Abstract
Aim:
To discover novel hepatitis C virus (HCV) inhibitors and elucidate the mechanism of action of the active compounds.
Methods:
HCV subgenomic replicon-based luciferase reporter cell line was used to screen 1200 synthetic compounds with novel structures. Huh7.5.1 cell line stably transfected with HCV NS3/4A protease reporter was established to investigate the anti-HCV mechanism of the active compounds. The active compounds were further examined in an in vitro HCV infection assay to confirm their anti-HCV activity.
Results:
After two-round screening in the anti-HCV replicon assay, some 2,4-diaminoquinazoline derivatives and carboxamide analogues were found to possess anti-HCV replicon activities (the IC50 values were less than 5 μmol/L). Among them, two representative compounds HZ-1157 and LZ-110618-6 inhibited HCV NS3/4A protease with IC50 values of 1.0 and 0.68 μmol/L, respectively. Furthermore, HZ-1157 and LZ-110618-6 inhibited HCV infection in vitro with IC50 values of 0.82 and 0.11 μmol/L, respectively.
Conclusion:
Some 2,4-diaminoquinazoline derivatives and carboxamide analogues have been identified as novel anti-HCV compounds.
Keywords: hepatitis C virus; NS3/4A protease; secreted embryonic alkaline phosphatase (Seap); 2,4-diaminoquinazoline; carboxamide; telaprevir
Introduction
Hepatitis C virus (HCV), a major cause of chronic liver disease, has infected approximately 130–210 million people worldwide. In most infected individuals, HCV establishes itself as a chronic infection that may lead to liver fibrosis, liver cirrhosis and hepatocellular carcinoma1,2. The genome of HCV is a single-stranded positive-sense RNA of approximately 9.6 kb, which has an open reading frame (ORF) flanked by untranslated regions (UTRs) at both the 5′ and 3′ ends. The ORF encodes a polyprotein precursor. Viral and host proteases will cleave the polyprotein precursor into the core structural proteins, E1 and E2, as well as the non-structural (NS) proteins p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B3.
Current standard treatment with polyethylene glycol decorated interferon-α (PEG-IFNα) and ribavirin (PR) is only effective for 40%–50% patients infected with genotype 1, the most prevalent HCV subtype in the United States, Europe, and Japan4,5. Moreover, the known side effects of PR, such as anemia, flu-like symptoms, fatigue and depression, make the 48-week treatment difficult for patients to tolerate6,7. Thus, there is an unmet medical need to develop therapeutic measures with high potency and improved tolerance. In 2011, two first-generation NS3/4A protease inhibitors (PIs), telaprevir (Incivek, NDC Code: 51167-100-01) and boceprevir (Victrelis, NDC Code: 0085-0314-02), were approved. Combined with PR, they improved sustained virological response rates among genotype 1 patients8,9,10. At the same time, a better understanding of the HCV life cycle has led to the development of direct antiviral agents (DAA). Aside from NS3/4A PIs, DAAs also comprise NS5B polymerase inhibitors, such as the newly approved drug sofosbuvir (Sovaldi, NDC Code: 61958-1501-1)11, and NS5A inhibitors, which are currently in development.
However, as is the case for HIV, there is a continuous need to improve and optimize current therapy and develop novel agents and drug classes. During DAA monotherapy, viral breakthrough with genotypic resistance occurs rapidly9,12. With triple therapy using combined PI and PR for genotype 1 infection, IFN remains an important and necessary component to avoid the emergence of resistant viral variants13. The triple therapy means that the adverse events brought by IFN remain and may be even increased by PI8,10,12,14,15. A combination therapy containing DAAs with different targets and substitutes with the same target but different resistance profiles may help solve the problem.
For the purpose of developing and studying novel anti-HCV drugs, we screened 1200 compounds with novel structures. A second-round assay discovered two compound series, 2,4-diaminoquinazoline derivatives and carboxamide analogues16,17, as potential active anti-HCV compounds. To investigate the action mechanism, we then established an efficient and sensitive cell-based HCV NS3/4A protease assay and tested those active compounds. Results revealed that several active compounds inhibit HCV replication by targeting the HCV NS3/4A protease. Further experiments using an infectious HCV system (J399EM) confirmed their anti-HCV activity.
Materials and methods
Cell culture and virus
The Huh 7.5.1 cell line and the HCV replicon cell line (J399LM) were kindly provided by Prof Xin-wen CHEN, Wuhan Institute of Virology, Chinese Academy of Sciences. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum at 37 °C with 5% CO2.
J399EM is an infectious HCV virus derived from the JFH1 virus (genotype 2a). The establishment of the infectious J399EM HCV virus has been described previously18. Virus was propagated in Huh7.5.1 cells. Briefly, Huh7.5.1 cells were infected with J399EM at MOI=0.1 and cultured for 4 d. Culture supernatant was collected and filtered through a 0.45 μm membrane and stored at -80 °C as virus stock. Virus titer was tested in Huh7.5.1 cells. Stocks were serially diluted, and the plaque-forming units (PFU) were determined as the number of fluorescent colonies formed in infected Huh7.5.1.
Luciferase activity assays of the HCV subgenomic replicon
The HCV replicon cell line was obtained from Huh7 cells stably transfected with HCV subgenomic viral RNA, J399LM, which was made by replacing the EGFP gene in the J399EM genome with the Renilla luciferase gene. Cells were seeded in a 96-well plate. Compounds were added to the culture medium at various concentrations. After 72 h of culture, the expression levels of the HCV replicon were measured using the luciferase assay system (Promega, Madison, Wisconsin, USA) following the manufacturer's instructions. The half maximal effective concentration (IC50) was calculated as the compound concentration that inhibited HCV RNA replication activity by 50%19.
Plasmid construction
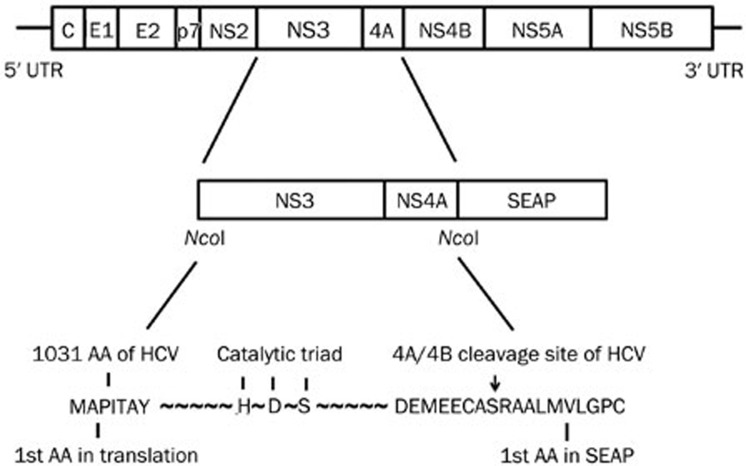
For the pSelect-NS3/4A-Seap plasmid, the NS3/4A coding sequence, including the cleavage site, was amplified from the full length JFH1 strain plasmid pJFH1T (genotype 2a) by PCR with the forward primer 5′-ccatggTTGCTCCCATCACTGCTT-3′ and the reverse primer 5′-ccatggGCATTCCTCCATCTCATCAA-3′ and cloned into the NcoI site of the pSELECT-zeo-Seap vector (InvivoGen, San Diego, California, USA). The schematic diagram of this plasmid is shown in Figure 1. For the pSelect-NS3/4A(mut)–Seap plasmid, the serine of the HDS catalytic triad was changed to alanine (S139A).
Figure 1.
The schematic diagram of HCV NS3/4A-Seap expression plasmids. The Seap gene with an N-terminal signal was fused in-frame to the HCV NS3/4A coding sequence. NS3/4A and Seap amino acids in the fusion regions are shown by capitalized characters. HDS is the catalytic triad of the HCV NS3 protease. The S to A mutation in the catalytic triad results in a disabled NS3 protease. The NS4A/4B cleavage site is indicated by an arrow.
Secreted embryonic alkaline phosphatase (Seap) assay and cell viability assay
The Seap activity in the culture supernatants was measured by the chemiluminescent method (Roche, Shanghai, China) following the manufacturer's instructions. Briefly, 12.5 μL supernatant samples were diluted with 37.5 μL dilution buffer. Then, they were incubated with 50 μL inactivation buffer at room temperature for 5 min. After adding 50 μL freshly prepared substrate reagent, the mixtures were incubated at room temperature for 10–20 min. Finally, the luminescence was measured with a luminometer.
Cell viability was measured by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich] method. Briefly, MTT (1 mg/mL) was dissolved in PBS. Cell culture supernatants were removed and replaced with MTT and further cultured for 4 h; cells were then lysed with 10% sodium dodecyl sulfate (SDS), 50% N,N-dimethylformamide, pH 7.2. OD570 values were read. The percentage of cell death was calculated against the control well without compounds20.
Western blot analysis
Cells were lysed in 200 μL RIPA buffer (Beyotime, Haimen, Jiangsu, China) containing a cocktail protease inhibitor (Roche). After centrifugation, equal amounts of total extract protein (15 μg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 5% BSA for 1 h at room temperature, the membrane was incubated with mouse anti-NS3 monoclonal antibody (Abcam, Shanghai, China) at 4 °C overnight. On the second day, the membrane was washed in TBST and incubated with peroxidase-conjugated goat anti-mouse secondary antibody (Beyotime). After a final treatment with enhanced chemiluminescence assay (ECL) reagents (GE Healthcare, Pittsburgh, Pennsylvania, USA), samples were exposed to X-ray film21.
Antiviral assay using infectious HCV virus
Huh 7.5.1 cells were seeded at a density of 1×104 cells per well of a 96-well plate with black walls and a clear bottom (Corning, Costar 3904). Twenty-four hours later, the medium was inoculated with J399EM virus at an MOI of 0.3. Four hours later, the virus inoculum was replaced by medium containing different concentrations of compounds in triplicate, cells were further incubated for an additional 72 h, and the EGFP fluorescence was measured directly in the plate by using a luminometer with excitation at 488 nm and emission at 516 nm19. The percentage of the inhibition effect of compounds was calculated against the control well. After the EGFP signal was read, cell viability was measured in the same plate by the MTT method as described above.
Results
Screening and discovery of synthetic compounds in anti-HCV replicon activities
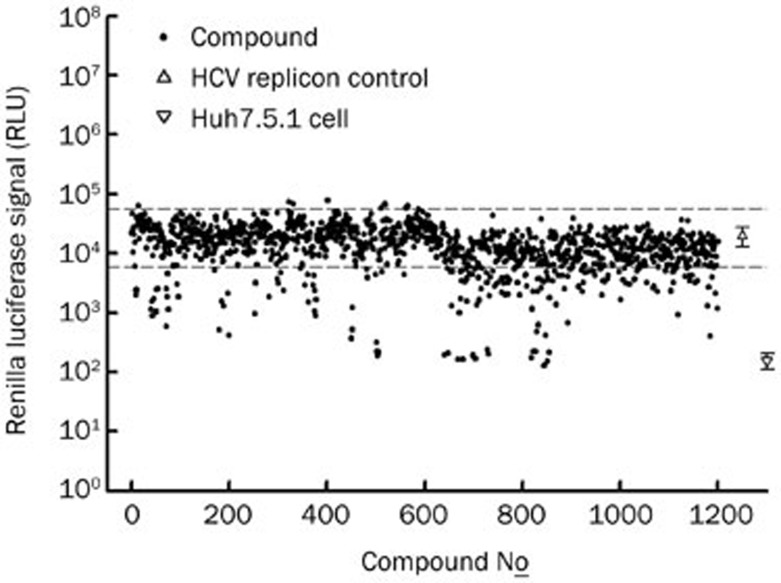
In total, 1200 synthetic compounds were screened in a HCV replicon reporter system, which was a Huh7 harboring a HCV subgenome with a Renilla luciferase reporter. The results of the preliminary screening are shown in Figure 2, with the luciferase signal of each compound plotted. Of the 1200 compounds screened, those with luciferase signals lower than the threshold (Mean-3×SD of the control well, lower dashed line, Figure 2) were considered to be anti-HCV active compounds and selected for the second-round test. In the second-round test, candidate compounds were serially diluted and tested by the same HCV replicon assay. IC50 values of the selected compounds are shown in Table 1. We discovered two categories of compounds, 2,4-diaminoquinazoline derivatives and carboxamide analogues, that exhibited anti-HCV activity at reasonable concentrations (IC50<5 μmol/L). These compounds were selected for further investigation of their anti-HCV mechanisms.
Figure 2.
The screening results of the anti-HCV replicon assay. 1200 synthetic compounds with novel structure were screened by using the HCV subgenomic replicon-based reporter cell line (J399LM, genotype 2a). Compounds were co-cultured with cells for 48 h. The luciferase signal was read and compared to control wells without compounds. Compounds with reporter signal lower than the mean-3xSD (lower dashed line) of control wells (without compound) were considered to be active compounds and selected for further tests. Normal Huh7.5.1 cells served as the blank.
Table 1. Anti-HCV replicon activities of compounds.
| Compound | HCV Replicon assay |
|||
|---|---|---|---|---|
| CC50 (μmol/L) | IC50 (μmol/L) | SI | ||
| 2,4-Diaminoquinazoline derivatives | HZ-1031 | >10 | 0.037±0.013 | >136 |
| HZ-1157 | >10 | 0.73±0.084 | >13 | |
| HZ-1327 | 6.3±0.53 | 4.7±0.54 | 1.3 | |
| HZ-1600 | 5.3±0.48 | 2.9±0.22 | 1.8 | |
| Carboxamide analogues | LZ-110618-1 | >10 | 0.018±0.0071 | >555 |
| LZ-110618-6 | >10 | 0.056±0.0093 | >187 | |
| LZ-110618-9 | >10 | 0.024±0.011 | >416 | |
| LZ-110618-24 | >10 | 0.032±0.012 | >312 | |
| VX-950 | >10 | 0.67±0.057 | >15 | |
Establishment of a transiently transfected HCV NS3/4A Seap reporter in Huh7.5.1 cells
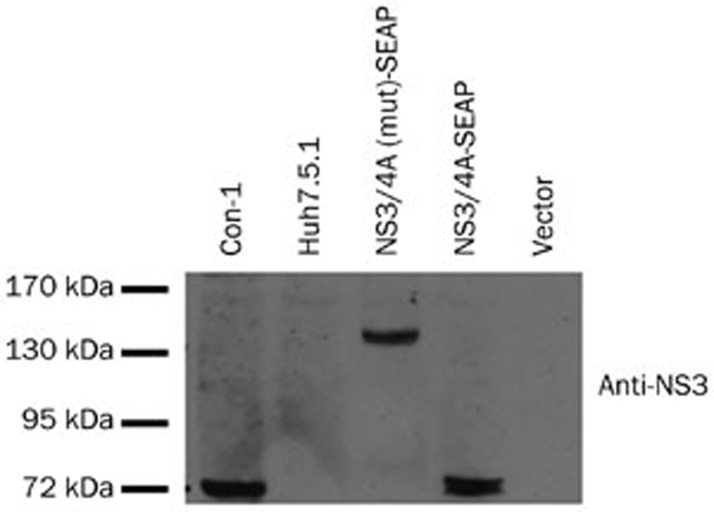
To identify whether these anti-HCV active compounds act against the HCV NS3/4A protease, a HCV NS3/4A protease reporter plasmid was constructed. The construct expresses a HCV NS3/4A-Seap fusion protein linked by a natural NS3/4A cleavage site (Figure 1). Successful self-cleavage by the NS3/4A protein releases the Seap protein, which is then secreted into the culture supernatant due to the exposed N-terminal signal peptide. In a mutated plasmid, pSelect-NS3/4A(mut)Seap, the serine in the catalytic triad was changed to alanine, making the mutated NS3 non-functional. The pSelect-NS3/4A-Seap plasmid and its mutated version were transiently transfected into Huh7.5.1 cells to validate their function by Western blot. As expected, a band of 70 kDa, corresponding to the NS3/4A fusion protein, was detected in cells transfected with the pSelect-NS3/4A-Seap plasmid (Figure 3, lane 4). Also as expected, a band of 130 kDa was detected in cells transfected with the pSelect-NS3/4A(mut)-Seap plasmid, corresponding to the NS3/4A-Seap fusion peptide with the Seap protein still attached (Figure 3, lane 3). This result indicated that the cleavage of NS3/4A-Seap fusion protein and the subsequent Seap secretion were fully dependent on HCV NS3 protease activity. Telaprevir (VX-950), a known inhibitor of the HCV NS3/4A serine protease22, was able to decrease the Seap level in the supernatant by 74% at 10 μmol/L (Figure 4A). The free form of NS3 also decreased by western blot analysis (Figure 4B).
Figure 3.
HCV NS3/4A expression of the pSelect-NS3/4A-Seap construct. Cells were lysed 72 h after transient transfection for western blotting. For the western blot, 15 μg total protein samples were loaded, and the NS3/4A fusion protein was detected with an anti-NS3 antibody. Con-1, infectious HCV virus (J399EM) infected Huh7.5.1 cell lysate; Huh7.5.1, normal Huh7.5.1 cell lysate; Vector, pSELECT-zeo-Seap plasmid (Invivogen). The representative result is from two independent experiments with the same results.
Figure 4.
The inhibitory activity of telaprevir on HCV NS3/4A protease by transient transfection. (A) The Seap activity in the supernatant of cells transfected with pSelect-NS3/4A-Seap. Huh7.5.1 cells were transiently transfected with the plasmid and co-cultured with telaprevir for 72 h. Culture supernatant was collected and subjected to the Seap assay. Representative data are from three independent experiments with the same results. (B) Western blot analysis for telaprevir inhibiting NS3 cleavage. pSelect-NS3/4A-Seap-transfected cells were lysed after 72 h co-cultured with telaprevir (as above). The NS3/4A fusion peptide was detected by a specific anti-NS3 antibody; GAPDH is shown here as the control. The representative result is from two independent experiments with the same results.
Establishment of the stably transfected HCV NS3/4A reporter cell line in Huh7.5.1 cells
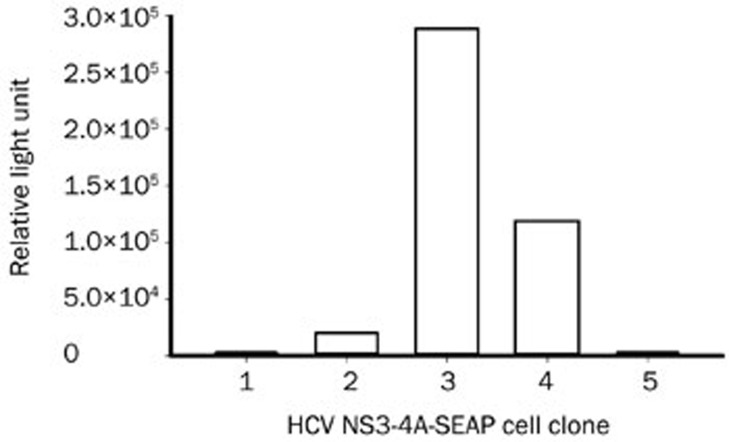
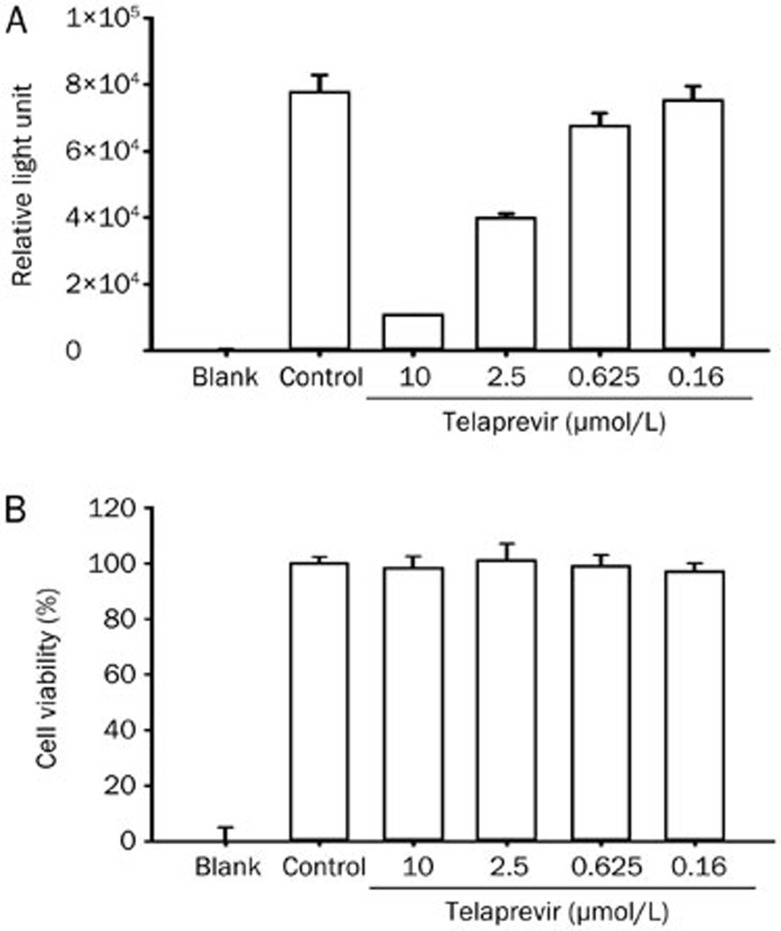
To establish a stably transfected HCV NS3/4A-Seap reporter cell line, Huh7.5.1 cells were transfected with pSelect-NS3/4A-Seap and selected by 500 mg/L zeocin for four weeks. We obtained five colonies of cells. The Seap levels in supernatants were tested, and clone 3, which had the highest Seap activity, was chosen and propagated (Figure 5). The inhibitory efficacy of telaprevir was tested in clone 3. The results showed that the IC50 of telaprevir was 0.931 μmol/L (Figure 6A). The CC50 was higher than 10 μmol/L (Figure 6B).
Figure 5.
The reporter activities of HCV NS3/4A-Seap stably transfected clones. Huh7.5.1 cells were transfected with the pSelect-NS3/4-Seap plasmid using Lipofectamine 2000 transfection reagent and selected with 500 mg/L zeocin for 4 weeks. Cell colonies were picked and propagated to the proper confluency, and the Seap activities in culture supernatants were assayed.
Figure 6.
The activity of telaprevir in the HCV NS3/4A-Seap stably transfected cell line. Seventy-two hours after telaprevir administration, supernatant was collected, and the Seap activity was tested. At the same time, cell viability was measured by the MTT method. The representative result is from two independent experiments with the same results.
To further validate the HCV NS3/4A-Seap reporter cell line, several reference compounds were used (Table 2). The anti-HCV activity of these compounds was also determined in parallel using an HCV infection assay and a replicon assay. As expected, BMS-790052 (daclatasvir), a NS5A inhibitor13,23, showed no inhibitory activity on the NS3/4A protease (Table 2). Ribavirin showed neither anti-HCV activity nor a protease inhibitory effect when it was used without IFN-α. The inhibition of HCV replication by mycophenolic acid (MPA) was primarily due to its depletion of guanosine, which is required for viral RNA synthesis24; therefore, it was inactive in the HCV NS3/4A protease assay. The reference compound, VX-950, was the only compound that was active in all three systems as a HCV NS3/4A protease inhibitor. These results validated the stably transfected HCV NS3/4A-Seap reporter cell line as a highly specific HCV NS3/4A protease inhibitor screening and target validating system.
Table 2. Test of reference compounds in anti-HCV assays.
| Compound | CC50 (nmol/L) | IC50 (nmol/L) |
||
|---|---|---|---|---|
| HCV infection assay | HCV replicon assay | NS3/4A-Seap | ||
| VX-950 | >10000 | 528±103 | 613±96 | 931±153 |
| BMS-790052 | >1000 | 0.054±0.010 | 0.078±0.015 | 1000 |
| Mycophenolic acid | >10000 | 520±29 | 468±103 | >10000 |
| Ribavirin | >100000 | >100000 | >100000 | >100000 |
Identification of anti-HCV active compounds that target the HCV NS3/4A protease
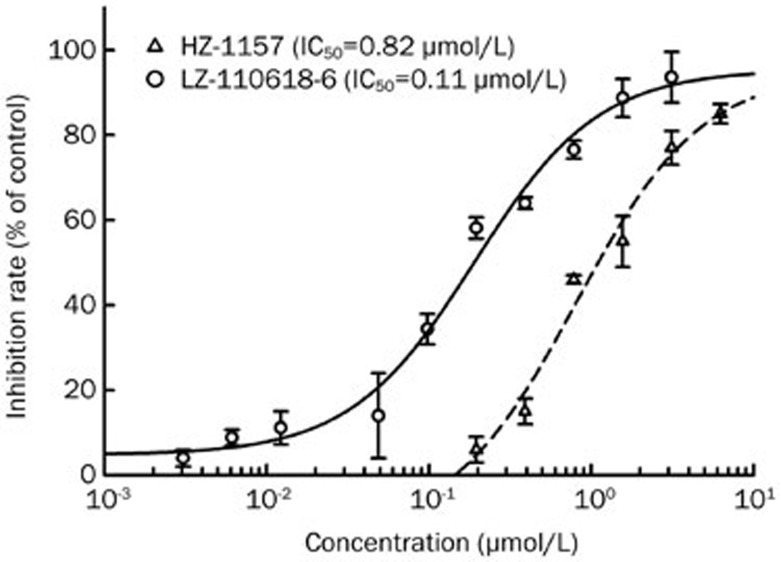
Afterwards, those anti-HCV active compounds selected from the HCV replicon assay were tested in the HCV NS3/4A-Seap reporter cell line. The results are shown in Table 3 and Figure 7. Of these compounds, HZ-1157 and LZ-110618-6 showed inhibitory effects on the HCV NS3/4A protease with IC50 values of 1.0 μmol/L and 0.68 μmol/L, respectively. Comparing their IC50 values in the HCV NS3/4A protease assay and HCV replicon assay, the IC50 value of HZ-1157 (1.0 μmol/L) in the HCV NS3/4A protease assay was close to its IC50 value in the HCV replicon assay (0.73 μmol/L). This result indicated that HZ-1157 inhibited HCV replication by targeting the viral NS3/4A protease. However, the IC50 value of LZ-110618-6 in the HCV NS3/4A protease assay (0.68 μmol/L) was much higher than its IC50 value in the HCV replicon assay (0.06 μmol/L). This suggests that LZ-110618-6 most likely has multiple targets in the HCV life cycle, and its anti-HCV activity could be partially due to the inhibition of the HCV NS3/4A protease.
Table 3. Anti-HCV-NS3/4A protease activities of compounds.
| Compound | NS3/4A-Seap assay |
|||
|---|---|---|---|---|
| CC50 (μmol/L) | IC50 (μmol/L) | SI | ||
| 2,4-Diaminoquinazoline derivatives | HZ-1031 | >10 | 0.037±0.013 | >136 |
| HZ-1031 | >10 | >10 | ||
| HZ-1157 | 14±0.15 | 1.0±0.92 | 14 | |
| HZ-1327 | 5.5±0.26 | 4.2±0.16 | 1.3 | |
| HZ-1600 | >10 | >10 | ||
| Carboxamide analogues | LZ-110618-1 | 9.3±0.87 | 9.1±0.51 | 1.0 |
| LZ-110618-6 | 15±0.93 | 0.68±0.071 | 22 | |
| LZ-110618-9 | 10±1.2 | 6.1±0.71 | 1.6 | |
| LZ-121012-24 | 8.6±0.62 | 5.5±0.52 | 1.5 | |
| VX-950 | >10 | 1.0±0.33 | >10 | |
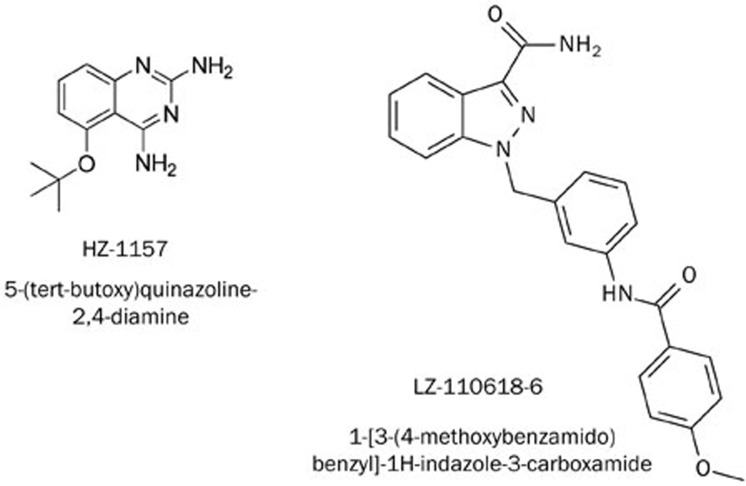
Figure 7.
Chemical structures of HZ-1157 and LZ-110618-6.
Confirmation of anti-HCV active compounds in an HCV infection assay
HZ-1157 and LZ-110618-6 were further tested against an infectious HCV virus (J399EM) which can infect and replicate in Huh7.5.1 cells in vitro. The results are shown in Figure 8. HZ-1157 and LZ-110618-6 effectively inhibited J399EM virus replication in Huh7.5.1 cells with IC50 values of 0.82 μmol/L and 0.11 μmol/L, respectively. Comparing the results of HZ-1157 and LZ-110618-6 in three assay systems (HCV replicon, infectious HCV and HCV NS3/4A protease), HZ-1157 showed similar IC50 values across all tests (0.73 μmol/L vs 0.82 μmol/L vs 1.0 μmol/L), indicating that HZ-1157 is a specific inhibitor of the HCV NS3/4A protease. However, the IC50 values of LZ-110618-6 differed across the three assay systems (0.06 vs 0.11 vs 0.68 μmol/L), as LZ-110618-6 was more active in assay systems that involve virus replication. This may indicate that LZ-110618-6 inhibits viral components other than the NS3/4A protease (see the Discussion section below). The exact anti-HCV mechanism through which LZ-110618-6 acts remains to be further investigated.
Figure 8.
The effects of LZ-110618-6 and HZ-1157 in an HCV infection assay. Compounds HZ-1157 and LZ-110618-6 were tested for their anti-HCV activities using an infectious HCV virus (J399EM) in Huh7.5.1 cells. Cells were first infected with J399EM virus, and then compounds were added and co-cultured for 72 h. The inhibition rate and cytotoxicity rate were calculated against control wells without compounds. The representative result is from two independent experiments with the same results.
Discussion
The development of anti-HCV drugs targeting multiple aspects of infection is a healthcare imperative. In 2005, the development of robust in vitro HCV infection models made it possible to screen anti-HCV compounds that inhibit the viral replication cycle25,26,27. A screen for anti-HCV agents usually utilizes the HCV replicon or infectious assay systems to cover the entire or, at least, most aspects of HCV propagation. However, these assays cannot differentiate the action mechanism of the compound and usually result in many false positives, and most of the valid active candidates turn out to be targeting host cellular components, making them unsuitable for further development. Therefore, using a system to study the inhibitory efficacy of the compounds on a specific HCV target is very useful. Another obvious advantage of using a target-specific assay system to help to identify the novel anti-HCV compound is the guarantee of high specificity. This could pave the way for further development with respect to increasingly strict regulations regarding drug safety and potential toxicity tests.
The NS3 protease of HCV is a prime target for the development of anti-HCV agents because it cleaves the viral polyprotein and liberates NS3, NS4A, NS4B, NS5A, and NS5B, allowing them to function normally in viral RNA replication, and it deactivates many host proteins involved in innate immunity to foster a favorable cellular environment for HCV replication28. The NS3 protease is most active when complexed with its cofactor NS4A29,30. For the in vitro evaluation of HCV NS3/4A protease inhibitors, there are generally two types of methods that can be used. One is to express and purify the NS3 protease in vitro, using a synthetic peptide as its substrate31. The other alternative method is a cell-based system, as we created here, which is rapid and easy to operate and does not require conventional protein expression and purification. In our system, the Seap activity in the supernatant can be monitored continuously. In addition, with the help of adenovirus delivery, the NS3/4A-Seap construct can be used in evaluations utilizing animal models. The Seap protein will be released into the blood by an active HCV protease, thus indicating the potency of an agent in vivo22. Telaprevir, a novel small-molecule peptidomimetic inhibitor of the HCV NS3/4A protease, was used here to verify the feasibility of the system. The IC50 of telaprevir in genotype 1b HCV replicon cells was 354 nmol/L22. To our knowledge, this is the first time that the inhibitory efficacy of telaprevir has been shown in a cell-based system that monitors only the HCV NS3/4A protease activity (genotype 2a). The IC50 of telaprevir in our system was approximately 931 nmol/L. The difference in IC50 between these two systems may due to the genotype difference or the difference between the replicon system and the single target system.
In Table 1, in addition to compounds of HZ-1157 and LZ-110618-6, we also identified other active anti-HCV compounds in our replicon assays, but they showed no specific inhibitory effect on HCV NS3/4A protease activity (Table 3). This does not rule out their potential activity against the HCV virus. They could be active anti-viral agents that target other viral components, and their anti-viral mechanism remains to be further investigated.
We also identified HZ-1157 and many other 2,4-diaminoquinazoline derivatives as anti-Dengue active compounds16. Dengue virus is an RNA virus, and both Dengue and HCV belong to the Flaviviridae family. They are both positive-sense RNA viruses and thus share a similar genome structure and life cycle. We propose that this compound could also be a Dengue viral protease inhibitor, which requires further investigation.
Moreover, LZ-110618-6, which showed better activity in the HCV replicon and infectious systems than in the HCV NS3/4A protease-specific assay, may possess the ability to inhibit other viral replication steps, including viral and cellular protease that are necessary for HCV replication. The possible multi-function mechanisms of this compound make it very interesting for further investigation.
Taken together, the present study discovered novel anti-HCV compounds using a combination of anti-viral assay systems, including the viral replicon, infectious virus and viral protease-specific assays. The results suggested that anti-HCV active compounds have been developed from 2,4-diaminoquinazoline derivatives and carboxamide analogues. HZ-1157 was identified as a potent inhibitor of HCV NS3/4A protease, and LZ-110618-6 was identified as a pan-inhibitor against various viral proteases, including HCV NS3/4A. Therefore, these active compounds could act as lead compounds for further anti-HCV drug development.
Author contribution
Ye YU, Jing-feng JING, and Xian-kun TONG performed all the experiments. Pei-lan HE technically assisted in performing part of the experiments. Yuan-chao LI and You-hong HU contributed synthetic compounds and reagents. Wei TANG were in charge of the data analysis. Xian-kun TONG and Jian-ping ZUO designed and supervised the project and wrote the manuscript.
Acknowledgments
This work was sponsored by the Ministry of Science and Technology of PRC (National Basic Research Program of China, 973 Program, 2013CB911104). The HCV infectious virus J399EM and the HCV replicon cell line J399LM were kindly provided by Professor Xin-wen CHEN, Wuhan Institute of Virology, Chinese Academy of Sciences.
References
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. Molecular biology of hepatitis C virus. J Gastroenterol. 2007;42:411–23. doi: 10.1007/s00535-007-2030-3. [DOI] [PubMed] [Google Scholar]
- Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–41. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350–9. doi: 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman KE, Flamm SL, Afdhal NH, Nelson DR, Sulkowski MS, Everson GT, et al. Response-guided telaprevir combination treatment for hepatitis C virus infection. N Engl J Med. 2011;365:1014–24. doi: 10.1056/NEJMoa1014463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–16. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- Hsu CS. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;369:678. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207–17. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbouseh NM, Jensen DM. Future therapies for chronic hepatitis C. Nat Rev Gastroenterol Hepatol. 2013;10:268–76. doi: 10.1038/nrgastro.2013.17. [DOI] [PubMed] [Google Scholar]
- Yang L, Shi LP, Chen HJ, Tong XK, Wang GF, Zhang YM, et al. Isothiafludine, a novel non-nucleoside compound, inhibits hepatitis B virus replication through blocking pregenomic RNA encapsidation. Acta Pharmacol Sin. 2014;35:410–8. doi: 10.1038/aps.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, et al. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417–28. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- Chao B, Tong XK, Tang W, Li DW, He PL, Garcia JM, et al. Discovery and optimization of 2,4-diaminoquinazoline derivatives as a new class of potent dengue virus inhibitors. J Med Chem. 2012;55:3135–43. doi: 10.1021/jm2015952. [DOI] [PubMed] [Google Scholar]
- Shi JJ, Ji FH, He PL, Yang YX, Tang W, Zuo JP, et al. Synthesis and hepatitis C antiviral activity of 1-aminobenzyl-1H-indazole-3-carboxamide analogues. ChemMedChem. 2013;8:722–5. doi: 10.1002/cmdc.201300083. [DOI] [PubMed] [Google Scholar]
- Han Q, Xu C, Wu C, Zhu W, Yang R, Chen X. Compensatory mutations in NS3 and NS5A proteins enhance the virus production capability of hepatitis C reporter virus. Virus Res. 2009;145:63–73. doi: 10.1016/j.virusres.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Shi JJ, Ji FH, He PL, Yang YX, Tang W, Zuo JP, et al. Synthesis and hepatitis C antiviral activity of 1-aminobenzyl-1H-indazole-3-carboxamide analogues. ChemMedChem. 2013;8:722–5. doi: 10.1002/cmdc.201300083. [DOI] [PubMed] [Google Scholar]
- Tong XK, Qiu H, Zhang X, Shi LP, Wang GF, Ji FH, et al. WSS45, a sulfated alpha-D-glucan, strongly interferes with Dengue 2 virus infection in vitro. Acta Pharmacol Sin. 2010;31:585–92. doi: 10.1038/aps.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SJ, Lin ZM, Wu YW, Bai BX, Yang XQ, He PL, et al. Therapeutic effects of DZ2002, a reversible SAHH inhibitor, on lupus-prone NZBxNZW F1 mice via interference with TLR-mediated APC response. Acta Pharmacol Sin. 2013;35:219–29. doi: 10.1038/aps.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni RB, Almquist SJ, Byrn RA, Chandorkar G, Chaturvedi PR, Courtney LF, et al. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2006;50:899–909. doi: 10.1128/AAC.50.3.899-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Li J, Zhang T, Wang X, Wang Y, Zhou Y, et al. Mycophenolate mofetil inhibits hepatitis C virus replication in human hepatic cells. Virus Res. 2012;168:33–40. doi: 10.1016/j.virusres.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–6. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Gale M., Jr Regulation of innate immunity against hepatitis C virus infection. Hepatol Res. 2008;38:115–22. doi: 10.1111/j.1872-034X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–55. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–60. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Steinkuhler C, Taliani M, Urbani A, Francesco RD, Pessi A. Synthetic depsipeptide substrates for the assay of human hepatitis C virus protease. Anal Biochem. 1996;237:239–44. doi: 10.1006/abio.1996.0235. [DOI] [PubMed] [Google Scholar]