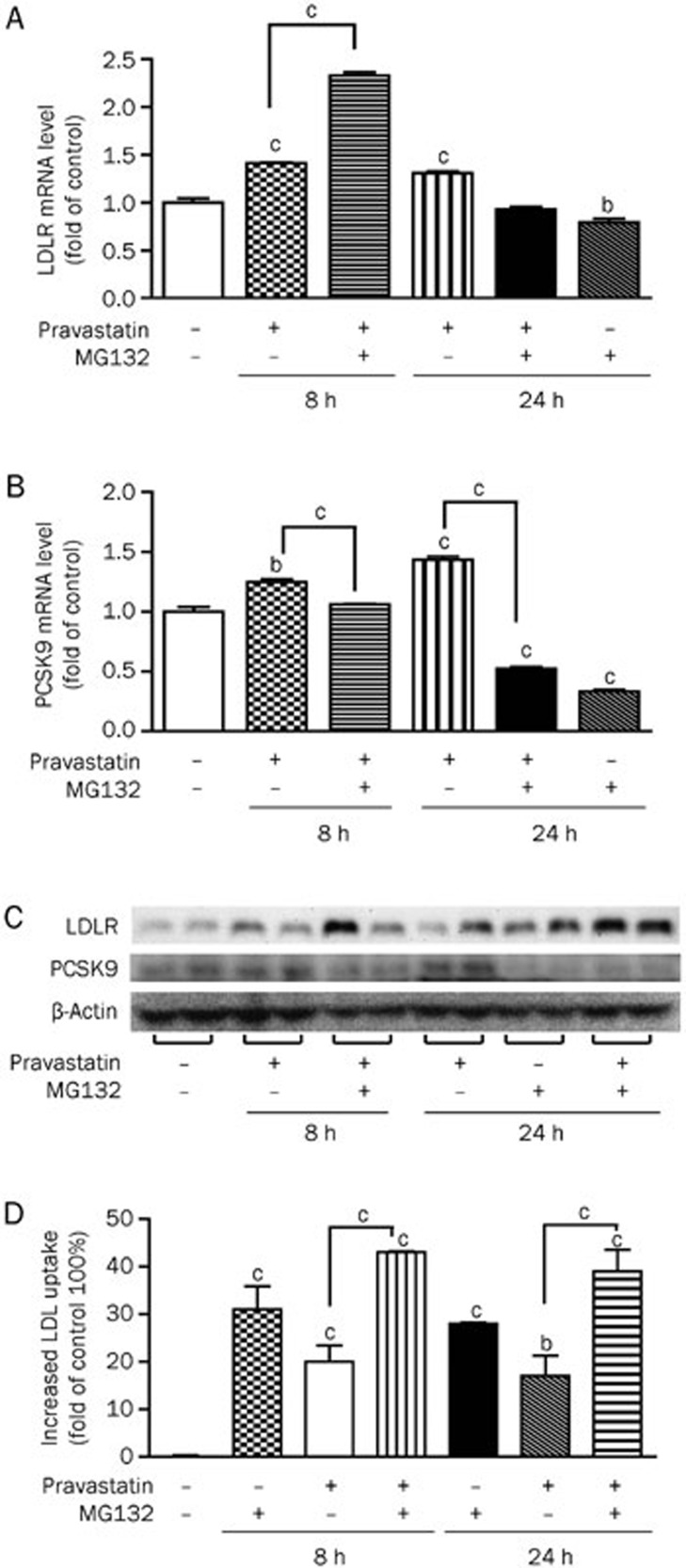
In view of the finding that MG132 upregulates LDLR but suppresses PCSK9 expression, we investigated whether a combination of MG132 and statins has a concomitant stimulatory effect on LDLR expression while suppressing the PCSK9 expression induced by statins alone. The HepG2 cells were treated with MG132 (0.3 μmol/L) and pravastatin (5 μmol/L) either alone or in combination. The combination treatment led to an increase in the LDLR mRNA level by 2.3-fold after 8 h, while pravastatin alone enhanced the transcript level by ∼1.4-fold. The combination additionally suppressed the increase in the PCSK9 mRNA induced by pravastatin by 63% at 24 h (P<0.01, Figure 6A, 6B). Western blot analysis revealed a robust increase in the LDLR expression in cells receiving both MG132 and pravastatin treatment for 8 or 24 h and a significant decrease in PCSK9 expression after 24 h (Figure 6C). The combined MG132 and pravastatin treatment additionally augmented the LDL uptake by 43% and 39%, while pravastatin alone increased LDL uptake by 20% and 17% after 8 and 24 h, respectively (Figure 6D).
Figure 6.
A combination of MG132 and pravastatin enhances LDLR while suppressing PCSK9 expression in HepG2 cells. Real-time PCR quantification (A, B) and Western blot analysis (C) of LDLR and PCSK9 expression levels and DiI-LDL uptake (D) in cells treated with MG132 (0.3 μmol/L), pravastatin (5 μmol/L), or both for the indicated times. The data are presented as the mean±SEM of three or more independent experiments. bP<0.05, cP<0.01 vs the vehicle-treated groups.
Discussion
Accumulating evidence supports the theory that elevated LDL-c plays a central role in the pathogenesis of coronary heart diseases. Therapies for lowering plasma cholesterol can effectively reduce the risk of cardiovascular events. However, there remains an urgent need for new approaches to reduce LDL-c in patients who cannot achieve their LDL-c goals or tolerate current drugs. In view of the finding that more than 70 % LDL-c clearance from the bloodstream is mediated by liver LDLR, the modulation of LDLR is considered to be an attractive strategy for reducing cholesterol2.
Here, we demonstrate that the proteasome-specific inhibitor MG132 increases LDLR transcription via a PKC-related pathway. MG132 additionally suppresses LDLR degradation by decreasing PCSK9 expression in a SREBP-1c-related manner in HepG2 cells. The two pathways act in concert to elevate the LDLR protein level and increase the LDL uptake by liver cells.
Chemicals reported to regulate the LDLR mRNA level act primarily through two pathways. Statins, inhibitors of the rate-limiting enzyme in cholesterol synthesis-HMG-CoA reductase, reduce cellular cholesterol levels. Low cellular cholesterol levels activate SREBP-2, which increases LDLR transcription and hepatic LDL uptake. On the other hand, berberine increases the LDLR mRNA level through enhancing the transcript stability mediated by the activation of the ERK pathway18.
Here, we observed that MG132 induces LDLR transcription following short-term incubation. Moreover, this effect does not appear to be mediated by activated SREBPs or enhanced ERK phosphorylation but is instead PKC-dependent. PKC is a protein kinase enzyme family that phosphorylates the serine and threonine residues of a number of enzymes. PKC has been shown to exert a protective effect in ischemia and increase cerebral blood flow, but its role in cholesterol homeostasis is unclear19,20. The specific PKC activator PMA (phorbol-12-myristate-13-acetate) stimulates LDLR transcription through PKC-dependent mechanisms21,22. Kamal et al showed that PKCε plays a role in sterol depletion-induced LDLR upregulation23. However, the precise mechanism remains unknown at present. These lines of evidence suggest that PKC exerts beneficial effects on LDL-c clearances through an LDLR modulation-based pathway. As proteasome degradation is a major pathway for PKC elimination, we speculate that proteasome inhibition can lead to the reduced degradation of specific PKCs and increased LDLR expression24.
The LDLR mRNA level returned to normal after 12 h of MG132 treatment, but the protein level remained enhanced throughout 24 h of incubation. The continued increase in the LDLR protein implies the involvement of post-translational modulation. In view of the earlier finding that the degradation of wild-type LDLR is not mediated by the proteasome pathway, diminished LDLR degradation by the proteasome does not appear to be the underlying cause for protein augmentation25,26. Thus, other pathways mediating the stabilization of LDLR protein possibly exist.
Inhibition lysosome degradation of LDLR is a key post-translational method to increase the LDLR level. PCSK9, which functions by promoting LDLR degradation in the lysosome, has emerged as an effective modulator of LDLR degradation. Our experiments revealed that long-term treatment with MG132 (8–24 h) down-regulates PCSK9 expression, which is associated with reduced LDLR degradation and increased LDL uptake in HepG2 cells. PCSK9 is primarily controlled by cellular cholesterol levels through SREBP-2, but other modulators exist. For instance, berberine reduces PCSK9 expression through decreasing HNF-1α, while bile acids and fibrates suppress PCSK9 transcription by activating FXR and PPARα, respectively27,28. In our experiments, the decrease in PCSK9 transcription induced by MG132 was independent of the cellular cholesterol levels, and no obvious change in the mature SREBP-2 was observed, thus excluding the involvement of SREBP-2 in this process. However, a specific SRE site mutation of the PCSK9 promoter blocked the effect of MG132, suggesting the involvement of other SREBP types. SREBP-1c has been reported to mediate insulin-induced PCSK9 upregulation, and its activity is primary controlled at the transcriptional level, despite cholesterol-sensitive cleavage29. Accordingly, we analyzed the mRNA levels of SREBP-1c as well as its target genes, FAS and SCD. Interestingly, the transcript levels were suppressed by MG132 in a similar pattern as PCSK9. Thus, the MG132 suppression of PCSK9 mRNA may be related to the SREBP-1c decrease. The observed reduced transcription of FAS and SCD supports the potential utility of MG132 in limiting fatty acid synthesis. SREBP-1c transcription has been reported to be modulated by a series of modulators, such as insulin, LXR and AMPK, but the modulators related to MG132 mediated SREBP-1c modulation need to be explored30,31,32,33.
PCSK9 inhibition represents a promising means of reducing LDL-c. However, statins increase the PCSK9 levels along with LDLR enhancement, which compromises the effects of LDLR augmentation. Thus, the development of effective dual modulators of LDLR and PCSK9 may be a more effective way to control blood cholesterol. A number of dual modulators have been reported to date, including berberine and Oncostatin M. They increase LDLR expression by stabilizing its mRNA through an ERK-dependent pathway18,34, but berberine decreases PCSK9 expression by suppressing HNF-1α, while Oncostatin M exerts this effect through a JAK-dependent pathway12,15. Berberine is poorly absorbed in vivo18, and Oncostatin M is a pleiotropic cytokine synthesized by stimulated T-cells and monocytes, which may not be suitable for clinical use. Therefore, the discovery of novel LDLR-PCSK9 dual modulators is essential.
Because MG132 effectively reduces PCSK9 expression, we hypothesized that a combination of MG132 and statins may block the increase in PCSK9 caused by statins alone. In the current study, this assumption was confirmed by treating HepG2 cells with a combination of MG132 and pravastatin. The LDLR expression was further stimulated, while the PCSK9 expression induced by pravastatin alone was suppressed. Moreover, the combination treatment greatly increased the DiI-LDL uptake by the HepG2 cells. These results collectively support the potential of MG132 as an effective supplement to statins for better efficacy in LDL-c suppression.
Proteasome inhibitors have attracted considerable attention as anti-tumor agents. Alternative applications for proteasome inhibitors have additionally been reported, such as heart ischemia/reperfusion, neuron degeneration, cardiomyocyte hypertrophy, and restenosis prevention35,36,37. In our research, proteasome inhibitors, such as lactacystin and PS-341 (an FDA-approved drug for multiple myeloma treatment), had similar effects on LDLR and PCSK9 modulation (Supplementary Figure 4), suggesting that proteasome inhibition plays a role in cholesterol homeostasis. In addition to the reported LDLR increase, proteasome inhibition exerts other beneficial effects on atherosclerosis. Wilck et al reported that low-dose proteasome inhibition has anti-oxidative and anti-inflammatory effects and attenuates the development of atherosclerotic lesions in low-density lipoprotein receptor-deficient mice38. In a recent article, chronic proteasome inhibition by MLN-273 (an inhibitor similar to PS-341) led to an improvement in blood lipid levels (decreased LDL-c, lipids, and increased HDL-c), although the differences recorded were not significant. However, the chronic application of MLN-273 caused endothelium oxidative stress, which impaired endothelium function and contributed to coronary atherosclerosis in pigs39. Notably, the dosage is critical for proteasome inhibitors to confer protective effects, and the partial inhibition of proteasome activity while preserving basal function is preferred. Thus, further research is warranted to establish the role of proteasome inhibition in cholesterol homeostasis in vivo.
In conclusion, we report that MG132, a specific proteasome inhibitor, exerts a promising dual-modulating effect on LDLR and PCSK9 expression, ultimately leading to an increased LDL uptake in HepG2 cells. Our findings support the potential relevance of proteasome inhibition in cholesterol homeostasis.
Author contribution
Hong YAN, Yan-ling MA, and Yi-ping WANG designed the research; Hong YAN and Yan-ling MA performed the research; Hong YAN, Shu-mei WANG, and Xin-bo WANG, and Fei GAO contributed new analytical tools and reagents; Hong YAN, Yan-ling MA, and Yi-ping WANG performed data analysis; Hong YAN, Yan-ling MA, Yu-zhou GUI, and Yi-ping WANG wrote or contributed to the writing of the manuscript.
Acknowledgments
We are grateful to Dr Jay D HORTON (Texas, Canada) and Dr Sahng Wook PARK (Seoul, Korea) for generously providing the CMV-PCSK9-Flag and PCSK9 promoter (D1, D4, and D4-mut) construct plasmids, respectively.
Footnotes
Supplementary figures are available at APS's website.
Supplementary Information
Supplementary Figures 1–4
References
- Kong WJ, Liu J, Jiang JD. Human low-density lipoprotein receptor gene and its regulation. J Mol Med. 2006;84:29–36. doi: 10.1007/s00109-005-0717-6. [DOI] [PubMed] [Google Scholar]
- Micheal SB, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Costet P, Krempf M, Cariou B. PCSK9 and LDL cholesterol: unravelling the target to design the bullet. Trends Biochem Sci. 2008;33:426–34. doi: 10.1016/j.tibs.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Tarugi P, Averna M, Di Leo E, Cefalu AB, Noto D, Magnolo L, et al. Molecular diagnosis of hypobetalipoproteinemia: an ENID review. Atherosclerosis. 2007;195:e19–27. doi: 10.1016/j.atherosclerosis.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Stein EA, Swergold GD. Potential of proprotein convertase subtilisin/kexin type 9 based therapeutics. Curr Atherosclerosis Reports. 2013;15:310. doi: 10.1007/s11883-013-0310-3. [DOI] [PubMed] [Google Scholar]
- Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, et al. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24:1454–9. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- Giugliano RP, Desai NR, Kohli P, Rogers WJ, Somaratne R, Huang F, et al. Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 in combination with a statin in patients with hypercholesterolaemia (LAPLACE-TIMI 57): a randomised, placebo-controlled, dose-ranging, phase 2 study. Lancet. 2012;380:2007–17. doi: 10.1016/S0140-6736(12)61770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kem DC. Proteasome inhibition during myocardial infarction. Cardiovasc Res. 2010;85:312–20. doi: 10.1093/cvr/cvp309. [DOI] [PubMed] [Google Scholar]
- Dreger H, Westphal K, Weller A, Baumann G, Stangl V, Meiners S, et al. Nrf2-dependent upregulation of antioxidative enzymes: a novel pathway for proteasome inhibitor-mediated cardioprotection. Cardiovasc Res. 2009;83:354–61. doi: 10.1093/cvr/cvp107. [DOI] [PubMed] [Google Scholar]
- Ogura M, Ayaori M, Terao Y, Hisada T, Iizuka M, Takiguchi S, et al. Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and in vivo. Arteriosclerosis Thromb Vasc Biol. 2011;31:1980–7. doi: 10.1161/ATVBAHA.111.228478. [DOI] [PubMed] [Google Scholar]
- Stephan ZF, Yurachek EC. Rapid fluorometric assay of LDL receptor activity by DiI-labeled LDL. J Lipid Res. 1993;34:325–30. [PubMed] [Google Scholar]
- Li H, Dong B, Park SW, Lee HS, Chen W, Liu J. Hepatocyte nuclear factor 1alpha plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J Biol Chem. 2009;284:28885–95. doi: 10.1074/jbc.M109.052407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Kanade S, Han B, Eckert RL. A proteasome inhibitor-stimulated Nrf1 protein-dependent compensatory increase in proteasome subunit gene expression reduces polycomb group protein level. J Biol Chem. 2012;287:36179–89. doi: 10.1074/jbc.M112.359281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Yoshida M, Shimizu M, Sato R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J Biol Chem. 2001;276:36431–7. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- Cao A, Wu M, Li H, Liu J. Janus kinase activation by cytokine oncostatin M decreases PCSK9 expression in liver cells. J Lipid Res. 2011;52:518–30. doi: 10.1194/jlr.M010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MM, Strickland DK, Bakillah A. The mammalian low-density lipoprotein receptor family. Ann Rev Nutr. 1999;19:141–72. doi: 10.1146/annurev.nutr.19.1.141. [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–48. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10:1344–51. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Churchill E, Mochly-Rosen D. Epsilon protein kinase C as a potential therapeutic target for the ischemic heart. Cardiovasc Res. 2006;70:222–30. doi: 10.1016/j.cardiores.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bright R, Sun GH, Yenari MA, Steinberg GK, Mochly-Rosen D. epsilonPKC confers acute tolerance to cerebral ischemic reperfusion injury. Neurosci Lett. 2008;441:120–4. doi: 10.1016/j.neulet.2008.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson1 GM, Vasa MZ, Deeley RG. Stabilization and cytoskeletal-association of LDL receptor mRNA are mediated by distinct domains in its 3′ untranslated region. J Lipid Res. 1998;39:1025–32. [PubMed] [Google Scholar]
- Huang W, Mishra V, Batra S, Dillon I, Mehta KD. Phorbol ester promotes histone H3-Ser10 phosphorylation at the LDL receptor promoter in a protein kinase C-dependent manner. J Lipid Res. 2004;45:1519–27. doi: 10.1194/jlr.M400088-JLR200. [DOI] [PubMed] [Google Scholar]
- Mehta KD, Radominska-Pandya A, Kapoor GS, Dave B, Atkins BA. Critical role of diacylglycerol- and phospholipid-regulated protein kinase C in induction of low-density lipoprotein receptor transcription in response to depletion of cholesterol. Mol Cell Biol. 2002;22:3783–93. doi: 10.1128/MCB.22.11.3783-3793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin B, Maccario H, Thirion S, Junoy B, Boyer B, Enjalbert A, et al. Ubiquitination as a priming process of PKC alpha and PKC epsilon degradation in the alphaT3-1 gonadotrope cell line. Neuroendocrinology. 2009;89:252–66. doi: 10.1159/000164694. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu W, Schwartz AL, Bu G. Degradation of the LDL receptor class 2 mutants is mediated by a proteasome-dependent pathway. J Lipid Res. 2004;45:1084–91. doi: 10.1194/jlr.M300482-JLR200. [DOI] [PubMed] [Google Scholar]
- Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–4. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourimate S, Le May C, Langhi C, Jarnoux AL, Ouguerram K, Zair Y, et al. Dual mechanisms for the fibrate-mediated repression of proprotein convertase subtilisin/kexin type 9. J Biol Chem. 2008;283:9666–73. doi: 10.1074/jbc.M705831200. [DOI] [PubMed] [Google Scholar]
- Langhi C, Le May C, Kourimate S, Caron S, Staels B, Krempf M, et al. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 2008;582:949–55. doi: 10.1016/j.febslet.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Costet P, Cariou B, Lambert G, Lalanne F, Lardeux B, Jarnoux AL, et al. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J Biol Chem. 2006;281:6211–8. doi: 10.1074/jbc.M508582200. [DOI] [PubMed] [Google Scholar]
- Kohjima M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:5. [PubMed] [Google Scholar]
- Zhou R, Wang L, Xu X, Chen J, Hu LH, Chen LL, et al. Danthron activates AMP-activated protein kinase and regulates lipid and glucose metabolism in vitro. Acta Pharmacol Sin. 2013;34:1061–9. doi: 10.1038/aps.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Develop. 2000;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohjima M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:507–11. [PubMed] [Google Scholar]
- Li C. Induction of low density lipoprotein receptor (LDLR) transcription by oncostatin M is mediated by the extracellular signal-regulated kinase signaling pathway and the repeat 3 element of the LDLR promoter. J Biol Chem. 1999;274:6747–53. doi: 10.1074/jbc.274.10.6747. [DOI] [PubMed] [Google Scholar]
- Adams J. Proteasome inhibitors as therapeutic agents. Expert Opin Ther Patents. 2003;13:45–57. [Google Scholar]
- Meiners S. Ubiquitin-proteasome pathway as a new target for the prevention of restenosis. Circulation. 2002;105:483–9. doi: 10.1161/hc0402.102951. [DOI] [PubMed] [Google Scholar]
- Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, et al. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–8. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- Wilck N, Fechner M, Dreger H, Hewing B, Arias A, Meiners S, et al. Attenuation of early atherogenesis in low-density lipoprotein receptor-deficient mice by proteasome inhibition. Arteriosclerosis Thromb Vasc Biol. 2012;32:1418–26. doi: 10.1161/ATVBAHA.112.249342. [DOI] [PubMed] [Google Scholar]
- Herrmann J, Saguner AM, Versari D, Peterson TE, Chade A, Olson M, et al. Chronic proteasome inhibition contributes to coronary atherosclerosis. Circ Res. 2007;101:865–74. doi: 10.1161/CIRCRESAHA.107.152959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–4