Abstract
N-terminal acetylation (Nt-acetylation) occurs on the majority of eukaryotic proteins and is catalyzed by N-terminal acetyltransferases (NATs). Nt-acetylation is increasingly recognized as a vital modification with functional implications ranging from protein degradation to protein localization. Although early genetic studies in yeast demonstrated that NAT-deletion strains displayed a variety of phenotypes, only recently, the first human genetic disorder caused by a mutation in a NAT gene was reported; boys diagnosed with the X-linked Ogden syndrome harbor a p.Ser37Pro (S37P) mutation in the gene encoding Naa10, the catalytic subunit of the NatA complex, and suffer from global developmental delays and lethality during infancy. Here, we describe a Saccharomyces cerevisiae model developed by introducing the human wild-type or mutant NatA complex into yeast lacking NatA (NatA-Δ). The wild-type human NatA complex phenotypically complemented the NatA-Δ strain, whereas only a partial rescue was observed for the Ogden mutant NatA complex suggesting that hNaa10 S37P is only partially functional in vivo. Immunoprecipitation experiments revealed a reduced subunit complexation for the mutant hNatA S37P next to a reduced in vitro catalytic activity. We performed quantitative Nt-acetylome analyses on a control yeast strain (yNatA), a yeast NatA deletion strain (yNatA-Δ), a yeast NatA deletion strain expressing wild-type human NatA (hNatA), and a yeast NatA deletion strain expressing mutant human NatA (hNatA S37P). Interestingly, a generally reduced degree of Nt-acetylation was observed among a large group of NatA substrates in the yeast expressing mutant hNatA as compared with yeast expressing wild-type hNatA. Combined, these data provide strong support for the functional impairment of hNaa10 S37P in vivo and suggest that reduced Nt-acetylation of one or more target substrates contributes to the pathogenesis of the Ogden syndrome. Comparative analysis between human and yeast NatA also provided new insights into the co-evolution of the NatA complexes and their substrates. For instance, (Met-)Ala- N termini are more prevalent in the human proteome as compared with the yeast proteome, and hNatA displays a preference toward these N termini as compared with yNatA.
Up to 85% of soluble eukaryotic proteins carry an N-terminal acetyl group at their N terminus, which is the result of a co-translational protein modification referred to as N-terminal protein acetylation (Nt-acetylation) or Nα-acetylation (1). This presumed irreversible protein modification is catalyzed by a specific category of the GCN5-related N-acetyltransferase domain containing superfamily of acetyltransferases; the ribosome associated N-terminal acetyltransferases or NATs1 (2). NATs catalyze the acetyl transfer from acetyl coenzyme A (Ac-CoA) to a primary α-amine of the first amino acid residue of a nascent protein chain. In eukaryotes, NATs are composed of at least one catalytic subunit and mainly target different substrate N termini based on their N-terminal sequences (3).
To date, five human NATs hNatA, hNatB, and hNatC; constituting the major human NAT complexes, and hNatD and hNatF have been identified and their substrate specificity characterized (1, 4–8). In addition, a putative hNatE complex has been described (9–10). Except for NatF, which is only expressed in higher eukaryotes (1), the substrate specificity profiles of the NatA-E complexes seem to be conserved among eukaryotes (5–9, 11–13).
Contrary to the original assumption that Nt-acetylation protected proteins from degradation (14), it was more recently demonstrated that this modification creates specific degradation signals (termed Ac/N-degrons) in cellular proteins, thereby diversifying this original view substantially. These degrons target at least some Nt-acetylated proteins for the conditional degradation by a novel branch of the N-end rule pathway, an ubiquitin-dependent proteolytic system (15–16). In addition, numerous reports implicate Nt-acetylation in cellular differentiation, survival, metabolism, and proliferation, thereby linking it to cancer (17–18). As such, Nt-acetylation is now linked to a whole range of molecular implications including protein destabilization and degradation by the Nt-acetylation dependent recruitment of ubiquitin ligases (15–16), protein translocation (19), membrane attachment (20), and protein complex formation (21).
Among all characterized NATs, NatA displays the broadest substrate specificity profile and thus represents the primary NAT in terms of substrate N termini as it is responsible for the Nt-acetylation of the methionine aminopeptidase (MetAP) iMet-processed serine, threonine, alanine, glycine, and valine starting N termini (3). The human NatA complex is composed of two essential subunits; the catalytic subunit hNaa10 (hARD1) and the regulatory subunit hNaa15 (NATH/hNAT1) (4). Deregulations of hNaa10 and/or NatA expression have been linked to various signaling molecules including hypoxia inducible factor-1α, DNA methyltransferase1/E-cadherin, β-catenin/cyclin D1, and Bcl-xL, showing its involvement in hypoxia, tumorigenesis, cell cycle progression, and apoptosis (17, 22–26).
Recently, the first structures of NATs and a NAT-complex were solved, providing a molecular understanding of the sequence specific Nt-acetylation of protein N termini (27–30). Structural analyses of noncomplexed Naa10 and NatA from Schizosaccharomyces pombe reveal an allosteric modulator function of Naa15 in steering Naa10 specificity and provide a rational for the distinctive substrate specificity profiles observed when assaying non-complexed versus complexed Naa10 (10, 27), with both forms co-existing in cells (10). In particular, three essential catalytic Naa10 residues were found to be incorrectly positioned in non-complexed Naa10, while these shift into the active site in Naa15-complexed Naa10, thereby permitting canonical NatA-mediated Nt-acetylation. Interestingly, noncomplexed Naa10 was shown to efficiently Nt-acetylate glutamate and aspartate starting N termini, whereas poorly acetylating canonical NatA type N termini (10). The study of Liszczak et al. further showed that NatA substrate binding specificity was coupled to the catalytic mechanism being used (27). More specifically, an essential glutamate residue (Glu24 in the protein accession Q9UTI3 (Swiss-Prot)) involved in catalysis, precludes methionine from entering the specificity pocket, whereas cognate NatA substrate N-terminal residues can easily be accommodated. Interestingly, and in contrast to NatA, both wild-type Naa10 and Glu24 mutated Naa10 (Naa10 E24A) were still capable of Nt-acetylating acidic amino acid starting N termini, most likely because of the substrate side-chain carboxyl moiety acting as a functional replacement group in the process of catalysis, whereas essentially no activity could be observed when probing a cognate NatA substrate (27).
Early yeast studies demonstrated that strains with mutated or deleted NAT genes were viable, but displayed a number of different phenotypes (31). For NatA, the first phenotypes described were defects in sporulation, mating, and entry into stationary phase when NAA10 (ARD1) was mutated (32). Four years later, the overlapping phenotypes of NAA10 and NAA15 (NAT1) mutant strains, revealed, along with other data, that Naa10 and Naa15 are in fact components of the NatA acetyltransferase complex (33–34). As compared with NatA phenotypes, NatB phenotypes are more severe, including slow growth and defects in mitochondrial inheritance (35–36). NatC subunits were initially found to be essential for propagation of the l-A dsRNA virus, and further for growth on nonfermentable carbon sources (37–39). The first reports implicating NAT gene point mutations in human genetic disorders only recently emerged. More specifically, two different point mutations in the X-linked NAA10 gene were both found to cause developmental delays and were linked to the Ogden syndrome (S37P) (40) and intellectual disability (R116W) (41), highlighting the essential importance of NATs and protein Nt-acetylation in biology and disease. Further, in Caenorhabditis elegans (42), Drosophila melanogaster (43), and Trypanosoma brucei (44), Naa10 was proven to be essential and, strengthened by the observed detrimental effects of NAA10 mutations (40–41), the NAA10 gene function is also believed to be essential in human.
Ogden syndrome boys harboring the p.Ser37Pro variant in the gene encoding Naa10 are characterized by craniofacial abnormalities, failure to thrive, developmental delay, hypotonia, cardiac arrhythmias, cryptorchidism, and an aged appearance, ultimately resulting in mortality during infancy (40). Although this mutation was shown to significantly impair Naa10 catalytic activity in vitro, we here assessed the influence and functional in vitro and in vivo consequences of this mutation on NatA complex formation and NatA activity in a yeast model. By phenotypic screening in yeast, we show that hNaa10 S37P displays a significantly impaired functionality in vivo. Further, using immunoprecipitation, we show that the human Naa10-Naa15 complex formation is negatively affected by the S37P mutation, and that immunoprecipitated hNatA S37P also displays a reduced in vitro catalytic activity as compared with wild-type hNatA. Finally, quantitative Nt-acetylome analyses suggest that reduced Nt-acetylation of one or more target substrates contributes to the pathogenesis of the Ogden syndrome.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Creation and Growth of Yeast Strains
The following expression vectors were used to create the isogenic yeast strains, pBEVY-URA, pBEVY-URA-hNAA15-hNAA10 (5) and pBEVY-URA-hNAA15-hNAA10 S37P. The pBEVY-URA-hNAA15-hNAA10 S37P expression vector was created from the pBEVY-URA-hNAA15-hNAA10 by site directed mutagenesis according to the manufacturer's instructions (QuikChange Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA) using the following primers; hNAA10 T109C forward: 5′-CTTCTACCATGGCCTTCCCTGGCCCCAGCTC-3′ and hNAA10 T109C reverse: 5′-GAGCTGGGGCCAGGGAAGGCCATGGTAGAAG-3′. The correctness of all plasmids was verified by DNA sequencing. S. cerevisiae strains were made as follows. The haploid MATa strain BY4742 (Y10000, EUROSCARF) was transformed with an empty expression vector pBEVY-URA and used as a control strain termed yNatA. Y10976 (EUROSCARF) with NAA10::kanMX4 was transformed with an empty expression vector pBEVY-URA and used as a strain without yNatA termed yNatAΔ. Y10976 was transformed with an expression vector pBEVY-URA-hNAA15-hNAA10 and used as a strain expressing hNatA, but not yNatA termed y[hNatA], and finally Y10976 was transformed with an expression vector pBEVY-URA-hNAA15-hNAA10 S37P, and used as a strain expressing human NatA with a mutated hNaa10 S37P, but not expressing yeast NatA termed y[hNatA S37P]. Yeast strains were selected and grown at 30 °C on plates lacking uracil. After pre-culturing in SC-medium lacking uracil (SC-Ura) medium, strain phenotypes were determined on YPD and YPD containing 0.3% caffeine or 0.2 μg/ml cyclohemimide (CHX).
Immunoprecipitation of hNatA
Yeast cultures (100 ml cultures) were grown in SC-Ura at 30 °C and harvested in the exponential growth phase (OD600 nm = 3.0). Cells were pelleted, washed two times in PBS (135 mm NaCl, 2.7 mm KCl, 7 mm Na2HPO4), and dissolved in 500 μl yeast lysis buffer (50 mm Tris-HCl pH 7.6, 12 mm EDTA, 250 mm NaCl, 140 mm Na2HPO4 supplemented with EDTA-free protease inhibitor (Roche Diagnostics)). To lyse yeast cells, 0.3 g of acid washed 425–600 μm glass beads (Sigma) were added to each sample and vortexed for 10 times 30 s (placing them on ice for 30 s in between). The cell lysates were centrifuged at 7500 × g for 10 min at 4 °C to remove cell debris. 3 μg of anti-hNaa15 was added to the supernatant and incubated for 3 h on a rotating wheel at 4 °C. 100 μl of Pierce Protein AG Magnetic Beads (Thermo scientific) was added to the mixture and this mixture was then incubated overnight on a rotating wheel at 4 °C. The beads were isolated using a magnet, washed 3 times in yeast lysis buffer, 2 times in NAT buffer (50 mm Tris-HCl pH 7.4, 10% glycerol, 1 mm EDTA) and dissolved in 250 μl NAT buffer. For SDS-PAGE and Western blotting, SDS sample buffer was added and samples boiled for 10 min at 95 °C. Anti-Naa15, anti-Naa10 and anti-β-tubulin were used to visualize hNaa15, hNaa10 and β-tubulin (loading control) respectively. Protein bands imaged by a ChemiDoc XRS system (BioRad) were quantified by densitometric scanning using the ImageLab 3.0 software from BioRad.
Nt-acetyltransferase Assay
Beads containing hNatA complexes were mixed with 300 μm acetyl-CoA, 300 μm of the SESS-starting substrate polypeptide NH2-SESSSKSRWGRPVGRRRRPVRVYP-COOH and NAT-buffer and incubated for 30 min at 37 °C in a shaker. The reaction was stopped by adding 6 μl 10% TFA to the mixture and acetylated peptides were quantified by RP-HPLC as described previously (45). To calculate the NAT-activity per Naa10 molecule, the measured NAT activity of hNatA was set to 1, and the NAT-activity of hNatA S37P was correlated with the relative amount of hNaa10 present in each sample as determined by Western blotting (supplemental Fig. S1). Western blots were imaged by a ChemiDoc XRS from BioRad and the intensity of the hNaa10 bands was determined by relative densitometry scanning using the ImageLab 3.0 software (BioRad). To account for the variance in the determination of hNaa10 present in each sample, standard deviations were calculated by the equation where A and dA represent the measured product formation and standard deviation from the in vitro acetyltransferase assay, and B and dB the relative fold difference of the hNaa10 band intensities and the standard deviation from these measurements respectively.
Preparation of Yeast Cell Extracts for N-terminal COFRADIC
The yeast proteomes were prepared from 300 ml of culture at OD600 nm = 3.0 as described (1, 5). Cell lysis was essentially performed as described above. Here, one EDTA-free protease inhibitor mixture tablet was used per 100 ml of lysis buffer, and 1 ml of lysis buffer was used for a pellet resulting from 300 ml of yeast culture. The cleared lysates were subsequently analyzed by N-terminal COFRADIC analysis (1, 46). Briefly, solid guanidinium hydrochloride was added to a final concentration of 4 m to denature all proteins. Subsequently, proteins were reduced and alkylated simultaneously, using TCEP (1 mm final concentration (f.c.)) and iodoacetamide (2 mm f.c.) respectively, for 1 h at 30 °C in the dark. Subsequent steps of the N-terminal COFRADIC setups analyzed were performed as described previously (47). The proteome was digested overnight at 37 °C with sequencing-grade, modified trypsin (Promega, Madison, WI) (enzyme/substrate of 1/100, w/w).
LC-MS/MS Analysis and Data Processing
The obtained peptide mixtures were introduced into an LC-MS/MS system, the Ultimate 3000 (Dionex, Amsterdam, The Netherlands) in-line connected to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Samples were first loaded on a trapping column (made in-house, 100 μm internal diameter (I.D.) x 20 mm, 5 μm beads C18 Reprosil-HD, Dr. Maisch). After back-flushing from the trapping column, the sample was loaded on a reverse-phase column (made in-house, 75 μm I.D. × 150 mm, 5 μm beads C18 Reprosil-HD, Dr. Maisch). Peptides were loaded with solvent A (0.1% trifluoroacetic acid, 2% acetonitrile), and were separated with a linear gradient from 2% solvent A' (0.05% formic acid) to 55% solvent B' (0.05% formic acid and 80% acetonitrile) at a flow rate of 300 nL/min followed by a wash reaching 100% solvent B'. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the six most abundant peaks in a given MS spectrum. Full scan MS spectra were acquired in the Orbitrap at a target value of 1E6 with a resolution of 60,000. The six most intense ions were then isolated for fragmentation in the linear ion trap, with a dynamic exclusion of 60 s. Peptides were fragmented after filling the ion trap at a target value of 1E4 ion counts. From the MS/MS data in each LC run, Mascot Generic Files were created using the Mascot Distiller software (version 2.3.01, Matrix Science). Although generating these peak lists, grouping of spectra was allowed with a maximum intermediate retention time of 30 s and a maximum intermediate scan count of five was used where possible. Grouping was done with 0.005 Da precursor tolerance. A peak list was only generated when the MS/MS spectrum contained more than 10 peaks. There was no de-isotoping and the relative signal to noise limit was set at two. These peak lists were then searched with the Mascot search engine (Matrix Science) using the Mascot Daemon interface (version 2.3, Matrix Science).
Spectra were searched against the baker's yeast (S. cerevisiae) Swiss-Prot database (version 2011_04 of the UniProtKB/Swiss-Prot protein database containing 7346 sequence entries (526,969 sequences in total)). 13C2D3-acetylation of lysine side-chains, carbamidomethylation of cysteine and methionine oxidation to methionine-sulfoxide were set as fixed modifications for the N-terminal COFRADIC analyses. Variable modifications were 13C2D3-acetylation and acetylation of protein N termini. Pyroglutamate formation of N-terminal glutamine was additionally set as a variable modification. Mass tolerance on precursor ions was set to 10 ppm (with Mascot's C13 option set to 1) and on fragment ions to 0.5 Da. Endoproteinase semi-Arg-C/P (Arg-C specificity with arginine-proline cleavage allowed) was set as enzyme allowing no missed cleavages. The peptide charge was set to 1+, 2+, 3+ and instrument setting was put to ESI-TRAP. Only peptides that were ranked one and scored above the threshold score, set at 99% confidence, were withheld. The estimated false discovery rate by searching decoy databases (a shuffled version of the yeast Swiss-Prot database made by the DBToolkit algorithm (48)) was found to lie between 0.89% and 1.17% on the spectrum level. Quantification of the degree of Nt-Acetylation was performed as described previously (1). All data management was done in ms_lims (49). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (50) with the data set identifier PXD000316 and DOI 10.6019/PXD000316 and PRIDE accessions 29985–29988.
RESULTS
Human NatA S37P Only Partially Rescues Yeast NatA-Δ Phenotypes
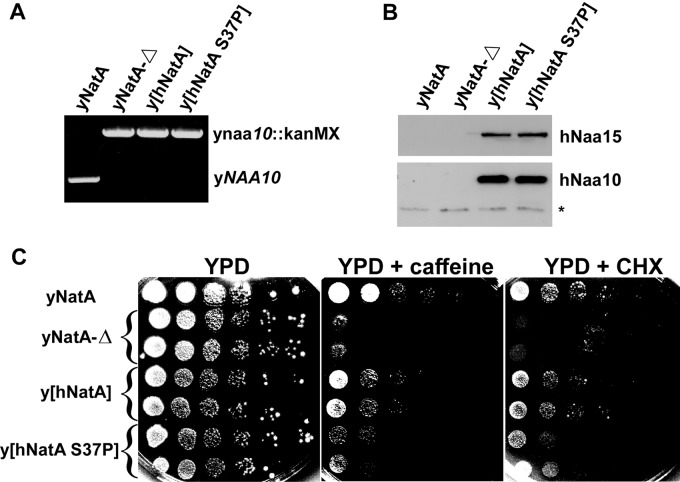
We previously showed that heterologous combinations of human and yeast NatA subunits neither produced active NatA nor complemented yNatA-Δ phenotypes (5). Therefore and to determine if ectopically expressed hNatA S37P is capable of suppressing the yeast NatA-Δ phenotypes, hNAA15 was expressed with hNAA10 or hNAA10 S37P in yeast, to respectively generate a yeast strain expressing hNatA (y[hNatA]) or mutant human NatA (y[hNatA S37P]) (Figs. 1A and 1B). We found that, in contrast to hNatA expression, phenotype profiling of hNatA S37P expression showed a reduced suppression of the observed NatA-Δ phenotypes (Fig. 1C). More specifically, sensitivity of the NatA-Δ yeast to caffeine and cycloheximide (CHX) was nearly completely suppressed by hNatA expression, whereas hNatA S37P expression resulted in a reduced complementation, suggesting a diminished functionality of hNatA S37P in vivo.
Fig. 1.
Human hNatA S37P expression in yeast only partially suppresses the yeast NatA-Δ Phenotypes. A, yNAA10-specific PCR confirming the ynaa10::kanMX deletion cassette in the yNatA-Δ, y[hNatA] and y[hNatA-S37P] strains. B, The yeast strains were analyzed by SDS-PAGE and Western blotting using anti-hNaa10 and anti-hNaa15 antibodies, confirming equal expression of the human NatA subunits in the y[hNatA] and y[hNatA S37P] strains (an unspecific band served as loading control (indicated by an asterisk)). C, Strains were grown in synthetic medium (-URA) overnight until the exponential phase of growth and equal cell numbers were inoculated in a YPD-pre-culture before plating of equal amounts of cells for growth in a 10-fold dilution series on YPD, YPD + 0.3% caffeine, or YPD + 0.2 μg/ml cycloheximide (CHX) for 3 days at 30 °C (yNatA, yNatA-Δ (2 clones), y[hNatA] (2 clones) and y[hNatA S37P] (2 clones)).
hNatA S37P Displays a Reduced hNaa10-hNaa15 Complex Formation, and a Reduced Catalytic Activity In Vitro
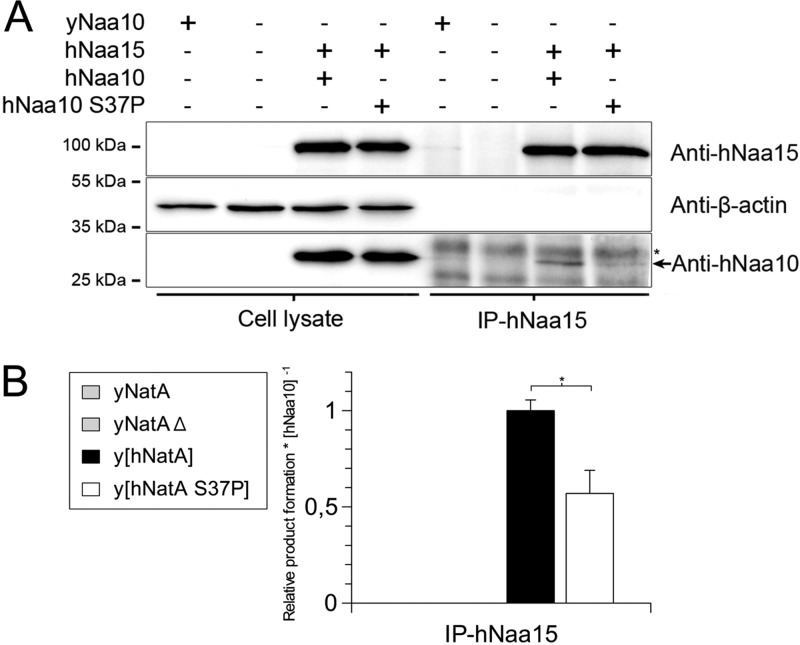
Yeast studies revealed that the catalytic subunit Naa10 associates with the ribosome through its interaction with the auxiliary subunit Naa15 where it is brought in close proximity to its nascent polypeptide substrates (51). Furthermore, the recent structure published by Liszczak et al. demonstrated that Naa10 binds in a tight binding pocket of Naa15 and that complex formation alters the catalytic site and activates Naa10's catalytic activity (27). To investigate whether the lack of full NatA-Δ phenotype complementation by hNaa10 S37P and hNaa15 was due to a lack of proper complex formation and/or NatA catalytic capacity, we performed immunoprecipitation of the hNatA complex from the yeast strains expressing the mutant or wild-type complex using anti-hNaa15 antibodies. Immunoprecipitation experiments clearly show that hNaa10 and hNaa15 are capable of forming a complex when overexpressed in yeast (Fig. 2A). However, the mutant hNaa10 S37P is more than twofold less capable of forming this functional hNatA complex as determined by anti-hNaa15 immunoprecipitation followed by Western blotting detecting co-immunoprecipitated hNaa10 (Fig. 2A and supplemental Fig. S1). Furthermore, we assayed the Nat-activity of the immunoprecipitated hNatA complexes (per Naa10 molecule) by an in vitro acetyltransferase assay. A significant decrease in product formation was measured for hNatA S37P (Fig. 2B) suggesting that in addition to hNaa10 S37P being less capable of forming a complex with hNaa15, the hNatA S37P complex also has a reduced catalytic activity in comparison to the hNatA complex.
Fig. 2.
Immunoprecipitated hNatA S37P shows a reduced complex formation and reduced NAT activity. Yeast strains were grown, harvested, lysed and subjected to hNatA immunoprecipitation using anti-hNaa15 antibodies and activity measurements. A, Immunoprecipitation of the hNatA complex with antibodies against the auxiliary subunit hNaa15. Immunoprecipitates and cell lysates were analyzed by SDS-PAGE and Western blotting. β-actin or an unspecific band (indicated with an asterisk) served as loading controls for the cell lysates and immunoprecipitates respectively. B, NAT-activity (per Naa10-molecule) of hNaa10 WT and S37P in complex with hNaa15. hNatA complexes were immunoprecipitated with anti-hNaa15 and used for in vitro NAT-activity measurements. The amount of Naa10 present in each sample was detected by Western blotting and densitometric scanning (Figure S1). NAT-activity measurements were performed in triplicates. Error bars include the standard deviation from both NAT-activity measurements and the densitometric scanning of Naa10 bands. Statistically significant results are indicated with an asterisk (p < 0.05). The p value was calculated using Student's t test.
Quantitative Nt-acetylome Analyses Reveal a Reduced Degree of Nt-acetylation for the Majority of NatA Substrates in the hNatA S37P Yeast Strain
Because phenotypic characterization in yeast disclosed a significantly impaired functionality of hNatA S37P in addition to a reduced complex formation and enzymatic activity when assaying the immunoprecipitated hNatA S37P, the in vivo outcome on the steady-state levels of protein Nt-acetylation was studied. Using N-terminal COFRADIC, the in vivo Nt-acetylomes of the y[hNatA] and y[hNatA S37P] yeast proteomes were quantitatively analyzed next to the yNatA and yNatA-Δ Nt-acetylomes. In addition to the study of species-specific NatA substrate repertoires at the proteome-wide level, this study points to potential differences in the in vivo substrate specificity profiles of the wild-type yNatA, hNatA and mutant hNatA complex. Here, in vitro 13C2D3-acetylation is used to discriminate between in vivo Nt-acetylated and free N termini as this introduces a 5 Da mass spacing between the free and Nt-acetylated form of an N-terminal peptide, enabling the calculation of the extent of Nt-acetylation (5, 52). Overall, in the four yeast strains analyzed, 1608 unique yeast N termini originating from 1277 yeast proteins were identified (supplemental Table S1). Here, an N terminus is defined as a peptide that is either in vivo Nt-acetylated or in vitro 13C2D3-acetylated (i.e. an in vivo free N terminus) and starts at position 1 or 2 of the Swiss-Prot annotated protein sequence, or an in vivo Nt-acetylated peptide with a starting position beyond 2. Of the 1,608 N termini, 1212 N termini were in compliance with the rules of Nt-acetylation (53) and initiator methionine (iMet) processing (supplemental Table S1). Of these, 1056 started at position 1 or 2 (1011 proteins), whereas 156 started beyond position 2 (145 proteins), the latter indicative of alternative translation initiation (54). 396 N termini did not comply with the rules of Nt-acetylation and iMet processing and thus indicate post-translational Nt-acetylation (54) or non-AUG translation initiation (55) among others. Interestingly, the majority of NatA type substrate N termini falling in this category were shown to be modified in a NatA dependent fashion (see below). 607 unique N termini were identified in all four setups analyzed (38% of all unique protein N termini identified) (supplemental Table S1). The difference in the degree of Nt-acetylation could be calculated for 97% to 99% of the N termini identified in the different yeast N-terminomes (supplemental Table S1). Considerable variations in the degree of Nt-acetylation, defined as a minimum difference of 10% (1), of yeast N termini with a database annotated start position were almost exclusively confined to the NatA type class of N termini. For the overall majority of these, Nt-acetylation falls to undetectable levels in the yNatA-Δ setup (i.e. the fraction of (partially) Nt-acetylated NatA type N termini drops from 75% to about 3%, (Table I and supplemental Table S1)), indicative for the fact that for most of these, no NatA-like redundant activity can be observed. In total, 576 database annotated NatA type N termini were identified (or 55% of all database annotated N termini identified). For 402 (or 70%) of these, the NatA dependence of Nt-acetylation could be determined pointing to 332 genuine NatA substrate N termini in yeast (i.e. 83% of all determined NatA type N termini represent yNatA and/or hNatA (S37P) substrate(s) and thus indicative for the fact that the vast majority of the nondetermined NatA type N termini also represent NatA substrate N termini (supplemental Table S1)). Further, and in compliance with the (X)PX rule of Nt-acetylation stating the prevention of Nt-acetylation under all circumstances (53), all determined NatA type X-Pro-starting N termini were found to be N-terminal free under all conditions analyzed (Table I and supplemental Table S1). Here, NatA substrate N termini were considered when corresponding N-terminal peptides were found to be completely nonacetylated in the yNatA-Δ strain as compared with the yNatA, y[hNatA] and/or y[hNatA S37P] strains, or displayed a shift in Nt-acetylation larger than 10% as compared with the control or hNatA versus hNatA S37P sample (supplemental Table S1). When correlating the degrees of Nt-acetylation of the NatA type N termini identified in the hNatA versus the hNatA S37P N-terminome data sets (Fig. 3), a general decrease in the degree of Nt-acetylation can be observed, indicative for the fact that the mutation reduces the efficiency of hNatA-dependent Nt-acetylation (Figs. 3 and 4), an observation matching the phenotypic analyses and reduced enzymatic activity of the complex in vitro (Figs. 1 and 2).
Table I. Overview of N-terminal acetylation in the yeast N-terminomes of yNatA, yNatA-Δ, y[hNatA] and y[hNatA S37P]. Only Swiss-Prot database annotated N-termini (start = 1 or 2) of which the degree of Nt-Acetylation could be univocally calculated/determined in the respective setups analyzed and were compliant with the rules of N-terminal acetylation and iMet processing (54) were used for the overall calculation of Nt-Acetylation.
| yNatA |
yNatA-Δ |
y[hNatA] |
y[hNatA S37P] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full (%) | Full + partial (%) | Total | Full (%) | Full + partial (%) | Total | Full (%) | Full + partial (%) | Total | Full (%) | Full + partial (%) | Total | |
| NatA | ||||||||||||
| A- | 20.9 | 64.2 | 67 | 0.0 | 0.0 | 63 | 1.5 | 69.2 | 65 | 0.0 | 69.2 | 65 |
| C- | 0.0 | 0.0 | 1 | 0.0 | 0.0 | 2 | 0.0 | 0.0 | 4 | 0.0 | 0.0 | 2 |
| G- | 0.0 | 14.3 | 28 | 0.0 | 0.0 | 29 | 0.0 | 27.6 | 29 | 0.0 | 16.7 | 30 |
| S- | 84.0 | 99.6 | 238 | 0.0 | 5.2 | 211 | 8.8 | 97.8 | 227 | 0.8 | 96.6 | 236 |
| T- | 13.7 | 68.6 | 51 | 0.0 | 0.0 | 52 | 0.0 | 60.0 | 45 | 0.0 | 60.4 | 48 |
| V- | 0.0 | 13.0 | 46 | 0.0 | 4.7 | 43 | 0.0 | 18.6 | 43 | 0.0 | 15.2 | 46 |
| Total | 51.2 | 75.2 | 431 | 0.0 | 3.2 | 400 | 5.1 | 74.9 | 413 | 0.5 | 73.4 | 427 |
| NatD | ||||||||||||
| S- (H2A) | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 |
| Total | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 | 100.0 | 0.0 | 1 |
| NatB | ||||||||||||
| MD- | 98.3 | 100.0 | 59 | 98.1 | 100.0 | 52 | 98.6 | 100.0 | 72 | 98.4 | 100.0 | 61 |
| ME- | 97.1 | 100.0 | 35 | 96.7 | 100.0 | 30 | 97.1 | 100.0 | 34 | 91.4 | 100.0 | 35 |
| MN- | 82.0 | 100.0 | 50 | 75.6 | 100.0 | 45 | 75.5 | 100.0 | 53 | 74.0 | 100.0 | 50 |
| MQ- | 38.5 | 84.6 | 13 | 18.8 | 81.3 | 16 | 25.0 | 85.0 | 20 | 33.3 | 86.7 | 15 |
| Total | 87.9 | 98.7 | 157 | 81.8 | 97.9 | 143 | 83.2 | 98.3 | 179 | 83.2 | 98.8 | 161 |
| NatC | ||||||||||||
| MF- | 54.5 | 72.7 | 11 | 42.9 | 42.9 | 7 | 45.5 | 72.7 | 11 | 40.0 | 60.0 | 10 |
| MI- | 5.3 | 52.6 | 19 | 5.6 | 38.9 | 18 | 10.0 | 50.0 | 20 | 5.0 | 45.0 | 20 |
| ML- | 29.0 | 38.7 | 31 | 25.0 | 30.0 | 20 | 32.4 | 48.6 | 37 | 26.7 | 36.7 | 30 |
| MY- | 20.0 | 50.0 | 10 | 28.6 | 57.1 | 7 | 20.0 | 60.0 | 10 | 10.0 | 70.0 | 10 |
| Total | 25.4 | 49.3 | 71 | 21.2 | 38.5 | 52 | 26.9 | 53.8 | 78 | 20.0 | 47.1 | 70 |
| None | ||||||||||||
| P- | 0.0 | 0.0 | 31 | 0.0 | 0.0 | 31 | 0.0 | 0.0 | 29 | 0.0 | 0.0 | 26 |
| MP- | 0.0 | 0.0 | 2 | 0.0 | 0.0 | 1 | 0.0 | 0.0 | 3 | 0.0 | 0.0 | 1 |
| Total | 0.0 | 0.0 | 33 | 0.0 | 0.0 | 32 | 0.0 | 0.0 | 32 | 0.0 | 0.0 | 27 |
| Other | ||||||||||||
| MA- | 0.0 | 25.0 | 4 | 0.0 | 0.0 | 2 | 0.0 | 66.7 | 3 | 0.0 | 33.3 | 3 |
| MG- | 0.0 | 25.0 | 4 | 0.0 | 33.3 | 3 | 0.0 | 50.0 | 2 | 0.0 | 66.7 | 3 |
| MH- | 0.0 | 0.0 | 1 | 0.0 | 0.0 | 1 | ||||||
| MK- | 2.6 | 20.5 | 39 | 0.0 | 18.2 | 33 | 2.6 | 26.3 | 38 | 2.8 | 33.3 | 36 |
| MM- | 0.0 | 100.0 | 5 | 0.0 | 80.0 | 5 | 0.0 | 80.0 | 5 | 0.0 | 100.0 | 4 |
| MS- | 0.0 | 81.8 | 11 | 0.0 | 80.0 | 5 | 0.0 | 88.9 | 9 | 0.0 | 75.0 | 4 |
| MT- | 0.0 | 40.0 | 10 | 0.0 | 28.6 | 7 | 0.0 | 55.6 | 9 | 0.0 | 57.1 | 7 |
| MV- | 0.0 | 50.0 | 10 | 0.0 | 40.0 | 5 | 0.0 | 37.5 | 8 | 0.0 | 33.3 | 6 |
| Total | 1.2 | 39.8 | 83 | 0.0 | 31.1 | 61 | 1.3 | 44.0 | 75 | 1.6 | 44.4 | 63 |
| Grand total | 48.8 | 70.6 | 776 | 18.7 | 27.9 | 689 | 24.8 | 72.1 | 778 | 20.3 | 71.3 | 749 |
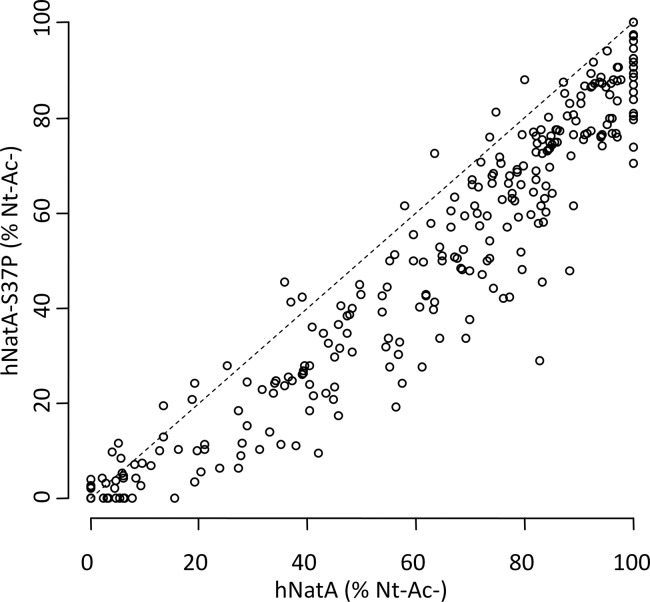
Fig. 3.
hNatA S37P expression in yeast significantly reduces the efficiency of NatA-mediated N-terminal acetylation at the proteome-wide level. Scatterplot displaying the correlation of the degrees of Nt-acetylation of the NatA type class N termini (i.e. Ser-, Ala-, Thr-, Val-, and Gly- starting N termini). Correlation of the degrees of Nt-acetylation of the determined NatA type class N termini identified in the yeast N-terminomes of a human NatA (hNatA) expressing strain (X-axis) and a S37P mutant human NatA (hNatA S37P) expressing strain (Y-axis) (n = 347). Only in 6.3% of the cases, the S37P mutation causes an increased degree of Nt-Ac as compared with the wild type hNatA whereas in 24.5% about equal levels of Nt-Ac (less than 10% difference in the degree of Nt-Ac) could be observed and thus for the majority of NatA-substrates (69.2% of NatA type N termini), the S37P mutation causes a decrease in the levels of Nt-Ac.
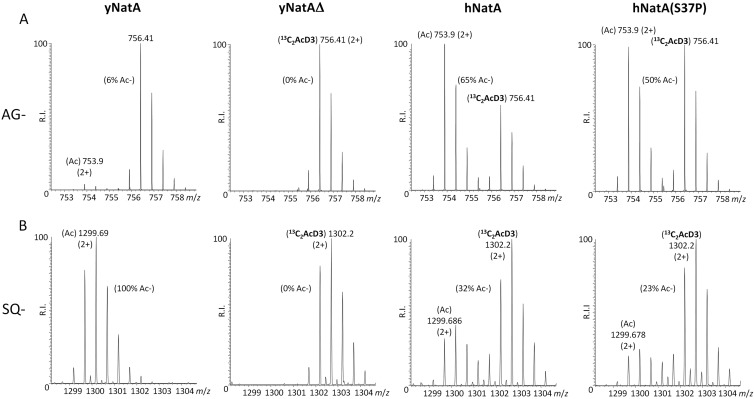
Fig. 4.
Nt-acetylome analyses reveal a reduced degree of Nt-acetylation for the majority of NatA substrates in the hNatA S37P as compared with the hNatA yeast setup. MS-spectra from the AG- (A) and SQ- (B) starting N-terminal peptides (doubly charged precursors) of the Pre-rRNA-processing protein ESF1 (2AGENPKKEGVDAR14) and the TPR repeat-containing protein associated with Hsp90 (2SQFEKQKEQGNSLFKQGLYR21) demonstrate these to be genuine yNatA substrate N termini as these were found to be partially (6%) or fully Nt-acetylated and 0% Nt-acetylated in the control and yNatA-Δ yeast proteomes, respectively. Two distinguishable isotopic envelopes could clearly be distinguished in the y[hNatA] and y[hNatA S37P] yeast strains [i.e. the in vivo acetylated (Ac) and in vitro 13C2 and trideutero-acetylated forms (13C2D3)] (right four panels) indicative for the fact that these N termini are only partially in vivo Nt-acetylated. The AG- and SQ- starting N-terminal peptides were found to be 65 and 50%, and 32 and 23% Nt-acetylated in the hNatA and hNatA S37P setups respectively, thus showing a reduced degree of Nt-acetylation of respectively 15 and 9%.
Species-specific NatA Nt-acetylation Substrate Specificity Profiles Indicate a Co-evolution of NatA Complexes With Their Species-matching Substrate Pools
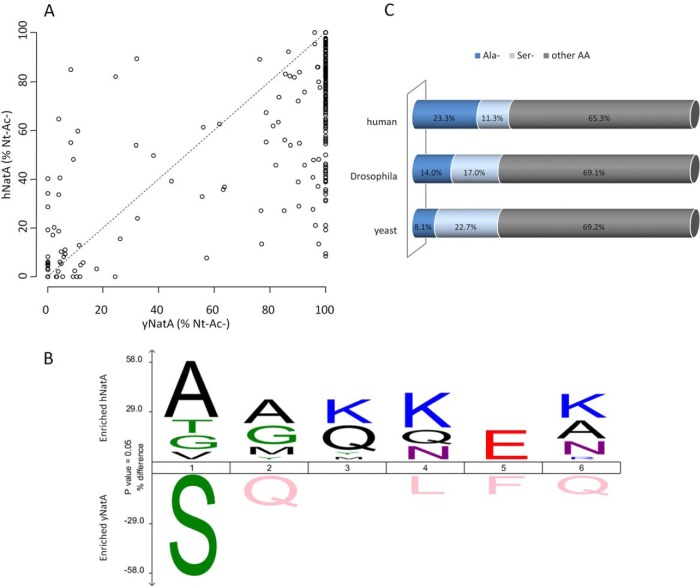
Related to the various orthologs/mutant NatA complexes (i.e. yNatA, hNatA versus hNatA S37P) studied alongside their substrate specificity profiles, and in analogy with what was reported previously (5), all yNatA substrate N termini displaying a degree of Nt-acetylation ≥20%, were also found to be Nt-acetylated in the yeast strain ectopically expressing hNatA. In general, 60% of the yNatA substrates displayed a higher degree of Nt-acetylation when compared with the Nt-acetylation levels of hNatA substrate N termini, whereas only 12% of the hNatA substrate N termini displayed an increased level of Nt-acetylation as compared with the Nt-acetylation levels observed for yNatA (supplemental Table S1, Figs. 4 and 5A). This indicates that despite the fact that the majority of substrates were found to be Nt-acetylated in both systems, differences in the efficiency of Nt-acetylation can be observed and that, under the conditions analyzed, in general yNatA seems to be the more efficient NatA complex in yeast as compared with hNatA, an observation in line with the phenotypic profiling of these strains. When taking two sets of NatA substrate N termini displaying the largest variation in Nt-acetylation observed in the systems analyzed (i.e. 30 N termini displaying the largest increase in Nt-acetylation in the hNatA system and 30 in the yNatA system), it is noteworthy that an increased Nt-acetylation potential of hNatA toward Ala-starting N termini is observed, whereas yNatA is more efficient in acetylating Ser-starting N termini (Fig. 5B). This observation is in line with our previously reported distribution analyses of Nat type substrates in the proteomes of yeast and human, that demonstrated that (Met-)Ala- N termini occur 3-fold more (up to 23%) in humans, whereas (Met-)Ser- N termini occurrence decreased from 23% to 11% when comparing yeast versus human proteomes (Fig. 5C) and (1). Overall, this hints to the fact that NatA substrate specificity/efficiency of Nt-acetylation has co-evolved with the repertoire of NatA type substrates expressed (Fig. 5).
Fig. 5.
Species-specific NatA substrate specificity profiles indicate a co-evolution of NatA complexes with their species-matching substrate pools. A, Scatterplot displaying the correlation of the degrees of Nt-acetylation of the NatA type class N termini (i.e. Ser-, Ala-, Thr-, Val-, and Gly- starting N termini). Correlation of the degrees of Nt-acetylation of the determined NatA type class N termini identified in the yeast N-terminomes of a yeast NatA (yNatA) (x-axis) and a human NatA (hNatA) expressing yeast strain (y-axis) (n = 333). In 11.7% of the cases, hNatA expression results in an increase of Nt-Ac as compared with the yNatA whereas in 27.6% of the cases about equal levels of Nt-Ac (less than 10% difference in the degree of Nt-Ac) could be observed. For 60.7% of identified NatA-substrates, hNatA expression resulted in a lower level of Nt-Ac as compared with the levels in control yeast (yNatA). B, A differential iceLogo (63) representation was created using two y/hNatA substrate subsets for which the hNatA (n = 30) or yNatA (n = 30) Nt-acetylation efficiency differed most notably (as deduced from the proteomics data and based on the degrees of Nt-acetylation calculated), thus representing the enriched and depleted residues in these respective NatA substrate sets. Statistically significant residues (p ≤ 0.05) are plotted with the size of the amino acid proportional to the difference observed in Nt-acetylation efficiency between human and yeast NatA. Multiple sequence alignments are given as 1 to 6 corresponding to the first 6 N-terminal residues of NatA substrates and with Nt-acetylation occurring at position 1. C, Stacked horizontal bar charts of the differential alanine and serine occurrence at position 2 of yeast (S. cerevisiae), fruit fly (D. melanogaster), and human (H. sapiens) iMet starting Swiss-Prot database protein entries (respectively 6620, 3168 and 20114 reviewed entries in the UniProtKB/Swiss-Prot protein database release 2013_07).
DISCUSSION
By introducing the human wild-type or mutant NatA S37P complex into yeast lacking NatA (NatA-Δ), we created a model system enabling the study of the Ogden syndrome mutant NatA, thereby revealing the functional impairment of hNatA S37P in vivo. Overall, the wild-type human NatA complex phenotypically complemented the NatA-Δ strain, whereas only a partial rescue was observed for the mutant NatA complex (Fig. 1C). Biochemical characterization of the mutant hNaa10 S37P previously showed a reduced NAT-activity as compared with hNaa10 WT (40). In this study, immunoprecipitation experiments revealed a significantly reduced hNaa10 S37P/hNaa15 complexation (as deduced from the difference in Naa10 present in anti-Naa15 immunoprecipitates), next to a reduced catalytic activity for the hNatA S37P complex in comparison with the wild type hNatA complex (Fig. 2). Overall this led us to postulate that multiple mechanisms (i.e. reduced complex formation and catalytic activity) might be at play lowering the ribosomal NatA-activity in the y[hNatA S37P] as compared with the y[hNatA] yeast strain. This postulation was strengthened by the quantitative Nt-acetylome analyses performed, revealing a reduced degree of Nt-acetylation for the majority of NatA substrates in the y[hNatA S37P] yeast strain (Fig. 3).
Noteworthy, and besides the hundreds of NatA substrate N termini identified, 12 database annotated NatA type substrate N termini were still found to be (partially) Nt-acetylated in the yNatA-Δ proteome sample analyzed (supplemental Table S1). When looking into their sequence profile it is of note that they predominantly display the NatD type substrate specificity (6). Of note, the SGGKG- starting N terminus of H2A was fully Nt-acetylated in all four setups analyzed in agreement with it being a NatD substrate (56). In contrast, four other SG-starting N termini were found to be partially Nt-acetylated in the yNatA-Δ setup with a reduced degree of Nt-acetylation as compared with the other setups, indicative for the fact that besides being Nt-acetylated by NatA, another Nat activity (presumably and most likely NatD) is responsible for these NatA redundant Nt-acetylation activities. Thus, some new NatD substrates might have been identified.
Further, next to the canonical NatA type substrate N termini identified, some N termini displaying an unknown Nat type substrate specificity (i.e. Met-starting N termini normally processed by MetAPs because their second amino acid has a small gyration radius (57–60)) also displayed a significant change in their degree of Nt-acetylation in one or more of the setups analyzed (supplemental Table S1). More specifically it seems that Met-Ser- and Met-Thr-starting N termini, whereas having a yNatA redundant yNAT capable of Nt-acetylating these (i.e. these were partially Nt-acetylated in the yNatA-Δ setup), hNatA and/or hNatA S37P expression increased their degree of Nt-acetylation. In fact, often these N termini showed an increased degree of Nt-acetylation in the hNatA setup(s) as compared with the yNatA setup. This could suggest that hNatA has evolved to more efficiently Nt-acetylate these types of N termini. In this respect, it is noteworthy that recombinant Naa10 and immunoprecipitated hNatA (from human HeLa cells) were found to inefficiently Nt-acetylate Met-starting oligopeptides (10). However another plausible though not mutually exclusive explanation might be a perturbed NatE activity, the second catalytic subunit enclosed in the NatA complex. First, human Naa50 was found to Nt-acetylate Met-Leu- starting N termini in vitro, a substrate specificity distinct from that of the NatA complex, and therefore considered a separate NAT type, NatE (9). Second, a proteome derived peptide library Nt-acetylation screen making use of recombinant hNaa50 confirmed and diversified this initial substrate specificity profile to hold Met-Lys, Met-Met and Met-Ala next to Met-Leu- oligopeptide hNaa50 substrates (10). Moreover, because siNatA knockdown studies in human HeLa cells previously demonstrated a reduced level of hNaa50 and the fact that in addition to the reduced degree of Nt-acetylation of proteins matching the NatA substrate specificity; siNatA knockdown showed the reduced acetylation of the MLGP-starting hnRNP F N terminus (5). Thus, it is not unlikely that in vivo Naa50 substrates might include Met-Ser- and Met-Thr- starting N termini and that the presence or absence of Naa10 and Naa15 affects Naa50 substrate acetylation. Interestingly, in Ogden syndrome cells, this implies that next to a decrease in NatA activity, hNaa10 S37P might indirectly affect NatE activity and potentially a reduction in Nt-acetylation of NatE substrates.
Further, besides the database annotated N termini identified, these datasets also holds numerous N termini not compliant with the current rules for N-terminal modifications; either no iMet is present and/or the NAT rules of Nt-acetylation do not match the canonical Nt-acetylation patterns (54). Various of these N termini belonging to the NatA type class of N termini and display a NatA dependence of Nt-acetylation, hinting to the fact that these N termini could represent either post-translational Nt-acetylation events (54), co-translational acting (di-)aminopeptidase activity, non-cognate (non-AUG) co-translational translation initiation events (55) or simply wrong database annotations. In this context it is interesting to note that, on proteolytic loss of their transit/signal sequence, the mature N termini of various mitochondrial localized proteins (such as the N termini of the mitochondrial ATPase-stabilizing factor 9 kDa (loss of 1–23 transit sequence) and the acyl carrier protein (loss of 1–36 transit sequence)) were here found to be (partially) Nt-acetylated in a NatA dependent manner (i.e. found to be 62 and 7% Nt-acetylated in the control setup, while being free in the yNatA-Δ setup) (supplemental Table S1) and thus indicative of the fact that NatA or Naa10 might act post-translationally in mitochondria on import, reminiscent of the post-translational NatA-like activities observed in plant plastids (11) and other eukaryotes with complex plastids (61), or alternatively act post-translationally in the cytosol on mitochondrial export. Because none of the eight proteins encoded by the mitochondrial genome were identified (62), their N-terminal modification status could not be assessed to ascertain the existence of mitochondrial Nat activity.
In conclusion, our yeast model supports a reduced NatA complex formation and catalytic capacity, and thereby a reduction of Nt-acetylation of NatA substrates as a cause for the Ogden syndrome.
Supplementary Material
Acknowledgments
We thank Professor Fred Sherman (1932–2013), University of Rochester, for his generosity and his support to establish S. cerevisiae as a model system. PVD is a Postdoctoral Fellow of the Research Foundation - Flanders (FWO-Vlaanderen).
Footnotes
Author contributions: P.V.D., S.I.S., K.G., and T.A. designed research; P.V.D., S.I.S., N.G., and T.A. performed research; P.V.D., S.I.S., and T.A. analyzed data; P.V.D., wrote the paper.
* This work was supported by research grants from the Research Council of Norway (Grant 197136 to TA), the Norwegian Cancer Society, the Bergen Research Foundation (BFS), The Western Norway Regional Health Authority, the Fund for Scientific Research Flanders (Belgium) (project numbers G.0269.13N (to PVD) and G.0440.10 (to KG)) and PRIME-XS (grant agreement number 262067, funded by the European Union 7th Framework Program).
 This article contains supplemental Fig. S1 and Table S1.
This article contains supplemental Fig. S1 and Table S1.
1 The abbreviations used are:
- NAT
- N-terminal acetyltransferase
- Ac-CoA
- Acetyl coenzyme A
- iMet
- Initiator methionine
- MetAP
- Methionine aminopeptidase
- NAA#
- N-alpha acetyltransferase # (gene/protein)
- Nt-acetylation
- N-terminal acetylation.
REFERENCES
- 1. Van Damme P., Hole K., Pimenta-Marques A., Helsens K., Vandekerckhove J., Martinho R. G., Gevaert K., Arnesen T. (2011) NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS genetics 7, e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Starheim K. K., Gevaert K., Arnesen T. (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci. 37, 152–161 [DOI] [PubMed] [Google Scholar]
- 3. Polevoda B., Arnesen T., Sherman F. (2009) A synopsis of eukaryotic Nalpha-terminal acetyltransferases: nomenclature, subunits and substrates. BMC Proc 3, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnesen T., Anderson D., Baldersheim C., Lanotte M., Varhaug J. E., Lillehaug J. R. (2005) Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 386, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hole K., Van Damme P., Dalva M., Aksnes H., Glomnes N., Varhaug J. E., Lillehaug J. R., Gevaert K., Arnesen T. (2011) The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PloS one 6, e24713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., Kim D. S., De Juan-Pardo E., Demeyer K., Hole K., Larrea E., Timmerman E., Prieto J., Arnesen T., Sherman F., Gevaert K., Aldabe R. (2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. U.S.A. 109, 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Starheim K. K., Gromyko D., Evjenth R., Ryningen A., Varhaug J. E., Lillehaug J. R., Arnesen T. (2009) Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell. Biol. 29, 3569–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evjenth R., Hole K., Karlsen O. A., Ziegler M., Arnesen T., Lillehaug J. R. (2009) Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 284, 31122–31129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Damme P., Evjenth R., Foyn H., Demeyer K., De Bock P. J., Lillehaug J. R., Vandekerckhove J., Arnesen T., Gevaert K. (2011) Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(alpha)-acetyltransferases and point to hNaa10p as the post-translational actin N(alpha)-acetyltransferase. Mol. Cell. Proteomics 10, M110 004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bienvenut W. V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C. (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol. Cell. Proteomics 11, M111 015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Starheim K. K., Arnesen T., Gromyko D., Ryningen A., Varhaug J. E., Lillehaug J. R. (2008) Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem. J. 415, 325–331 [DOI] [PubMed] [Google Scholar]
- 13. Polevoda B., Norbeck J., Takakura H., Blomberg A., Sherman F. (1999) Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18, 6155–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jornvall H. (1975) Acetylation of Protein N-terminal amino groups structural observations on alpha-amino acetylated proteins. J. Theor. Biol. 55, 1–12 [DOI] [PubMed] [Google Scholar]
- 15. Hwang C. S., Shemorry A., Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shemorry A., Hwang C. S., Varshavsky A. (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yi C. H., Pan H., Seebacher J., Jang I. H., Hyberts S. G., Heffron G. J., Vander Heiden M. G., Yang R., Li F., Locasale J. W., Sharfi H., Zhai B., Rodriguez-Mias R., Luithardt H., Cantley L. C., Daley G. Q., Asara J. M., Gygi S. P., Wagner G., Liu C. F., Yuan J. (2011) Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 146, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalvik T. V., Arnesen T. (2013) Protein N-terminal acetyltransferases in cancer. Oncogene 32, 269–276 [DOI] [PubMed] [Google Scholar]
- 19. Forte G. M., Pool M. R., Stirling C. J. (2011) N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 9, e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann I., Munro S. (2006) An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119, 1494–1503 [DOI] [PubMed] [Google Scholar]
- 21. Scott D. C., Monda J. K., Bennett E. J., Harper J. W., Schulman B. A. (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee C. F., Ou D. S., Lee S. B., Chang L. H., Lin R. K., Li Y. S., Upadhyay A. K., Cheng X., Wang Y. C., Hsu H. S., Hsiao M., Wu C. W., Juan L. J. (2010) hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest. 120, 2920–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gromyko D., Arnesen T., Ryningen A., Varhaug J. E., Lillehaug J. R. (2010) Depletion of the human Nalpha-terminal acetyltransferase A induces p53-dependent apoptosis and p53-independent growth inhibition. Int. J. Cancer 127, 2777–2789 [DOI] [PubMed] [Google Scholar]
- 24. Fisher T. S., Etages S. D., Hayes L., Crimin K., Li B. (2005) Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J. Biol. Chem. 280, 17749–17757 [DOI] [PubMed] [Google Scholar]
- 25. Lim J. H., Park J. W., Chun Y. S. (2006) Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 66, 10677–10682 [DOI] [PubMed] [Google Scholar]
- 26. Lim J. H., Chun Y. S., Park J. W. (2008) Hypoxia-inducible factor-1alpha obstructs a Wnt signaling pathway by inhibiting the hARD1-mediated activation of beta-catenin. Cancer Res. 68, 5177–5184 [DOI] [PubMed] [Google Scholar]
- 27. Liszczak G., Goldberg J. M., Foyn H., Petersson E. J., Arnesen T., Marmorstein R. (2013) Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 20, 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grauffel C., Abboud A., Liszczak G., Marmorstein R., Arnesen T., Reuter N. (2012) Specificity and versatility of substrate binding sites in four catalytic domains of human N-terminal acetyltransferases. PLoS One 7, e52642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liszczak G., Arnesen T., Marmorstein R. (2011) Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 286, 37002–37010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liszczak G., Marmorstein R. (2013) Implications for the evolution of eukaryotic amino-terminal acetyltransferase (NAT) enzymes from the structure of an archaeal ortholog. Proc. Natl. Acad. Sci. U.S.A. 110, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polevoda B., Sherman F. (2003) Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 308, 1–11 [DOI] [PubMed] [Google Scholar]
- 32. Whiteway M., Szostak J. W. (1985) The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 43, 483–492 [DOI] [PubMed] [Google Scholar]
- 33. Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. (1989) Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8, 2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park E. C., Szostak J. W. (1992) ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 11, 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polevoda B., Cardillo T. S., Doyle T. C., Bedi G. S., Sherman F. (2003) Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J. Biol. Chem. 278, 30686–30697 [DOI] [PubMed] [Google Scholar]
- 36. Hermann G. J., King E. J., Shaw J. M. (1997) The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J. Cell Biol. 137, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polevoda B., Sherman F. (2001) NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J. Biol. Chem. 276, 20154–20159 [DOI] [PubMed] [Google Scholar]
- 38. Tercero J. C., Wickner R. B. (1992) MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267, 20277–20281 [PubMed] [Google Scholar]
- 39. Lee Y. J., Wickner R. B. (1992) MAK10, a glucose-repressible gene necessary for replication of a dsRNA virus of Saccharomyces cerevisiae, has T cell receptor alpha-subunit motifs. Genetics 132, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rope A. F., Wang K., Evjenth R., Xing J., Johnston J. J., Swensen J. J., Johnson W. E., Moore B., Huff C. D., Bird L. M., Carey J. C., Opitz J. M., Stevens C. A., Jiang T., Schank C., Fain H. D., Robison R., Dalley B., Chin S., South S. T., Pysher T. J., Jorde L. B., Hakonarson H., Lillehaug J. R., Biesecker L. G., Yandell M., Arnesen T., Lyon G. J. (2011) Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 89, 28–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N., Dufke A., Cremer K., Hempel M., Horn D., Hoyer J., Joset P., Ropke A., Moog U., Riess A., Thiel C. T., Tzschach A., Wiesener A., Wohlleber E., Zweier C., Ekici A. B., Zink A. M., Rump A., Meisinger C., Grallert H., Sticht H., Schenck A., Engels H., Rappold G., Schrock E., Wieacker P., Riess O., Meitinger T., Reis A., Strom T. M. (2012) Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682 [DOI] [PubMed] [Google Scholar]
- 42. Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., Hewitson M., Holz C., Khan M., Lazik S., Martin C., Nitzsche B., Ruer M., Stamford J., Winzi M., Heinkel R., Roder M., Finell J., Hantsch H., Jones S. J., Jones M., Piano F., Gunsalus K. C., Oegema K., Gonczy P., Coulson A., Hyman A. A., Echeverri C. J. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 [DOI] [PubMed] [Google Scholar]
- 43. Wang Y., Mijares M., Gall M. D., Turan T., Javier A., Bornemann D. J., Manage K., Warrior R. (2010) Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev. Dyn. 239, 2813–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ingram A. K., Cross G. A., Horn D. (2000) Genetic manipulation indicates that ARD1 is an essential N(alpha)-acetyltransferase in Trypanosoma brucei. Mol. Biochem. Parasitol. 111, 309–317 [DOI] [PubMed] [Google Scholar]
- 45. Evjenth R., Hole K., Ziegler M., Lillehaug J. R. (2009) Application of reverse-phase HPLC to quantify oligopeptide acetylation eliminates interference from unspecific acetyl CoA hydrolysis. BMC Proc 3, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K. (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 47. Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. (2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362–1370 [DOI] [PubMed] [Google Scholar]
- 48. Martens L., Vandekerckhove J., Gevaert K. (2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584–3585 [DOI] [PubMed] [Google Scholar]
- 49. Helsens K., Colaert N., Barsnes H., Muth T., Flikka K., Staes A., Timmerman E., Wortelkamp S., Sickmann A., Vandekerckhove J., Gevaert K., Martens L. (2010) ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 50. Wang R., Fabregat A., Rios D., Ovelleiro D., Foster J. M., Cote R. G., Griss J., Csordas A., Perez-Riverol Y., Reisinger F., Hermjakob H., Martens L., Vizcaino J. A. (2012) PRIDE Inspector: a tool to visualize and validate MS proteomics data. Nat. Biotechnol. 30, 135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gautschi M., Just S., Mun A., Ross S., Rucknagel P., Dubaquie Y., Ehrenhofer-Murray A., Rospert S. (2003) The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23, 7403–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Damme P., Van Damme J., Demol H., Staes A., Vandekerckhove J., Gevaert K. (2009) A review of COFRADIC techniques targeting protein N-terminal acetylation. BMC Proc. 3, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goetze S., Qeli E., Mosimann C., Staes A., Gerrits B., Roschitzki B., Mohanty S., Niederer E. M., Laczko E., Timmerman E., Lange V., Hafen E., Aebersold R., Vandekerckhove J., Basler K., Ahrens C. H., Gevaert K., Brunner E. (2009) Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 7, e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Helsens K., Van Damme P., Degroeve S., Martens L., Arnesen T., Vandekerckhove J., Gevaert K. (2011) Bioinformatics analysis of a Saccharomyces cerevisiae N-terminal proteome provides evidence of alternative translation initiation and post-translational N-terminal acetylation. J. Proteome Res. 10, 3578–3589 [DOI] [PubMed] [Google Scholar]
- 55. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Polevoda B., Hoskins J., Sherman F. (2009) Properties of Nat4, an N(alpha)-acetyltransferase of Saccharomyces cerevisiae that modifies N termini of histones H2A and H4. Mol. Cell. Biol. 29, 2913–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiao Q., Zhang F., Nacev B. A., Liu J. O., Pei D. (2010) Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49, 5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arfin S. M., Bradshaw R. A. (1988) Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry 27, 7979–7984 [DOI] [PubMed] [Google Scholar]
- 59. Moerschell R. P., Hosokawa Y., Tsunasawa S., Sherman F. (1990) The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes c created by oligonucleotide transformation. J. Biol. Chem. 265, 19638–19643 [PubMed] [Google Scholar]
- 60. Tsunasawa S., Stewart J. W., Sherman F. (1985) Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J. Biol. Chem. 260, 5382–5391 [PubMed] [Google Scholar]
- 61. Huesgen P. F., Alami M., Lange P. F., Foster L. J., Schroder W. P., Overall C. M., Green B. R. (2013) Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS One 8, e74483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Borst P., Grivell L. A. (1978) The mitochondrial genome of yeast. Cell 15, 705–723 [DOI] [PubMed] [Google Scholar]
- 63. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.