Abstract
The development of the neuromuscular synapse depends on signaling processes that involve protein phosphorylation as a crucial regulatory event. Muscle-specific kinase (MuSK) is the key signaling molecule at the neuromuscular synapse whose activity is required for the formation of a mature and functional synapse. However, the signaling cascade downstream of MuSK and the regulation of the different components are still poorly understood.
In this study we used a quantitative phosphoproteomics approach to study the phosphorylation events and their temporal regulation downstream of MuSK. We identified a total of 10,183 phosphopeptides, of which 203 were significantly up- or down-regulated. Regulated phosphopeptides were classified into four different clusters according to their temporal profiles. Within these clusters we found an overrepresentation of specific protein classes associated with different cellular functions. In particular, we found an enrichment of regulated phosphoproteins involved in posttranscriptional mechanisms and in cytoskeletal organization. These findings provide novel insights into the complex signaling network downstream of MuSK and form the basis for future mechanistic studies.
The formation of the neuromuscular synapse (NMS)1 is crucially linked to signal transduction events induced by the receptor tyrosine kinase MuSK. MuSK is activated by the heparansulfate proteoglycan agrin (1). Agrin is produced by motor neurons and deposited in the basal lamina of the synaptic cleft. Agrin does not bind MuSK directly but interacts with Lrp4, a member of the LDL receptor family (2, 3). Upon binding, the Lrp4–MuSK complex presumably undergoes a structural rearrangement that leads to dimerization and subsequent autophosporylation of MuSK. The resulting activation of the MuSK kinase induces a signaling cascade leading to the formation of the NMS, including postsynaptic differentiation characterized by the accumulation of acetylcholine receptors (AChRs) at synaptic sites and presynaptic differentiation as depicted by the development of active zones (4).
Consistent with this model, agrin, MuSK and lrp4 mutant mice fail to form NMSs and consequently die at birth as a result of respiratory failure (5–7). Their levels of AChR expression are normal, but very few nerve–muscle contacts (in agrin−/− mice) or no contacts (in MuSK−/− and lrp4−/− mice) are accompanied by AChR clusters or other postsynaptic specializations. In addition, motor axons continue to grow and fail to form arborized nerve terminals. Agrin, MuSK, and Lrp4 also regulate the distribution of other synaptic proteins such as acetylcholinesterase, rapsyn, neuregulin-1, and its receptor erythroblastosis oncogene B. These proteins, usually concentrated at NMSs, are uniformly distributed in muscle from mice lacking agrin, MuSK or Lrp4. Likewise, genes, which are normally transcribed selectively in nuclei underlying NMSs, are transcribed equally in synaptic and non-synaptic nuclei of muscle in these mice.
Lrp4 lacks a kinase domain and its role as a potential docking molecule remains controversial (2, 3, 8). In contrast, the crucial role of MuSK kinase activity and MuSK scaffolding ability has been demonstrated in numerous experimental approaches. (I) Expression of MuSK mutants with a defective kinase domain inhibits agrin-induced AChR clustering (9). (II) Tyrosine kinase inhibitors block agrin-induced AChR clustering (10). (III) Specific residues in the MuSK cytoplasmic domain, in particular an NPXY motif in the juxtamembrane region, are required for downstream signaling (11, 12). (IV) Several molecules including scaffolding proteins, adaptor proteins, kinases and phosphatases have been identified that are downstream of MuSK (13). These results together with the above-mentioned data put MuSK at the center of signal transduction events that result in the formation of mature and functional NMSs. Despite the description of several targets of MuSK signaling, the current depiction of the agrin–Lrp4/MuSK signaling axis remains incomplete (13). In particular, the temporal and spatial regulation of MuSK signaling during NMS formation is unknown.
Here, we used quantitative phosphoproteomics to study the dynamics of MuSK signaling. We identified 203 phosphopeptides, representing 152 unique proteins, that were at least 2-fold up- or down-regulated. These peptides fell into different clusters according to their temporal profiles. In addition, clusters were enriched for specific protein classes or pathways. We found an overrepresentation of cytoskeletal proteins, which sustains and extends data demonstrating that cytoskeletal rearrangements and dynamics are crucial for MuSK-dependent AChR clustering.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were purchased from commercial sources: anti-phosphotyrosine PY99 (Santa Cruz Biotechnology, Dallas, TX), PY-100 (Cell Signaling Technology, Leiden, The Netherlands), and anti-AChR β (Sigma-Aldrich). Antibodies against the extracellular domain (Ig1–2) of MuSK were produced in rabbits.2 Antibodies against the C-terminal sequence of MuSK were described previously (11). Biotin-conjugated α-bungarotoxin was obtained from Invitrogen (Carlsbad, CA). Horseradish-peroxidase-coupled secondary antibodies were purchased from Jackson ImmunoResearch Europe, Suffolk, UK. Streptavidin and Protein A agarose beads were obtained from Novagen (Merck MGaA, Darmstadt, Germany) and Pierce (Thermo Fisher Scientific, Rockford, IL), respectively. Soluble neural agrin (isoform A4B8) was prepared from transiently transfected HEK 293T cells as described by Herbst and Burden (11).
Cell Culture
C2 myoblasts were propagated and differentiated into myotubes as described previously (14).
Immunoprecipitation, AChR Pulldown, and Immunoblotting
C2 myotubes were starved for 2 h in DMEM and then stimulated with agrin for 15, 60, or 240 min. Cells were lysed in RIPA buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 20 mm HEPES, pH 7.4, 150 mm NaCl, 2.5 mm EDTA) supplemented with protease (1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and 0.2 mm PMSF) and phosphatase inhibitors (1 mm sodium orthovanadate, 50 mm natrium fluoride, 5 mm sodium molybdate, 5 mm sodium pyrophosphate, and 1 mm β-glycerophosphate). Cleared lysates were precipitated with polyclonal affinity-purified antibodies directed to MuSK (C terminus). The next day protein A agarose (Pierce) was added for 1 to 3 h, the beads were washed three times with lysis buffer, and precipitated protein complexes were subjected to SDS-PAGE. AChRs were isolated from cleared lysates as described previously (15). Following SDS-PAGE, proteins were transferred to PVDF membrane (Millipore, Darmstadt, Germany). The membrane was incubated overnight with anti-phosphotyrosine antibodies, anti-MuSK (extracellular domain), or anti-AChR β antibodies. After 1 h of incubation with secondary antibodies, signals were detected via chemiluminescence (Roche) on a ChemiDoc XRS+ System (Bio-Rad, Hercules, CA).
Sample Preparation and Digestion
Agrin-induced C2 cells were washed with cold PBS and lysed in 50 mm HEPES, pH 7.5, 5 mm EDTA, 150 mm KCl, 10% glycerol, 1% Triton X-100, 1 mm DTT, 20 mm β-glycerophosphate, 10 mm sodium fluoride, 1 mm sodium orthovanadate, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and 0.2 mm PMSF. Protein lysates were passed through a 27-gauge syringe needle, cleared via centrifugation, and precipitated with 5 volumes of acetone. Protein pellets of the four samples were suspended in 0.5 m triethylammonium bicarbonate and 8 m urea. Protein concentration was determined via Bradford assay (Bio-Rad). For each sample, 1 mg of protein was reduced by a 1:10 (v:v) addition of 50 mm tris-(2-carboxyethyl) phosphine at 60 °C and alkylated by the 1:20 (v:v) addition of 200 mm methyl methanethiosulfonate for 15 min. Samples were diluted with 0.5 m triethylammonium bicarbonate to a urea concentration of 6 m and digested for two hours with lysozyme C at an enzyme-to-protein ratio of 1:40. After the lysozyme C digest, samples were diluted with 0.5 m triethylammonium bicarbonate to a urea concentration of 0.8 m and digested with trypsin at an enzyme-to-protein ratio of 1:20 overnight.
iTRAQ Labeling
600 μg of tryptic peptides were used, and their volumes were reduced on a SpeedVac to ∼60 μl. Each sample was mixed with two vials of iTRAQ 4-plex reagents (AB Sciex, Foster City, CA) supplemented with 70 μl of ethanol each and incubated for 2 h. Untreated sample was labeled with iTRAQ114 and further timepoints in increasing order (15 min = iTRAQ115, 60 min = iTRAQ116, 240 min = iTRAQ117). Labeling efficiency was determined in a 90-min gradient LC-MS/MS analysis to ensure a labeling efficiency of 99%. Each sample was measured individually, and iTRAQ was defined as a variable modification in the search. One-third of the samples were mixed, and the bias was determined to ensure a 1:1:1:1 mixing ratio of the samples. After this analysis the rest of the sample was mixed to a total sample amount of 2 mg of peptide. This final sample was diluted with water to an ethanol concentration of less than 20% and then lyophilized. The dry sample was dissolved in 0.1% trifluroacetic acid (TFA) (Pierce), and the pH was adjusted to 2 with 10% TFA. The sample was desalted using a Strata-X solid-phase extraction cartridge (Phenomenex, Aschaffenburg, Germany; 200 mg). Peptides were eluted with 70% acetonitrile (ACN) and 0.1% formic acid (FA). Afterward the sample was diluted with 0.1% FA to a final ACN concentration of less than 35% and lyophilized.
Phosphopeptide Enrichment by IMAC and TiO2
The IMAC protocol was adapted from Ref. 16. Dried sample was dissolved in 1 ml of IMAC loading buffer (40% ACN, 0.1% FA) and incubated with 40 μl of IMAC beads (PHOS-Select Iron Affinity Gel, Sigma) on a rotary wheel for 60 min at room temperature. The suspension was then transferred into a Mobicol (MoBiTec, Göttingen, Germany) spin column with a 10-μm pore filter. The flow-through of unbound peptides was collected and dried by dry freezing for TiO2 enrichment.
IMAC beads were washed 10 times using 400 μl of IMAC loading buffer and twice with 5% ACN, 0.1% FA. The first two washes were pooled with the flow-through of the IMAC enrichment. Peptides were eluted from the IMAC resin with 4 × 100 μl NH4OH. The eluate was set to a pH of 2 using 10% FA and stored at −80 °C until unification with the TiO2 eluate. For the TiO2 enrichment, the dried IMAC flow-through was dissolved in 0.1% TFA, desalted as described above, and freeze dried. Peptides were dissolved in 1 ml of TiO2 loading buffer (300 mg/ml lactic acid, 80% ACN, 0.1% TFA) and incubated with 4 mg of TiO2 resin (Titansphere bulk media, 5 μm, GL Science, Torrance, CA) with overhead rotation at room temperature. The suspension was then transferred into a Mobicol spin column with a 10-μm pore filter. The TiO2 resin was washed with 8 × 500 μl of TiO2 loading buffer, 8 × 500 μl of 80% ACN, 0.1% TFA, 4 × 500 μl of 1% ACN, 0.1% TFA, and 4 × 500 μl of 1% ACN, 0.1% FA. Bound peptides were eluted from the TiO2 resin by the addition of 4 × 100 μl of 0.3 m NH4OH and 4 × 100 μl of 0.7 m NH4OH. TiO2 eluate was acidified with FA, mixed with IMAC elution, and lyophilized. The flow-through of the experiment was stored and later used to perform protein quantification in a whole proteome analysis.
Separation via High-resolution Strong Cation Exchange Chromatography
Dried peptides were dissolved in 50 μl of SCX Buffer A (5 mm NaH2PO4, pH 2.7, 15% ACN), and the pH was adjusted to a pH greater than 3 with 8.5% H3PO4.
SCX was performed on an UltiMate 3000 nanoLC system (Thermo Fisher Scientific) equipped with a 25 cm × 1 mm polysulfoethyl-A (3 μm) column (PolyLC) at flow rate of 25 μl/min. The sample was applied to the column and separated with a combined pH and salt gradient of 100% buffer A (5 mm NaH2PO4, pH 2.7, 15% ACN) at 10 min, 10% buffer B (5 mm NaH2PO4, pH 2.7, 1 m NaCl, 15% ACN) and 50% buffer C (5 mm NaH2PO4, pH 6, 15% ACN) at 90 min, 25% buffer B and 50% buffer C at 100 min, 50% buffer B and 50% buffer C at 105 min, 50% buffer B and 50% buffer C at 120 min, and 100% buffer A at 125 min. Following 5 min of flow-through, 130 fractions were collected (1 min/fraction) and stored at −80 °C. The SCX gradient for whole proteome analysis was 100% buffer A at 10 min, 10% buffer B and 50% buffer C at 40 min, 25% buffer B and 50% buffer C at 50 min, 50% buffer B and 50% buffer C at 55 min, 50% buffer B and 50% buffer C at 70 min, and 100% buffer A at 75 min. Here 70 fractions were collected (1 min/fraction) and stored at −80 °C.
Mass Spectrometry
SCX fractions were separated on an Ultimate 3000 nanoLC system (Thermo Fisher Scientific) equipped with a 25 cm × 75 μm inner diameter Acclaim PepMap RSLC C18 column (5 μm, 100 Å). For all LC-MS experiments, a linear gradient from 100% solvent A (5% ACN (Sigma-Aldrich), 0.1% FA (Merck, Germany)) to 70% solvent B (30% ACN, 0.08% FA) in 60 min was applied for peptide separation. An LTQ Velos Orbitrap ETD (Thermo Fisher Scientific) mass spectrometer was coupled on-line with the Ultimate 3000 via a nano-electrospray source (Proxeon, Thermo Fisher Scientific). The mass spectrometer was operated in data-dependent mode. For the analysis of the SCX fractions, one full scan (m/z 350–2000) was acquired in the Orbitrap (resolution 60,000, target value 1,000,000, maximal fill time of 400 ms), followed by MS/MS scans of the five most abundant ions in the LTQ (target value 10,000, maximal fill time of 200 ms, isolation at 1.9 Th with the ion trap, CID fragmentation with normalized collision energy of 35) and in the HCD cell (resolution 7500, target value 100,000, maximal fill time of 600 ms, isolation at 1.6 Th with the ion trap, HCD fragmentation with normalized collision energy of 50). CID was performed enabling multistage activation fragmentation on neutral loss masses of −32.6, −49, and −98 Da from the precursor m/z. Selected ions were excluded for 60 s with an exclusion mass tolerance of 5 ppm. Polydimethylcyclosiloxane (m/z 445.12) was used as a lock mass for internal recalibration. Singly charged ions were excluded from selection, and the monoisotopic precursor selection feature was enabled.
Data Analysis
Raw files were processed with Proteome Discoverer (version 1.4.0.282, Thermo Fisher Scientific, Bremen, Germany). Database searches were performed using Mascot (version 2.2, Matrix Science, London, UK) (17) against a concatenated target-decoy database based on the mouse UniProt database (version 2012_11). MaxQuant SequenceReverser (version 1.0.13.13) was used to generate the decoy database and append contaminants (18) (101,726 sequences in total). Oxidation of methionine and phosphorylation of serine, threonine and tyrosine were set as dynamic modifications and methylthio-cysteine and iTRAQ at the N terminus and lysine were specified as fixed modifications. Trypsin was defined as the proteolytic enzyme, cleaving after lysine or arginine, except when followed by proline, and up to two missed cleavages were allowed. A mass tolerance of 7 ppm was set as the precursor ion tolerance. The fragment ion tolerance values for HCD and CID spectra were set to 0.03 and 0.5 Da, respectively. Reporter ion intensities were extracted in Proteome Discoverer from the closest centroid mass within an integration tolerance of 5 mmu. PhosphoRS (version 3.0) was employed to determine the localization of phosphorylated residues (19). Phosphosites with a site probability greater than 75% were regarded as confidently localized. Only peptide spectrum matches with search engine ranks of one and minimum peptide lengths of eight amino acids were exported.
Bioinformatics Analysis
The R environment (version 2.14.1) was used to analyze, plot and cluster the data. Peptide spectrum matches originating from HCD and CID scans were filtered separately to achieve an overall false discovery rate of 1% at the peptide level. Peptide spectrum matches with an isolation interference of more than 25% were removed to avoid ratio distortion. The entire dataset was converted in ibspectra format and imported to Isobar to derive a common protein grouping across phosphopeptide-enriched and global proteome samples (20, 21). Based on this protein grouping, protein regulations in the flow-through sample were calculated (supplemental Fig. S1 and supplemental Table S1) and used to correct peptide ratios determined for the phosphopeptide enriched sample. Both analyses were also performed using Isobar, which allows one to determine the statistical significance of protein/peptide regulation based on reporter intensity (supplemental Fig. S2). Phosphopeptides of proteins that were not quantified in the flow-through sample were left unchanged. Reporter log-ratios were normalized to a median of zero per channel, corresponding to a 1:1 ratio between channels. Phosphopeptides that were regulated at least 2-fold and found significantly regulated in Isobar with a p value of less than 0.05 after Benjamini–Hochberg correction were subjected to k-means clustering (22). The Euclidean distance of log-ratios normalized to their respective maxima per peptide was used to measure the distance between regulated phosphopeptides. Normalization to the maximum per peptide was done in order to group peptides based on their temporal response profile instead of the magnitude of induction. The resulting clusters were checked for enrichment in IPA (Ingenuity® Systems, Redwood City, CA), and subnetworks densely populated with regulated proteins of individual clusters were extracted (23). The interaction data underlying this network analysis originated from the manually curated Ingenuity Knowledge Base. Overrepresented patterns of regulated phosphosites were identified by Motif-X (24, 25), using the IPI mouse database as background and a minimum number of 10 occurrences per motif. The significance threshold was left at the default setting of 1e-6. NetworKIN was used to predict potential kinases from substrates (26). Because kinase motifs for murine kinases are not available, human kinases were used for the prediction. Mascot search results were converted to pride.xml format using PRIDE Converter (Version 2.0.20) (27, 28). Data are available via ProteomeXchange with the identifiers PXD000558 (phosphopeptide enrichment) and PXD000878 (whole proteome analysis).
RESULTS
Agrin-induced MuSK Stimulation and MS Experimental Strategy
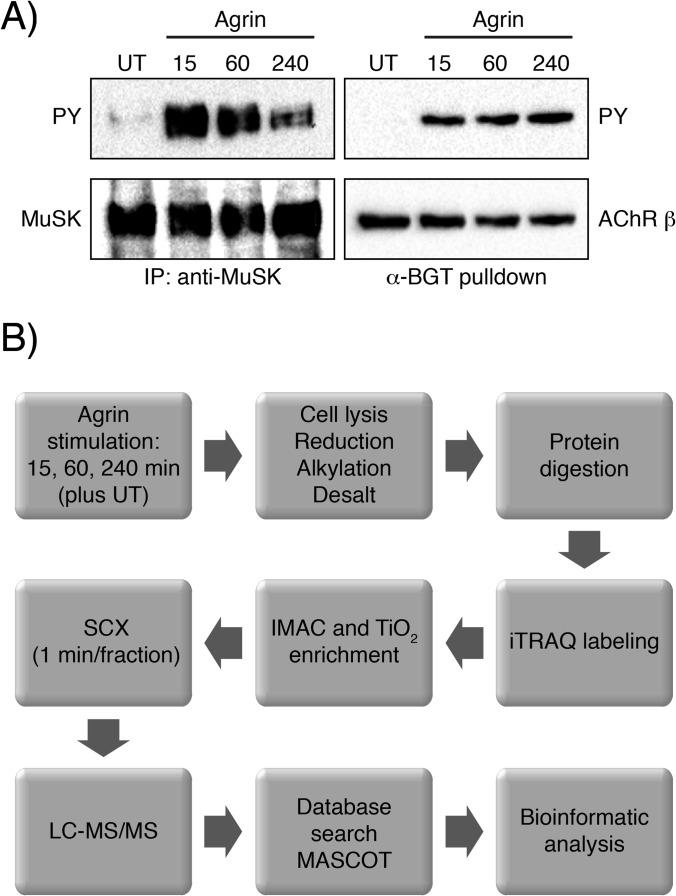
MuSK signal transduction and subsequent NMS formation can be recapitulated in so-called myotubes, differentiated muscle cells that express synaptic proteins such as MuSK and Lrp4. Myotubes, when treated with soluble agrin, form aggregates of AChRs, reminiscent of plaque-like NMSs found in prenatal mouse embryos. Further, agrin-treated myotubes can be used to study MuSK activation and downstream signaling events. We stimulated myotubes with agrin and found rapid and transient tyrosine phosphorylation of MuSK as previously reported (Fig. 1A). A consequence of MuSK activation is the tyrosine phosphorylation of AChR β subunits. Accordingly, AChR β showed strong and sustained tyrosine phosphorylation (Fig. 1A).
Fig. 1.
MuSK and AChR β phosphorylation in muscle cells and MS experimental strategy. A, cell lysates from agrin-induced muscle cells were subjected to an immunoprecipitation with anti-MuSK antibodies or a pulldown using biotinylated α-BGT. Samples were analyzed by immunoblotting using antibodies against phosphotyrosine, MuSK or AChR. B, experimental strategy for phosphoproteomic analysis. Muscle cells unstimulated (UT) or stimulated across three time points were lysed. Proteins were extracted, digested and isotopically labeled with the iTRAQ 4-plex reagent. Global phosphorylation was assessed via TiO2 and IMAC enrichment. Eluted peptides were then analyzed via LC-MS/MS and the resulting data were processed for phosphopeptide identification and quantification as described in “Experimental Procedures.”
For MS analysis we stimulated C2 myotubes with agrin for 15, 60 or 240 min (Fig. 1B). Unstimulated cells were used as a reference sample. Cells were lysed and the samples were prepared as described in “Experimental Procedures.” Peptides were stably isotope-labeled with iTRAQ isobaric reagent, mixed, loaded on a TiO2 column and phosphopeptides were eluted. Enriched phosphopeptides as well as the unphosphorylated flow-through were subjected to high-resolution SCX fractionation to reduce the complexity of the samples and SCX was followed by LC-MS/MS.
Samples were analyzed using a hybrid CID/HCD acquisition scheme (29). Acquired spectra were searched against a concatenated target/decoy murine UniProt database using Mascot (17). Spectra were filtered to a false discovery rate of 1% at the peptide level. Quantification of peptides was inferred by reporter ion intensities recorded in HCD scans only and transferred to the corresponding CID scan identifications.
Grouping peptides in the phosphopeptide-enriched sample allowed the quantification of 15,533 peptides based on sequence and confidently localized phosphosites (phosphoRS > 75%) in Isobar (supplemental Table S1). 10,183 (66%) had at least one confidently localized phosphosite. Whole proteome analysis of the phosphopeptide enrichment flow-through allowed the quantification of 3867 protein groups (supplemental Table S1). Determined protein ratios were used to correct phosphopeptide regulation for changes in protein abundance with the built-in functionality of Isobar. The distribution of phosphorylated residues among serines (86%), threonines (13%) and tyrosines (0.95%) was similar to what has been reported in the past (30).
Among the more than 10,000 quantified phosphopeptides, 203 were regulated more than 2-fold in at least one time point with significant reporter ion intensities, as determined by Isobar, at the level of p ≤ 0.05 after Benjamini–Hochberg correction (supplemental Table S1). These regulated peptides originated from 152 distinct proteins. Submitting phosphosites of the 203 regulated phosphopeptides to Motif-X (24, 25) revealed two motifs that were significantly enriched among the regulated phosphoserine residues (Fig. 2A).
Fig. 2.
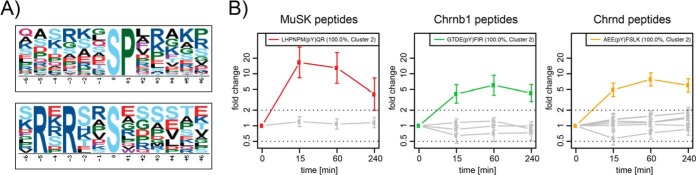
Depth and validation of the detected phosphoproteome. A, analysis of regulated phosphopeptides in Motif-X (24, 25) revealed two overrepresented motifs around phosphoserine. B, regulation of phosphosites in MuSK and AChR. Quantification of phosphopeptides on these proteins showed regulation of MuSK Y553 (red), AChR β Y390 (green) and AChR δ Y393 (orange). Furthermore, phosphorylation of MuSK S678; AChR β S378 and T387; and AChR δ S381, S382, and S383 was detected but not significantly regulated (all gray). Phosphosites of regulated peptides could be localized with high confidence (phosphoRS site probabilities are indicated in brackets). Error bars, S.D.
Temporal MuSK and AChR Phosphorylation
MuSK tyrosine phosphorylation sites have previously been mapped to Y553, Y576, Y750, Y754 and Y755 in vitro and in vivo (31). Y553 represents the major phosphorylation site in MuSK, accounting for more than 50% of all phosphorylation events (32). Likewise, phosphopeptides carrying Y553 were detected in this quantitative MS analysis (Fig. 2B). Y553 was rapidly and transiently phosphorylated. Phosphorylation peaked at 15 min (17-fold induction) and was reduced to close to basal level at 240 min. Similarly, we detected up-regulated phosphopeptides of the AChR β and δ subunits (Fig. 2B). Previous reports state that Y390 in AChR β is phosphorylated in response to agrin (33). We found the same site phosphorylated, displaying a temporal regulation with a peak at 60 min (6-fold induction) and a sustained phosphorylation even at 240 min. AChR δ phosphorylation showed a similar temporal regulation (8-fold induction at 60 min). The site of phosphorylation was mapped to Y393. Several other phosphosites were identified, but their appearance was not altered by agrin stimulation.
We also detected a MuSK phosphopeptide carrying S678 phosphorylation. This site has previously been mapped using in vitro phosphorylated MuSK (31). Our quantitative analysis demonstrated stable phosphorylation that was not regulated by agrin stimulation. In addition, we identified a novel phosphorylation site on S751. This site was removed from the quantitative analysis because of high isolation interference. Nevertheless, identification and site localization were achieved with high confidence (supplemental Fig. S3, phosphoRS site probability = 99.8%).
Cluster Analysis of the MuSK Signaling Network
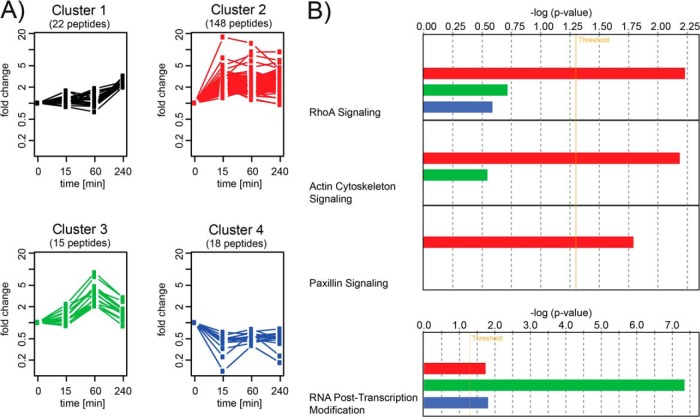
Grouping the 203 regulated phosphopeptides into four clusters by k-means clustering (22) revealed different temporal regulation patterns of the underlying phosphosites (Fig. 3A). Therefore, phosphopeptide fold changes were log-transformed and normalized to their respective maximum absolute values before clustering (see also “Experimental Procedures”). The resulting clusters allowed us to discriminate groups of phosphopeptides responding to agrin stimulation at different time points. Cluster 2 comprised sites that became phosphorylated soon upon agrin stimulation. Clusters 3 and 1 encompassed delayed phosphorylation events occurring at the one-hour and four-hour time points. Cluster 4 represented phosphosites that were down-regulated at least 2-fold upon MuSK activation. In addition to their temporal profiles, the clusters also differed in their content of specific protein classes. In particular, Cluster 3 was highly enriched in RNA binding proteins and transcription factors. Clusters 2 and 4, which both responded early to agrin stimulation, contained the greatest amounts of cytoskeletal proteins (supplemental Fig. S4). Cluster 2 contained 20 cytoskeletal proteins (26 found in total), of which 15 were actin-associated proteins, 3 were microtubule-binding proteins and 1 was an intermediate filament protein.
Fig. 3.
Grouping of phosphopeptides into clusters with different temporal profiles. A, clustering of regulated phosphopeptides revealed four clusters of different temporal regulation patterns. B, pathway and functional enrichment analysis in IPA® showed overrepresentation of pathways related to cytoskeletal function in Cluster 2 (red) in the three most significantly enriched pathways, as well as function enrichment for posttranscriptional-modification-related proteins in Cluster 3 (green). Only pathways with more than two proteins with regulated phosphosites were considered in the enrichment analysis.
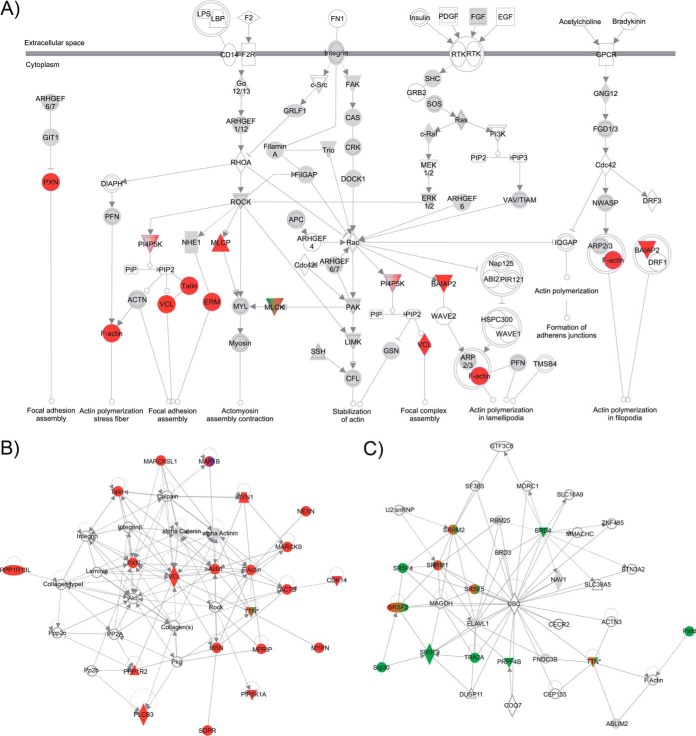
Cluster 2, which showed a rapid increase in phosphorylation and which also included MuSK Y553, AChRβ Y390 and AChR δ Y393, represented the largest group, with 148 peptides. Pathway enrichment analysis using IPA on the proteins contained in the determined clusters revealed an overrepresentation of proteins involved in actin cytoskeleton and RhoA signaling in Cluster 2 (Fig. 3B). RhoA signaling has previously been implicated in agrin-induced AChR clustering (33). In addition, proteins involved in Paxillin signaling were enriched. Consistent with these data, a significant correlation with proteins involved in cytoskeletal function was observed (Fig. 4A). Here, phosphatidylinositol 4-phosphate 5-kinase (PIP5K or PI4P5K), which has been implicated as a downstream effector of the Rho family in actin organization, was identified together with players of the same pathway: Moesin, a member of the ERM protein family, and Vinculin (34). Potentially affected pathways included focal adhesion and complex assembly and actin polymerization. This finding was further supported by Network Analysis in IPA. Here, based on interactomic data stored in the manually curated Ingenuity Knowledge Base, subnetworks densely populated with regulated phosphoproteins of Cluster 2 were extracted. The subnetwork containing the greatest number of regulated phosphoproteins (30 out of 35) was enriched for proteins involved in “cellular assembly and organization” (supplemental Fig. S5A). It included proteins involved in microtubule function such as CLIP-associating proteins 1 and 2, the serine/threonine-protein kinase MARK2, the microtubule-associated proteins MAPRE3 and MAP4, and the actin cross-linker brain-specific angiogenesis inhibitor 1-associated protein 2. The second-best hit (21 out of 35) revealed a network specifically associated with “developmental disorder,” “neurological disease,” and “skeletal and muscular disorders” (supplemental Fig. S5B). This network contained MuSK and AChR β and δ. Most interesting, we identified a network enriched in regulated phosphoproteins (21 out of 35) that has been linked to “skeletal and muscular disorders” as well as “nervous system development and function” (Fig. 4B). Proteins within this network were functionally involved in actin rearrangements. Most prominently, Talin, Vinculin and Paxillin as components of synaptic podosomes at the site of AChR clusters (35, 36); actin, F-actin and the actin binding protein MARCKS; and Myopaladin as an actin-cross-linking protein and cytoskeletal scaffolding protein (37, 38) take part or assist in the network. Further, we detected a regulated phosphorylation for PDZRN3 (Cluster 2) and LL5β (Cluster 4), which have been functionally associated with AChR cluster formation (35, 36, 39). Our data now implicate phosphorylation as an important regulatory event during their cooperative action in the formation of AChR clusters.
Fig. 4.
Enriched pathways and functions in Cluster 2 and Cluster 3. A, proteins in Cluster 2 (in red) are mapped on the actin cytoskeleton signaling pathways. B, network enrichment of Cluster 2 constituents (in red) reveals a subnetwork of the protein interactome that is enriched for “skeletal and muscular disorders” and “nervous system development and function.” C, network enrichment of Cluster 3 constituents (in green) reveals a subnetwork that is enriched in proteins involved in RNA posttranscriptional functions. A detailed description of the network shapes is provided in the supplemental material. Regulated phosphoproteins are color-coded as in Fig. 3 (Cluster 1 = black, Cluster 2 = red, Cluster 3 = green, Cluster 4 = blue). Proteins corresponding to detected unregulated phosphopeptides are shown in gray. Proteins found in more than one cluster are colored accordingly.
To our surprise, we found strong enrichment of proteins involved in RNA posttranscriptional modifications for Cluster 3 (Fig. 3B). This unraveled a function of MuSK signaling in the regulation of gene expression that has not been characterized so far (Fig. 4C).
Aiming to further delineate the signaling downstream of MuSK stimulation, we sought to identify which kinases induced phosphorylation changes observed in Cluster 2 and consequently trigger these effects on the actin cytoskeleton. Therefore, we further analyzed the phosphomotifs within Cluster 2 using Motif-X and identified an SP motif (Fig. 5A). Interestingly, this SP motif was significantly enriched in Cluster 2, as it was based on 23 motif hits in Cluster 2, compared with 25 motif hits of the closely related SP motif in Fig. 2A that was derived from the entire dataset of regulated phosphopeptides. Although the minimum number of occurrences in Motif-X was lowered to 10, it was not possible to extract significant motifs for the other clusters. Subjecting regulated phosphosites as kinase substrates to NetworKIN (26) revealed enrichment of cdk5, CKII, and p38-MAPK kinase families in Cluster 2 (Fig. 5B).
Fig. 5.
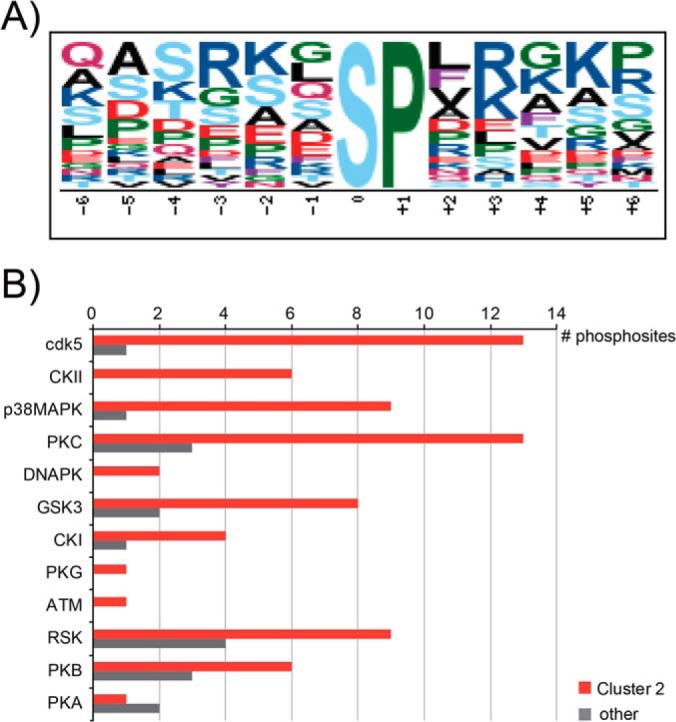
Bioinformatics analysis of potential kinases involved in Cluster 2 phosphorylation. A, analysis of phosphorylation sites in Motif-X (24, 25) revealed an overrepresentation of an SP motif. B, prediction of upstream kinase families in NetworKIN (26) showed an overrepresentation of several kinase families in Cluster 2 relative to Clusters 1, 3, and 4 (other).
DISCUSSION
Induction of MuSK kinase activity and subsequent downstream signaling represent crucial events during the formation of the NMS. This suggests that direct and indirect phosphotargets of MuSK signaling activity critically regulate the molecular mechanisms during NMS formation. We therefore used a muscle cell culture model system to identify and characterize the phosphoproteome during NMS formation. Here we show that activation of MuSK induces the phosphorylation of a large set of proteins with a distinct temporal manner of regulation. The phosphorylation of 152 different proteins was at least 2-fold up- or down-regulated. Grouping them according to their temporal phosphorylation rate produced four distinct clusters. These clusters can be distinguished not only by their phosphorylation kinetics, but also by their characteristic content of specific protein classes. In particular, we found an overrepresentation of proteins involved in cytoskeletal rearrangements in the cluster of proteins responding early to agrin stimulation, which consolidates the current opinion that synapse formation requires a variety of cytoskeletal changes and adjustments. Furthermore, our results suggest that MuSK also targets mechanisms that lead to posttranscriptional modifications.
Tyrosine phosphorylation is by far the least abundant type of phosphorylation, accounting for about 1% to 2% of all phosphorylation events (30). In comparison, serine and threonine phosphorylation have relative abundance values of 80% to 90% and 10% to 20%, respectively. Consistent with these reported numbers, we found similar relative abundances for phosphotyrosine, phosphothreonine and phosphoserine (1%, 13% and 86%, respectively) in agrin-treated samples. Indeed, we were able to identify MuSK Y553, AChR β Y390 and AChR δ Y393. Y553 is the major phosphorylation site in MuSK, and the tyrosines in AChR β and δ are also prominent because AChR subunits are abundant in muscle cells (31, 32). However, proteins such as docking protein-7, CT10-regulated kinase or Src (40, 41) that are known to be tyrosine phosphorylated downstream of MuSK were missing from our dataset. Using an IMAC approach to characterize global phosphorylation events favors the enrichment of phosphoserine and phosphothreonine peptides because MS analysis of complex samples is biased toward more abundant peptides and is therefore likely to miss phosphotyrosine peptides, which occur at low abundance (42–44). This challenge can be overcome by immunoprecipitations using antibodies against phosphotyrosine to specifically enrich tyrosine phosphorylated peptides or proteins. Therefore, use of a modified phospho-enrichment protocol that includes an immunoprecipitation with anti-phosphotyrosine antibodies in combination with TiO2-based IMAC is necessary in order to achieve a complete picture of phosphotyrosine, -serine and -threonine changes. Although regulated tyrosine phosphorylation also has been found further downstream of receptor tyrosine kinase–induced signaling, as in the case of STAT proteins and MAPKs (45, 46), it is generally thought that the benefit of analyzing phosphoserine and -threonine changes lies in their more downstream position in the signaling cascade. In our case this enabled us to detect target proteins that were not directly associated with or targeted by MuSK. Thus we identified proteins that act further downstream in the signaling cascade and therefore contribute more to the final effects of this pathway.
A more in-depth analysis of the composition of clusters revealed that phosphorylation changes at later time points affected RNA binding proteins and transcription factors more strongly, whereas the regulation of cytoskeletal proteins could be found earlier, with the exception of Palladin, found in Cluster 3. In particular, some of the cytoskeletal proteins in Cluster 2 such as Talin, Paxillin, Vinculin, Moesin, Myosin IXb and CLIP-associating protein 2 are thought to be important for cytoskeletal rearrangements based on their function in podosomes and axon growth cones, both structures associated with dynamic morphological changes, and most important in AChR insertion in agrin-induced clusters (35, 47–49).
Conceptually, it has been suggested that changes in actin dynamics can be communicated to transcriptional genome activity via complex mechanisms (50). Most likely, this could be an explanation for the delayed activation/kinetics of Clusters 3 and 1, which included a high percentage of transcription factors and RNA binding proteins.
Cluster 3 contained proteins implicated in posttranscriptional modifications, in particular mRNA splicing. We found increased phosphorylation of several members (serine/arginine-rich splicing factors 2, 4, and 9 and Tra2A) of the SR protein family. These are phosphorylated at numerous serines in the C-terminal RS domain by the SR-specific protein kinase family. RS domain phosphorylation is necessary for entry of SR proteins into the nucleus, and it might also play important roles in alternative splicing, mRNA export, and other processing events (51). Likewise, phosphorylation of serines in the RS domains of the above-mentioned SR proteins is up-regulated upon MuSK activation. This strongly suggests that MuSK signaling initiates a signaling network that controls posttranscriptional mechanisms such as mRNA splicing and export. It has been reported that AChR gene expression is increased in a MuSK-dependent manner (52–54). However, unlike for other receptor tyrosine kinases, a direct connection between MuSK activation and induction of gene expression has not been established yet. Our data now demonstrate that MuSK signaling also reaches the cell nucleus, thereby affecting gene expression.
A key finding from our study was the identification of an abundant number of regulated proteins that are implicated in cytoskeletal rearrangements. Our bioinformatical analysis put Rho GTPase-dependent signaling at the center of MuSK-induced downstream signaling. Consistent with this, we found regulated Paxillin, which has been implicated in focal adhesion and complex assembly (55). Paxillin serves as a central scaffolding protein at cell adhesion sites to the extracellular matrix. Tyrosine phosphorylation of Paxillin activates Rac1 and, at the same time, inactivates RhoA. It also enhances FAK binding, thereby increasing focal adhesion turnover. Together with the well-characterized focal adhesions, other adhesive structures known as podosomes and invadopodia also exist in cells including osteoclasts, macrophages, and neurons, as well as in myotubes and invasive cancer cells. Because most of the same proteins are found at these adhesion sites, podosome/invadopodia dynamics and functions are similarly regulated by Wiskott–Aldrich syndrome protein, actin related protein 2/3 complex, and the Rho family GTPases (35, 56). Feeding into the same pathway, we also found MAPK14, a member of the p38 MAPKs, among the regulated phosphopeptides. It has been shown that p38 kinases act downstream of Rac1 and upstream of changes in actin dynamics regulating cytoskeletal rearrangements such as induction of lamellipodia (57, 58).
Consistent with our findings, it is well established that NMS formation requires a complex set of morphological alterations at pre- and postsynaptic sites involving actin cytoskeleton reorganization. Likewise, agrin-induced AChR clustering and stabilization in muscle cells require cytoskeletal rearrangements and distinct actin cytoskeleton dynamics. More specifically, AChR cluster movement and formation depend on actin polymerization (47, 59). Numerous studies have shown that MuSK signaling triggers several mechanisms that regulate cytoskeletal dynamics, for example, via the Rho GTPases Rac1, Cdc42 and RhoA or via Src kinases (60). More recently, the actin nucleation factors neural Wiskott–Aldrich syndrome protein and actin related protein 2/3 have been implicated in agrin-induced AChR clustering (61). In addition, cluster maturation was associated with proteins like LL5β, Paxillin, Vinculin and Talin (35, 36). These proteins were also identified as regulated targets of MuSK signaling, uncovering a novel connection between agrin-induced phosphorylation and cytoskeletal dynamics during cluster development. Most interesting, tyrosine phosphorylation of Paxillin is thought to be Src- and FAK-dependent, and both have also been associated with AChR clustering and/or podosome function. These data put Paxillin at the center of cytoskeleton-driven events that regulate AChR clustering via Rho GTPases or podosomes (or both). In that respect, it will be of great interest to study the role of Paxillin and, above all, Paxillin phosphorylation during agrin-induced MuSK signaling.
Our finding that many cytoskeletal proteins are present in the set of regulated phosphopeptides is very much consistent with previous studies. For the first time we have now generated a global view of regulated targets and have placed them into a distinct network that clearly demonstrates the enrichment of cytoskeletal proteins and their potential interconnections.
Proteomics approaches have been used to examine the proteome in diseased muscle (62). In particular, numerous studies included proteomic profiling in neuromuscular diseases (63). Puente and colleagues used phosphoproteomics to identify phosphoproteins and kinases during the differentiation of muscle cells (64, 65). Proteomics was also used to characterize neuromuscular proteins from torpedo electric organ; that study identified several new components with potential signaling function but failed to identify essential components such as Lrp4 and MuSK (66). An approach for isolating secreted muscle proteins that are involved in NMS formation had limited success because of technical restrictions (67). Our study is the first quantitative phosphoproteomics attempt to analyze the global phosphoproteome during NMS formation. The experimental strategy using agrin-induced muscle cells allowed us to produce a comprehensive picture of the complex signaling network downstream of MuSK. These results provide a valuable resource for future studies to functionally characterize the MuSK interactome.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (68) with the dataset identifiers PXD000558 and PXD000878.
Supplementary Material
Acknowledgments
We thank colleagues from the Mechtler lab, the IMP/IMBA Bioinformatics Group, the PRIDE Team, Dorothea Anrather and Florian Breitwieser for valuable discussions and support.
Footnotes
Author contributions: K.M. and R.H. designed research; G.D., B.Z.C., M.S., E.R., and R.H. performed research; G.D., B.Z.C., M.T., O.H., K.M., and R.H. analyzed data; G.D., K.M., and R.H. wrote the paper.
* The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013)/Grant Agreement Nos. 262067 - PRIME-XS and 241548 - MeioSys and from the Austrian Science Fund SFB F3402, TRP 308-N15, and P24685-B24.
 This article contains supplemental material.
This article contains supplemental material.
2 Diederichs and R. Herbst, unpublished data.
1 The abbreviations used are:
- NMS
- neuromuscular synapse
- AChR
- acetylcholine receptor
- CID
- collision-induced dissociation
- FA
- formic acid
- HCD
- higher-energy collisional dissociation
- IMAC
- immobilized metal affinity chromatography
- iTRAQ
- isobaric tags for relative and absolute quantitation
- Lrp4
- low-density lipoprotein receptor-related protein 4
- MuSK
- muscle-specific kinase
- RhoA
- Ras homolog gene family, member A
- RS
- Arg-Ser
- SCX
- strong cation exchange
- Src
- neuronal proto-oncogene tyrosine-protein kinase Src.
REFERENCES
- 1. Glass D. J., DeChiara T. M., Stitt T. N., DiStefano P. S., Valenzuela D. M., Yancopoulos G. D. (1996) The receptor tyrosine kinase MuSK is required for neuromuscular junction formation and is a functional receptor for agrin. Cold Spring Harb. Symp. Quant. Biol. 61, 435–444 [PubMed] [Google Scholar]
- 2. Kim N., Stiegler A. L., Cameron T. O., Hallock P. T., Gomez A. M., Huang J. H., Hubbard S. R., Dustin M. L., Burden S. J. (2008) Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 135, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang B., Luo S., Wang Q., Suzuki T., Xiong W. C., Mei L. (2008) LRP4 serves as a coreceptor of agrin. Neuron 60, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanes J. R., Lichtman J. W. (2001) Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2, 791–805 [DOI] [PubMed] [Google Scholar]
- 5. DeChiara T. M., Bowen D. C., Valenzuela D. M., Simmons M. V., Poueymirou W. T., Thomas S., Kinetz E., Compton D. L., Rojas E., Park J. S., Smith C., DiStefano P. S., Glass D. J., Burden S. J., Yancopoulos G. D. (1996) The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell 85, 501–512 [DOI] [PubMed] [Google Scholar]
- 6. Gautam M., Noakes P. G., Moscoso L., Rupp F., Scheller R. H., Merlie J. P., Sanes J. R. (1996) Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell 85, 525–535 [DOI] [PubMed] [Google Scholar]
- 7. Weatherbee S. D., Anderson K. V., Niswander L. A. (2006) LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development 133, 4993–5000 [DOI] [PubMed] [Google Scholar]
- 8. Gomez A. M., Burden S. J. (2011) The extracellular region of Lrp4 is sufficient to mediate neuromuscular synapse formation. Dev. Dyn. 240, 2626–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glass D. J., Apel E. D., Shah S., Bowen D. C., DeChiara T. M., Stitt T. N., Sanes J. R., Yancopoulos G. D. (1997) Kinase domain of the muscle-specific receptor tyrosine kinase (MuSK) is sufficient for phosphorylation but not clustering of acetylcholine receptors: required role for the MuSK ectodomain? Proc. Natl. Acad. Sci. U.S.A. 94, 8848–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuhrer C., Sugiyama J. E., Taylor R. G., Hall Z. W. (1997) Association of muscle-specific kinase MuSK with the acetylcholine receptor in mammalian muscle. EMBO J. 16, 4951–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herbst R., Burden S. J. (2000) The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 19, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou H., Glass D. J., Yancopoulos G. D., Sanes J. R. (1999) Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J. Cell Biol. 146, 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghazanfari N., Fernandez K. J., Murata Y., Morsch M., Ngo S. T., Reddel S. W., Noakes P. G., Phillips W. D. (2011) Muscle specific kinase: organiser of synaptic membrane domains. Int. J. Biochem. Cell Biol. 43, 295–298 [DOI] [PubMed] [Google Scholar]
- 14. Nizhynska V., Neumueller R., Herbst R. (2007) Phosphoinositide 3-kinase acts through RAC and Cdc42 during agrin-induced acetylcholine receptor clustering. Dev. Neurobiol. 67, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 15. Mazhar S., Herbst R. (2012) The formation of complex acetylcholine receptor clusters requires MuSK kinase activity and structural information from the MuSK extracellular domain. Mol. Cell. Neurosci. 49, 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villen J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 18. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 19. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 20. Breitwieser F. P., Colinge J. (2013) Isobar(PTM): a software tool for the quantitative analysis of post-translationally modified proteins. J. Proteomics 90, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breitwieser F. P., Muller A., Dayon L., Kocher T., Hainard A., Pichler P., Schmidt-Erfurth U., Superti-Furga G., Sanchez J. C., Mechtler K., Bennett K. L., Colinge J. (2011) General statistical modeling of data from protein relative expression isobaric tags. J. Proteome Res. 10, 2758–2766 [DOI] [PubMed] [Google Scholar]
- 22. Hartigan J. A., Wong M. A. (1979) Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C Appl. Stat. 28, 100–108 [Google Scholar]
- 23. Calvano S. E., Xiao W., Richards D. R., Felciano R. M., Baker H. V., Cho R. J., Chen R. O., Brownstein B. H., Cobb J. P., Tschoeke S. K., Miller-Graziano C., Moldawer L. L., Mindrinos M. N., Davis R. W., Tompkins R. G., Lowry S. F., Inflamm, and Host Response to Injury Large Scale Collab. Res P. (2005) A network-based analysis of systemic inflammation in humans. Nature 437, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 24. Chou M. F., Schwartz D. (2011) Biological sequence motif discovery using Motif-X. Curr. Protoc. Bioinf. 13, 13.15–13.24 [DOI] [PubMed] [Google Scholar]
- 25. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 26. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jorgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barsnes H., Vizcaino J. A., Eidhammer I., Martens L. (2009) PRIDE Converter: making proteomics data-sharing easy. Nat. Biotechnol. 27, 598–599 [DOI] [PubMed] [Google Scholar]
- 28. Cote R. G., Griss J., Dianes J. A., Wang R., Wright J. C., van den Toorn H. W., van Breukelen B., Heck A. J., Hulstaert N., Martens L., Reisinger F., Csordas A., Ovelleiro D., Perez-Rivevol Y., Barsnes H., Hermjakob H., Vizcaino J. A. (2012) The PRoteomics IDEntification (PRIDE) Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteomics 11, 1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kocher T., Pichler P., Schutzbier M., Stingl C., Kaul A., Teucher N., Hasenfuss G., Penninger J. M., Mechtler K. (2009) High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J. Proteome Res. 8, 4743–4752 [DOI] [PubMed] [Google Scholar]
- 30. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 31. Watty A., Neubauer G., Dreger M., Zimmer M., Wilm M., Burden S. J. (2000) The in vitro and in vivo phosphotyrosine map of activated MuSK. Proc. Natl. Acad. Sci. U.S.A. 97, 4585–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Till J. H., Becerra M., Watty A., Lu Y., Ma Y., Neubert T. A., Burden S. J., Hubbard S. R. (2002) Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure 10, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 33. Weston C., Gordon C., Teressa G., Hod E., Ren X. D., Prives J. (2003) Cooperative regulation by Rac and Rho of agrin-induced acetylcholine receptor clustering in muscle cells. J. Biol. Chem. 278, 6450–6455 [DOI] [PubMed] [Google Scholar]
- 34. Oude Weernink P. A., Schmidt M., Jakobs K. H. (2004) Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 500, 87–99 [DOI] [PubMed] [Google Scholar]
- 35. Proszynski T. J., Gingras J., Valdez G., Krzewski K., Sanes J. R. (2009) Podosomes are present in a postsynaptic apparatus and participate in its maturation. Proc. Natl. Acad. Sci. U.S.A. 106, 18373–18378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Proszynski T. J., Sanes J. R. (2013) Amotl2 interacts with LL5beta, localizes to podosomes and regulates postsynaptic differentiation in muscle. J. Cell Sci. 126, 2225–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goicoechea S. M., Arneman D., Otey C. A. (2008) The role of palladin in actin organization and cell motility. Eur. J. Cell Biol. 87, 517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mosevitsky M. I. (2005) Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 245, 245–325 [DOI] [PubMed] [Google Scholar]
- 39. Lu Z., Je H. S., Young P., Gross J., Lu B., Feng G. (2007) Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 177, 1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hallock P. T., Xu C. F., Park T. J., Neubert T. A., Curran T., Burden S. J. (2010) Dok-7 regulates neuromuscular synapse formation by recruiting Crk and Crk-L. Genes Dev. 24, 2451–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mittaud P., Camilleri A. A., Willmann R., Erb-Vogtli S., Burden S. J., Fuhrer C. (2004) A single pulse of agrin triggers a pathway that acts to cluster acetylcholine receptors. Mol. Cell. Biol. 24, 7841–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nita-Lazar A., Saito-Benz H., White F. M. (2008) Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics 8, 4433–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thingholm T. E., Jensen O. N., Larsen M. R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 44. Zhang G., Neubert T. A. (2011) Comparison of three quantitative phosphoproteomic strategies to study receptor tyrosine kinase signaling. J. Proteome Res. 10, 5454–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reich N. C. (2013) STATs get their move on. Jak-Stat 2, e27080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katz M., Amit I., Yarden Y. (2007) Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta 1773, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmidt N., Basu S., Sladecek S., Gatti S., van Haren J., Treves S., Pielage J., Galjart N., Brenner H. R. (2012) Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J. Cell Biol. 198, 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe T., Noritake J., Kakeno M., Matsui T., Harada T., Wang S., Itoh N., Sato K., Matsuzawa K., Iwamatsu A., Galjart N., Kaibuchi K. (2009) Phosphorylation of CLASP2 by GSK-3beta regulates its interaction with IQGAP1, EB1 and microtubules. J. Cell Sci. 122, 2969–2979 [DOI] [PubMed] [Google Scholar]
- 49. McMichael B. K., Scherer K. F., Franklin N. C., Lee B. S. (2014) The RhoGAP activity of myosin IXB is critical for osteoclast podosome patterning, motility, and resorptive capacity. PLoS One 9, e87402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olson E. N., Nordheim A. (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghosh G., Adams J. A. (2011) Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 278, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu A. K., Cheung J., Smith F. D., Ip F. C., Ip N. Y. (2001) Overexpression of muscle specific kinase increases the transcription and aggregation of acetylcholine receptors in Xenopus embryos. Brain Res. Mol. Brain Res. 96, 21–29 [DOI] [PubMed] [Google Scholar]
- 53. Lacazette E., Le Calvez S., Gajendran N., Brenner H. R. (2003) A novel pathway for MuSK to induce key genes in neuromuscular synapse formation. J. Cell Biol. 161, 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strochlic L., Cartaud A., Mejat A., Grailhe R., Schaeffer L., Changeux J. P., Cartaud J. (2004) 14-3-3 gamma associates with muscle specific kinase and regulates synaptic gene transcription at vertebrate neuromuscular synapse. Proc. Natl. Acad. Sci. U.S.A. 101, 18189–18194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deakin N. O., Turner C. E. (2008) Paxillin comes of age. J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Albiges-Rizo C., Destaing O., Fourcade B., Planus E., Block M. R. (2009) Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122, 3037–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang C., Jacobson K., Schaller M. D. (2004) MAP kinases and cell migration. J. Cell Sci. 117, 4619–4628 [DOI] [PubMed] [Google Scholar]
- 58. Schindeler A., Lavulo L., Harvey R. P. (2005) Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp. Cell Res. 307, 367–380 [DOI] [PubMed] [Google Scholar]
- 59. Dai Z., Luo X., Xie H., Peng H. B. (2000) The actin-driven movement and formation of acetylcholine receptor clusters. J. Cell Biol. 150, 1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu H., Xiong W. C., Mei L. (2010) To build a synapse: signaling pathways in neuromuscular junction assembly. Development 137, 1017–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cartaud A., Stetzkowski-Marden F., Maoui A., Cartaud J. (2011) Agrin triggers the clustering of raft-associated acetylcholine receptors through actin cytoskeleton reorganization. Biol. Cell 103, 287–301 [DOI] [PubMed] [Google Scholar]
- 62. Ohlendieck K. (2011) Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skeletal Muscle 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ohlendieck K. (2010) Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Rev. Proteomics 7, 283–296 [DOI] [PubMed] [Google Scholar]
- 64. Puente L. G., Carriere J. F., Kelly J. F., Megeney L. A. (2004) Comparative analysis of phosphoprotein-enriched myocyte proteomes reveals widespread alterations during differentiation. FEBS Lett. 574, 138–144 [DOI] [PubMed] [Google Scholar]
- 65. Puente L. G., Voisin S., Lee R. E., Megeney L. A. (2006) Reconstructing the regulatory kinase pathways of myogenesis from phosphopeptide data. Mol. Cell. Proteomics 5, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 66. Mate S. E., Brown K. J., Hoffman E. P. (2011) Integrated genomics and proteomics of the Torpedo californica electric organ: concordance with the mammalian neuromuscular junction. Skeletal Muscle 1, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gajendran N., Frey J. R., Lefkovits I., Kuhn L., Fountoulakis M., Krapfenbauer K., Brenner H. R. (2002) Proteomic analysis of secreted muscle components: search for factors involved in neuromuscular synapse formation. Proteomics 2, 1601–1615 [DOI] [PubMed] [Google Scholar]
- 68. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.