Abstract
Circadian rhythms are self-sustained and adjustable cycles, typically entrained with light/dark and/or temperature cycles. These rhythms are present in animals, plants, fungi, and several bacteria. The central mechanism behind these “pacemakers” and the connection to the circadian regulated pathways are still poorly understood. The circadian rhythm of the cyanobacterium Synechococcus elongatus PCC 7942 (S. elongatus) is highly robust and controlled by only three proteins, KaiA, KaiB, and KaiC. This central clock system has been extensively studied functionally and structurally and can be reconstituted in vitro. These characteristics, together with a relatively small genome (2.7 Mbp), make S. elongatus an ideal model system for the study of circadian rhythms.
Different approaches have been used to reveal the influence of the central S. elongatus clock on rhythmic gene expression, rhythmic mRNA abundance, rhythmic DNA topology changes, and cell division. However, a global analysis of its proteome dynamics has not been reported yet.
To uncover the variation in protein abundances during 48 h under light and dark cycles (12:12 h), we used quantitative proteomics, with TMT 6-plex isobaric labeling. We queried the S. elongatus proteome at 10 different time points spanning a single 24-h period, leading to 20 time points over the full 48-h period.
Employing multidimensional separation and high-resolution mass spectrometry, we were able to find evidence for a total of 82% of the S. elongatus proteome. Of the 1537 proteins quantified over the time course of the experiment, only 77 underwent significant cyclic variations. Interestingly, our data provide evidence for in- and out-of-phase correlation between mRNA and protein levels for a set of specific genes and proteins. As a range of cyclic proteins are functionally not well annotated, this work provides a resource for further studies to explore the role of these proteins in the cyanobacterial circadian rhythm.
Circadian clocks or rhythms are widely observed in many species and throughout different kingdoms, from plants and animals to fungi and bacteria. Circadian rhythms are defined as self-sustained and adjustable cycles, typically entrained by light/dark and/or temperature cycles, occurring within 24-h periods (1).
Circadian rhythms were originally associated with more complex organisms, based on the notion that simple and fast-dividing organisms, such as bacteria, would not need a robust cyclic mechanism governing cellular processes (2, 3). More recently, cyclic rhythms have been shown to exist in evolutionarily very old systems such as cyanobacteria, which possess very robust circadian machinery (pacemaker) regulating gene expression, metabolism, and even the cell cycle (4, 5).
The cyanobacterium Synechococcus elongatus in particular has become a very useful model for studying circadian rhythms. Besides having a small genome (2.7 Mbp) (6, 7), this cyanobacterium also features a very simple and extremely robust core clock mechanism controlling its circadian rhythm. The core circadian system consists of just three proteins, KaiA, KaiB, and KaiC. KaiC undergoes a phosphorylation/dephosphorylation cycle, where KaiA is the promoter of its hyperphosphorylated state and KaiB opposes KaiA's action, promoting KaiC's hypophosphorylated state. Alongside this phosphorylation, the dynamic co-assembly of larger KaiABC complexes plays a controlling role in this circadian system (8). This relatively simple circadian pacemaker provides such a robust system that it can even be reproduced in vitro, requiring only the three Kai proteins and the addition of ATP/Mg2+ (9).
The connection of the central circadian mechanism (or the core post-translational oscillator) to its output is less well defined. However, it has been shown to encompass a transcription/translation feedback loop in which the KaiABC genes and their products participate in positive and negative autoregulatory feedback loops (10, 11), similar to what is observed in higher organisms (12). Different studies connect the KaiABC central clock with control over transcriptional activity through different effectors such as the histidine kinase Synechococcus adaptative sensor A, the putative transcription factor regulator of phycobilisome-associated A (13, 14), the regulator of phycobilisome-associated B regulator (15), low amplitude and bright A, the sensor histidine kinase circadian input kinase A (16, 17), and the input factors Pex and light-dependent period A. Others have proposed an oscilloid model in which gene expression is influenced by control over DNA topology (18, 19). These observations link the central clock to the global gene expression, including the expression of the kaiABC genes themselves (10).
Several bioluminescence studies have indicated a cyclic promoter activity of 100% (20), and microarray studies report a 30% to 60% rate of cyclic mRNA abundance in S. elongatus (21, 22). Despite the fact that these experiments were performed under different conditions and experimental setups, this discrepancy also might be caused by post-transcriptional regulation, and therefore protein abundance might likewise differ from observed mRNA abundance patterns.
In addition to extensive analyses of gene expression and mRNA levels in cyanobacteria, a limited number of proteome analyses have been performed. To our knowledge, a global proteome analysis of S. elongatus has not been published so far, although proteome analyses have been reported for some related species such as Synechocystis (23), Cyanothece (24–26), Prochlorococcus (27), and Anabaena (28).
To enable the global analysis of the S. elongatus proteome dynamics, we used high-resolution quantitative MS-based proteomics. To uncover the variation in protein abundances over 48 h under light/dark (LD)1 cycles (12:12 h), we employed quantitative shotgun proteomics using tandem mass tag (TMT) 6-plex isobaric labeling. These isobaric tags provide a sensitive labeling method for the analysis of several different experimental conditions (up to six at a time). We queried the S. elongatus proteome at 10 different time points spanning a single 24-h period, resulting in 20 time points over the full 48-h period.
With this approach we were able to detect the abundances of proteins covering 82% of the S. elongatus genome, within which we observed significant abundance changes of 544 proteins. Among these proteins, 77 proteins showed well-defined cyclic abundance profiles. The comparison of our results to previously published mRNA abundance profiles yielded a significantly lower degree of cyclic expression, pointing to the importance of post-transcriptional and/or post-translational regulatory mechanisms. Moreover, our data provide novel insights into the phasing of cyclic protein abundances relative to corresponding mRNA levels, as we observed that several protein abundance levels cycled, albeit out of phase, with their corresponding cycling mRNA levels.
EXPERIMENTAL PROCEDURES
Cyanobacteria Cell Culture
The wild-type strain of S. elongatus PCC 7942 was routinely grown photoautotrophically in BG11 medium (29) at 30 °C under continuous illumination with white light at 80 μm photons/m2s (Versatile Environmental Test Chamber, SANYO, Bensenville, IL) and a continuous stream of air. Cell concentrations were measured by determining the optical densities of the culture at 750 nm (OD750) (SPECORD®200 PLUS, Analytik Jena, Germany). The culture was kept in log growth phase (up to an OD750 of 1.0) by dilution up to a specific volume and subjected to a 12:12-h LD cycle for three days. Synchronized culture was finally diluted to an OD750 of ∼0.4 one day before the sampling started. At certain time points (Fig. 1A), which varied from 1-h to 3-h intervals, 40 ml of the culture was centrifuged at 15,000 × g for 10 min and the supernatant was removed. The cell pellet was resuspended in 1 ml of BG11 medium and centrifuged again for 5 min. The supernatant was removed and the pellet was washed with 1 ml of PBS buffer before the last round of centrifugation. Cell pellets were frozen in liquid nitrogen prior to storage at −20 °C.
Fig. 1.
High-resolution proteomics analysis of the S. elongatus proteome. A, S. elongatus cells were synchronized and cultured in light/dark (12:12 h) conditions. Samples were collected across a 48-h period at 20 independent time points with 1-h to 3-h intervals. Peptides were labeled with TMT 6-plex isobaric tags, with the sixth channel used to label a mixture of all the time points. Before LC-MS/MS analysis with an Orbitrap Velos, the four resulting sample mixtures were fractionated via SCX. B, logarithmic protein copy numbers of all 2179 proteins identified, as calculated using spectral counts (described in “Experimental Procedures”). Their abundances cover a dynamic range of 5 orders of magnitude. The most abundant proteins belong to the photosynthetic pathway (highlighted in green), and some proteins relevant to the circadian rhythm mechanism had medium to high abundances (central clock mechanism proteins—KaiA, KaiB, and KaiC—are highlighted in red, and the transcriptional regulator PadR family (Pex) is highlighted in purple).
Sample Preparation
Cyanobacteria pellets from samples of 20 time points were lysed in 8 m urea, 50 mm triethyl ammonium bicarbonate containing one tablet of EDTA-free protease inhibitor mixture (Sigma) and one tablet of PhosSTOP phosphatase inhibitor mixture (Roche). After three sonication cycles at 4 °C, total cell lysates were obtained through centrifugation at 14,000 rpm for 30 min at 4 °C. The supernatant was recovered and the protein concentration was determined via the Bradford method (Bio-Rad). The proteins were subjected to reduction and alkylation of the cysteine residues using 200 mm dithiotreitol (Sigma, Manchester, UK) and 200 mm iodoacetamide (Sigma). The proteins were first digested with Lys-C (Roche Diagnostics, Ingelheim, Germany) at an enzyme:protein ratio of 1:75 for 4 h at 37 °C, and this was followed by 4× dilution of the samples with 50 mm triethyl ammonium bicarbonate and digestion with trypsin (Roche Diagnostics), at an enzyme:protein ratio of 1:100, overnight at 37 °C. 40 μg from each sample and from a mixture of all samples were desalted using a 1-cc Sep Pack C18 columns (Waters, Etten-Leur, The Netherlands) and dried in vacuo.
Peptide Labeling
Peptides were labeled with TMTs using the TMT 6-plex labeling kit (Pierce). Two separate experiments, each consisting of two TMT 6-plex labeling experiments, were performed in total. Each experiment consisted of the use of five tags, one for each time point and the sixth for the mixture of all time points (internal standard). The manufacturer's protocol was followed, with a few adjustments. After desalting, 40 μg of peptides per channel were dissolved in 100 μl of 200 mm triethyl ammonium bicarbonate. The TMT labeling reagents were dissolved in 40 μl of acetonitrile (Biosolve, Valkenswaard, Netherlands) per vial and added to the samples in two steps to maximize the labeling efficiency. First, 10 μl of reagent solution were added to the sample; after 5 min, the other 10 μl were added and the reaction was incubated for 1 h at room temperature. In the quenching step, 4 μl of 5% hydroxylamine were added. After 15 min, the six channels were mixed in a 1:1 ratio and stored at −20 °C.
Strong Cation Exchange Fractionation
Peptides were fractionated using strong cation exchange (SCX) as described previously (30). In short, four SCXs were performed using a ZorbaxBioSCX-Series II column (0.8 mm × 50 mm, 3.5 μm). Solvent A consisted of 0.05% formic acid in 20% acetonitrile, and solvent B consisted of 0.05% formic acid and 0.5 m NaCl in 20% acetonitrile. The following gradient was used: 0–0.01 min (0%–2% B); 0.01–8.01 min (2%–3% B); 8.01–14.01 min (3%–8% B); 14.01–28 min (8%–20% B); 28–38 min (20%–40% B); 38–48 min (40%–90%B); 48–54 min (90% B); 54–60 min (0% B). The resulting 50 fractions were dried in vacuo, resuspended in 10% formic acid, and stored at −20 °C.
For the Sequential Windowed Acquisition of All Theoretical Spectra (SWATH, Ammerbuch-Entringen, Germany) approach, unlabeled samples from a 24-h time-series were pooled together and fractionated via SCX to create a spectral library.
Liquid Chromatography–Tandem Mass Spectrometry
Peptide fractions were analyzed on an Orbitrap Velos (Thermo Fisher Scientific, Bremen, Germany) that was coupled to an Agilent 1200 HPLC system (Agilent Technologies, Waldbronn, Germany). Both the trap (20 mm × 100 μm inner diameter) and the analytical column (35 cm × 50 μm inner diameter) were packed in-house using Reprosil-pur C18 3 μm (Dr. Maisch). Peptides were trapped at 5 μl/min in 100% solvent A (0.1 m acetic acid (Merck)). Elution was achieved with solvent B (0.1 m acetic acid in 80% acetonitrile) at 100 nl/min. The 180-min gradient used was as follows: 0–10 min, 100% solvent A; 10.1–115 min, 10%–24% solvent B; 115–160 min, 24%–50% solvent B; 160–163 min, 50%–100% solvent B; 163–164 min, 100% solvent B; 164–165 min, 0%–100% solvent A; 165–180 min, 100% solvent A. Nanospray was achieved using a coated fused silica emitter (New Objective, Cambridge, MA) (outer diamter, 360 μm; inner diameter, 20 μm; tip inner diameter, 10 μm) biased to 1.7 kV. The mass spectrometer was operated in the data-dependent acquisition mode. An MS2 method was used with a Fourier transform survey scan from 350 to 1500 m/z (resolution, 30,000; automatic gain control target, 5E5). The 10 most intense peaks were subjected to high-energy C-trap dissociation fragmentation (resolution, 7500; automatic gain control target, 3E4; normalized collision energy, 45%; maximum injection time, 500 ms; dynamic exclusion, 60 s). Predictive automatic gain control was enabled.
For the SWATH approach, samples were analyzed by a TripleTOF 5600 fitted with a Nanospray III source (AB SCIEX, Concord, Ontario, Canada) coupled to an Agilent 1290 Infinity ultra-high-pressure liquid chromatography system (Agilent Technologies). Briefly, for the spectral library data, the mass spectrometer was operated in data-dependent acquisition mode to obtain MS/MS spectra for the 20 most abundant parent ions following each survey MS1 scan. Additional datasets were recorded as triplicates in data-independent mode using SWATH MS2 acquisitions, essentially as described by Gillet et al. (31). In summary, a window of 26 m/z (containing 1 m/z for the window overlap) was passed in 32 incremental steps over the full mass range of 350–1250 m/z.
Database Search and Validation
Raw data were converted to .mgf files with Proteome Discoverer (version 1.3, Thermo, Boston, MA). Mascot (version 2.3.02, Matrix Science) was used to search the MS/MS data against the S. elongatus PCC 7942 UniProt database (version 4–2010) including a list of common contaminants and concatenated with the reversed versions of all sequences (5826 sequences). Trypsin was chosen as the cleavage specificity, with two missed cleavages allowed. Carbamidomethylation (C) was set as a fixed modification. The variable modifications used were oxidation (M), TMT 6-plex (K), and TMT 6-plex (N-term). The database searches were performed using a peptide tolerance of 50 ppm and a fragment mass tolerance of 0.05 Da (high-energy C-trap dissociation). The 50-ppm mass window was chosen to allow random assignment of false positives that were later removed by filtering using the instrument's actual mass accuracy (10 ppm). A .dat file of each of the four experiments was exported from Mascot and filtered with Rockerbox (32) to a false discovery rate of 1% using the concatenated database decoy method. Quantification was performed with the R package Isobar (33). A minimum peptide score of 20 and a minimum protein score of 40 were used as identifications thresholds. The peak intensities obtained were corrected for isotope impurity of the TMT labels and normalized with the median peak intensity. For all experiments, a minimum of one unique peptide was considered for protein quantification.
Raw data obtained with the TripleTOF 5600 were searched with ProteinPilot™, using the Paragon™ search engine (version 4.3, AB SCIEX). MS/MS data were searched against the same S. elongatus PCC 7942 UniProt database. Trypsin was chosen as the cleavage specificity. For the modifications, Cys alkylation was set to iodoacetamide and I.D. focus was set to biological modifications (i.e. phosphorylations, amidations, semitryptic fragments, etc.); as a special factor, urea denaturation was selected. The database search was performed with a thorough effort, with a detected protein threshold (unused protscore (confidence)) set to achieve 99% confidence. The false discovery rate analysis option was selected.
Datasets from SWATH MS2 acquisitions were processed using the full-scan MS/MS filtering module for data-independent acquisition within Skyline 1.3 (34). The .group file obtained from ProteinPilot was converted to .xml using the group2xml.exe script (version 4.3.0.1456, AB SCIEX), to create the spectral library (.blib). The top six peptides and fragment ions were extracted from SWATH MS2 acquisitions within Skyline using a fragment ion resolution setting of 15,000. Peak areas were normalized using the total area sums method.
Protein Copy Number Calculations
The sum of the number of peptide-spectrum matches obtained in all the experiments was normalized by the molecular weight of each corresponding protein and then divided by the total abundance factor calculated for all the identified proteins. This relative abundance factor was multiplied by the total amount of protein material used in all the experiments and divided by the protein molecular weights, leading to the protein copy number values. To determine the copy numbers of each protein in each cyanobacterium cell, we then divided this value by the number of cells used in the experiment.
Significance Analysis and Clustering
Quantitative data containing protein intensities from 20 time points, as obtained from the experiments described above, was analyzed as follows. The table containing protein names and respective intensities at all time points was loaded into R (version 3.0.0) (35) as a datamatrix (eset) object. Subsequently all rows containing proteins with their intensities over the 20 time points were filtered and processed in R using the following criteria: (i) No missing data (intensities) at any of the time points per protein were allowed. Proteins with missing data were removed from the matrix. (ii) All data in the matrix were scaled (Z transformed). (iii) All proteins that had an interquartile range variation less than 1 (e.g. those proteins that showed no real change at any of the time points) were removed. (iv) The data were exported to a tab-delimited file for subsequent analysis. The filtered protein data were loaded into the Multi Experiment Viewer software program (MeV v. 4.8.1) (36, 37). Based on the figure of merit, a method of determining the optimal number of k-means clusters, we chose to perform k-means clustering with six clusters. All clusters with their containing proteins were exported and used for further analysis.
Cyclic Profile Validation
The proteins considered significant and observed across all different time points were subjected to Pearson correlation analysis between the first 24-h ratios and the second 24-h ratios. For this purpose the statistical software IBM® SPSS® Statistics Data Editor was used. In order for a protein profile to be considered cyclic, a significant correlation with a p value less than 0.05 was required. Consequently, a heatmap was produced by hierarchical clustering of the protein abundance profiles, using Pearson correlation and complete linkage. The data were Z-score transformed for better visualization. A visual inspection of the profiles was performed to confirm their cyclic nature. The same procedure was applied for the analysis of the mRNA profiles from Ito et al. (21). The “raw” data (GSE14225) were extracted from GEO (Gene Expression Omnibus) with R (35), and an average of the two replicates was performed.
RESULTS
Global Proteome Analysis of the Cyanobacterium S. elongatus
Here we present the first in-depth quantitative proteome analysis of the cyanobacterium S. elongatus, conducted over a 48-h time span under LD conditions. This was accomplished by sampling 20 independent time points at 1-h to 3-h intervals. The sampling scheme consisted of 1-h intervals at the LD and dark-to-light transitions, whereas 2-h and 3-h intervals covered the respective day and night phases (Fig. 1A). The resulting 20 samples were quantified using the isobaric TMT labeling strategy and four 6-plex quantitation experiments with one pooled sample as an internal control. After cell lysis, protein digestion, and TMT labeling, the peptides were fractionated by means of SCX; this was done in order to decrease the complexity of the proteome and, at the same time, reduce precursor ion interference upon MS/MS analysis, which is inherent to isobaric quantification. This yielded an average of 20 fractions per experiment and ∼80 LC-MS/MS runs (Fig. 1A). The subsequent search analysis was performed using Mascot in combination with Isobar (33). Over 45,092 unique peptides (used for quantitation) and 2179 proteins were identified, covering 82% of the predicted S. elongatus proteome (2657 proteins). Of the identified proteins, 82% were represented by two or more peptides. The presented work constitutes one of the most complete coverages of a proteome reported to date.
The concentrations of each detected protein were evaluated using a well-established method based on spectral counts (38–40), as described in “Experimental Procedures.” In this experiment, we covered a dynamic range in protein abundance of over 5 orders of magnitude (Fig. 1B). The most abundant proteins found are mainly involved in photosynthesis, a crucial part of the energetic metabolism. However, proteins involved in this pathway are also spread throughout the whole dynamic range (Fig. 1B and supplemental Fig. S1A).
In spite of the high proteome coverage we obtained for S. elongatus, we observed that 768 (∼35%!) of the identified proteins are still annotated in the database as functionally uncharacterized. These proteins are distributed over the entire protein abundance range, even in the top 50 abundant proteins. These non-annotated proteins might be implicated in all kinds of processes, including specialized functions as well as “housekeeping” (supplemental Fig. S1B).
Abundance Profiles of the Kai Proteins and Their Putative Input and Output Channels
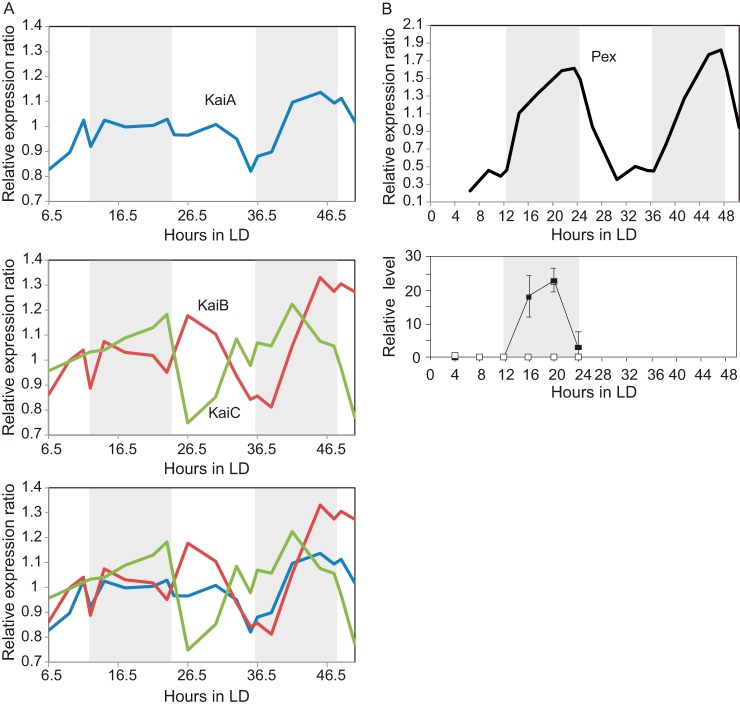
We first investigated the abundance profiles of some well-studied proteins in S. elongatus and compared these profiles to previously published data (Fig. 2). The main engine driving the circadian clock in S. elongatus is composed of the three proteins KaiA, KaiB, and KaiC. In accordance with a previous study (41), KaiA did not display a cyclic abundance profile (Fig. 2A). The profiles of KaiB and KaiC also did not show obvious cyclic abundances and, moreover, seemed to show opposite abundance profiles in our data (Fig. 2A). Cyclic abundances for KaiB and KaiC have been reported under continuous-light conditions (41) and synchronization in their total cellular abundances. However, in contrast, Qin et al. (10) reported that KaiC is not cyclic under LD conditions, which is in close agreement with our results.
Fig. 2.
Abundance profiles of key clock proteins. A, changes in the abundance of the central clock mechanism proteins in light/dark conditions (LD) across 48 h. The protein expression profiles obtained here seem to agree with previously published work (10, 41). KaiA's profile is represented in blue, KaiB's profile is in red, and KaiC's profile in green. B, comparison of the abundance profile of the transcriptional regulator, PadR family (Pex), obtained in this work via quantitative LC-MS/MS and across 48 h in LD (top profile), with the previously published profile obtained via Western blotting and across 24 h in LD (bottom profile adapted from Ref. 42). Both protein abundance profiles show an increase in abundance during the dark period. Gray areas represent the dark period, and white areas represent the light period.
Next we assessed proteins linked to circadian rhythm regulation or propagation for which abundance profiles had been published. Because current studies on cyanobacterial circadian rhythms consider transcript abundance levels rather than protein abundance, the number of available datasets is limited. One protein that fulfills these criteria is pex, a PadR family transcriptional regulator that is essential for circadian rhythm in S. elongatus. Pex is responsible for elongation and delay of the circadian rhythm in S. elongatus via the negative regulation of KaiA expression (42, 43). Takai et al. (42) demonstrated that the Pex protein exhibits cyclic abundance in LD conditions, with its maximum during the subjective night. This Western blot–based analysis agrees with the presented proteomics dataset (Fig. 2B). Another component of the circadian clock output pathways is the KaiC expression inhibitor low amplitude and bright A. Although low amplitude and bright A was found to exhibit cyclic mRNA abundance in continuous-light conditions (21), its protein abundance is also found to oscillate (not shown). However, most of the other reported components (e.g. regulator of phycobilisome-associated A, regulator of phycobilisome-associated B, Synechococcus adaptive sensor A, circadian input kinase A, light-dependent period A) were not found to have cyclic abundances at the protein level as revealed by our analysis.
In order to provide further confidence in the quantitative TMT data, an independent SWATH analysis was performed on a small set of selected proteins, namely, six cyclic, three non-significant and non-cyclic, and three significant but non-cyclic proteins (supplemental Fig. S4), according to our TMT data analysis. When we compared the protein expression profiles obtained via SWATH to the other data, we observed very good agreement.
Significance Analysis
Next, we conducted a global analysis of the abundance behavior of all proteins quantified in our analysis. With this approach we sought to find profiles that could be linked to circadian rhythm. We filtered our dataset, taking first only proteins that had quantitative information throughout the whole 48-h experiment (1537 proteins). Next we used interquartile range analysis to find significant variations in the 48-h profiles. Ratios were normalized and transformed to a z-score, and this was followed by removal of the proteins with an interquartile range variation of less than 1. From this evaluation we found 544 proteins that were significantly regulated at at least 1 out of 20 time points. Included in this set were the known clock pathway proteins KaiB, Pex, circadian input kinase A, regulator of phycobilisome-associated A, and low amplitude and bright A.
In our data analysis, we also observed ratio compression, which can be associated with the precursor ion interferences inherent to isobaric quantification. Nevertheless, this underestimation of the fold change had no influence on our analysis, as here we determined whether the ratio was significantly different from the background, which was similarly compressed.
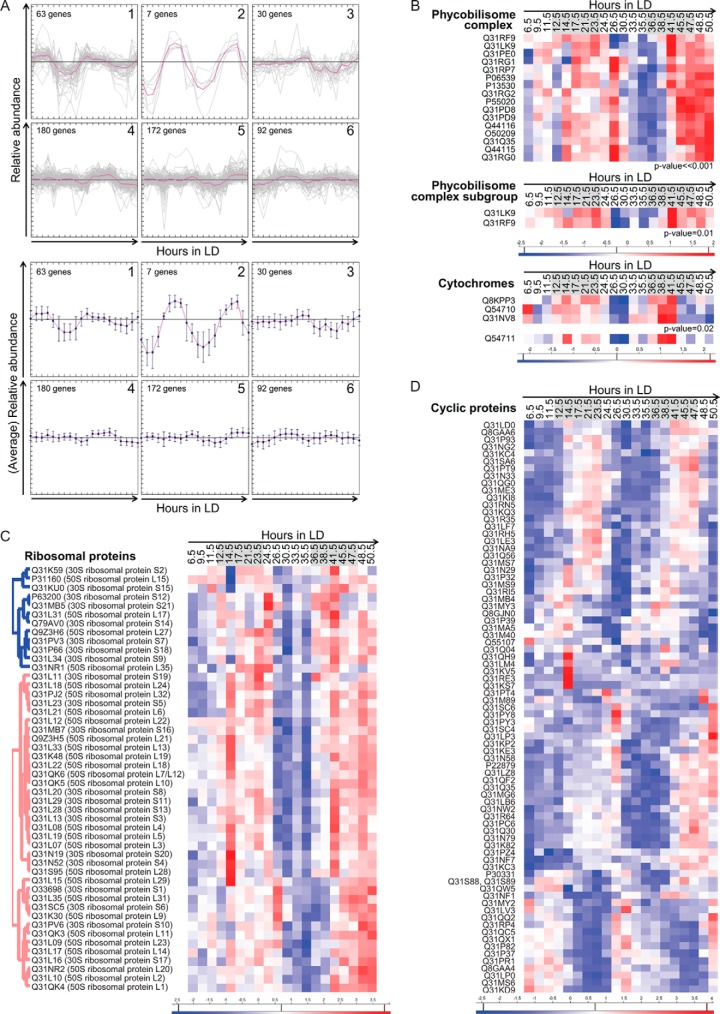
The resulting significant protein abundance profiles were clustered with K-means to facilitate the global analysis and the search for novel cyclic abundance profiles. This was done using MultiExperiment Viewer software. The 544 protein abundance profiles were divided into six clusters, as optimized with the figure of merit (Fig. 3A). Clusters 1 and 2 showed anti-correlated profiles and more pronounced differential abundance over time than the remaining clusters. Clusters 4 and 5 also exhibited anti-correlated cyclic profiles, but with less pronounced changes in abundance over time. Clusters 3 and 6 showed proteins with specific trends that had a noticeable relative abundance. However, they did not show cyclic profiles or correlation with the other clusters or with each other.
Fig. 3.
Clustering analysis on the significantly changing protein abundance profiles. A, K-means clustering analysis on the 544 protein abundance profiles found to change significantly across the 48-h LD experiment, as described in “Experimental Procedures.” At the top all the different protein profiles are visible in each corresponding cluster (the pink line represents the average profile of each cluster); at the bottom, only the averaged abundance profiles and their variance bars are visible for each cluster. B, hierarchical clustered heatmaps of three sets of proteins from the photosynthetic pathway. Each interest group of proteins demonstrated significantly similar abundance changes (p value < 0.05) across 48 h, as determined via Protein Profiles R package analysis. Red represents high abundance and blue low abundance of the proteins. All proteins are represented by their corresponding UniProt accession number. C, hierarchical clustered heatmap of ribosomal proteins. The type of analysis and representation is the same as that mentioned for B. D, hierarchical clustered heatmap of proteins with cyclic abundance profiles. These protein profiles were first analyzed with respect to their significance, as mentioned above, and then a Pearson correlation was performed, comparing the first 24 h of the profiles with the second 24 h, as explained in “Experimental Procedures.” The representation and clustering procedure was as mentioned above. The gray areas represent the dark period. The color scale is shown below each heatmap.
Functional Analysis of Proteins Co-occurring within Clusters
To investigate whether any of the clusters were associated with a specific pathway, protein interaction network, localization, or function, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to look for functional enrichments (44, 45). Although this analysis revealed no significant enrichment of specific functions in the clusters, we made some interesting observations using the Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway analysis. For cluster 1 we found six proteins to be associated with the two-component system and penicillin and cephalosporin biosynthesis, associated with signal transduction and the synthesis of secondary metabolites, respectively. An interesting example of the first pathway is the protein regulator of phycobilisome-associated A, which is known to be a transcription factor involved in genome-wide circadian gene expression and controlled by the kinase Synechococcus adaptive sensor A, which in turn is controlled by KaiC, indicating a possible link with circadian control. Cluster 2 had a lower number of associated proteins, but most of them displayed a quite prominent cyclic profile. As no specific functional association could be made with cluster 2, one could envision that the variety of pathways controlled by the circadian clock is reflected by the variety of proteins present in this cluster. Indeed, we found proteins involved in transcription, biotin metabolism, and response to stress. Cluster 3 contained proteins involved in amino acid, carbohydrate, and nucleotide metabolism, as well as DNA replication and repair and protein export. In cluster 4 the photosynthetic proteins were most prominent. Cluster 5 contained ribosomal proteins, and in cluster 6 proteins related to gene expression and translation were present. These results show that functionally related proteins do not necessarily associate with specific trends of protein abundance; however, some trends may be associated with pathway components or protein complexes.
Protein Profile Analysis in Selected Networks
Next, we sought to analyze selected pathways and/or protein complexes in more detail with the protein profile R package. This package looks for the significance between similar abundance profiles, independent of their intensities (46). We started with the photosynthetic pathway and its complexes, also well characterized in cyanobacteria. Here we could observe groups of proteins with very similar abundance profiles. One example is the phycobilisome complex, the components of which capture energy from light and transfer it to photosystem II through chlorophyll A. These proteins have significantly similar profiles (p value ≪ 0.001) (Fig. 3B and Table I), up-regulated in the light period, although not completely cyclic. Within this group of proteins we found an even more pronounced profile, namely, the phycobilisome core-membrane linker polypeptide and the phycobilisome rod-core linker polypeptide showing an increase in abundance during the dark period (p value = 0.01) (Fig. 3B and Table I). The two components serve as stabilizers at the core of the phycobilisome complex in collaboration with other proteins; therefore they are not directly involved in the light-capturing process. Another example is the cytochrome complex b6/f, which is responsible for the transfer of electrons from photosystem II to photosystem I. Although we were not able to cover all of its components, we observed very similar protein abundance profiles, specifically between apocytochrome f and cytochrome b6-f complex subunit 4. These two combined with the cytochrome b559, which is part of photosystem II, also show a similar trend (p value = 0.02) (Fig. 3B and Table I). All three of them were more abundant during the dark period. Another b6/f component, cytochrome b6, exhibited similar behavior despite the fact that its profile was not significantly similar to the ones mentioned above (Fig. 3B and Table I). This relationship was previously described for the cyanobacterium Cyanothece ATCC 51142 (26). Other cytochromes that are part of photosystem II, such as cytochrome 550, and part of the electron transport chain, such as cytochrome c6, have similar abundance profiles (p value = 0.02) but are more dynamic, as they have a maximum during the dark and during the light period.
Table I. Similarity of abundance profiles in related proteins. Three sets of proteins from the photosynthetic pathway with significantly similar abundance changes (p value < 0.05) across 48 h, as determined via Protein Profiles R package analysis.
| Accession | Protein name |
|---|---|
| Phycobilisome complex (p value ≪ 0.001) | |
| Q31RF9 | Phycobilisome core-membrane linker polypeptide |
| Q31LK9 | Phycobilisome rod-core linker polypeptide |
| Q31PE0 | Phycobilisome rod linker polypeptide |
| Q31RG1 | Allophycocyanin, β subunit |
| Q31RP7 | Putative uncharacterized protein |
| P06539 | C-phycocyanin β chain |
| P13530 | C-phycocyanin α chain |
| Q31RG2 | Phycobilisome 7.8-kDa linker polypeptide, allophycocyanin-associated core |
| P55020 | Plastocyanin |
| Q31PD8 | Phycocyanin linker protein 9K |
| Q31PD9 | Phycobilisome rod linker polypeptide |
| Q44116 | Phycobilisome maturation protein |
| O50209 | Allophycocyanin, β subunit |
| Q31Q35 | Allophycocyanin α chain-like |
| Q44115 | Phycocyanobilin lyase subunit α |
| Q31RG0 | Allophycocyanin α chain |
| Phycobilisome complex subgroup (p value = 0.01) | |
| Q31LK9 | Phycobilisome rod-core linker polypeptide |
| Q31RF9 | Phycobilisome core-membrane linker polypeptide |
| Cytochromes (p value = 0.02) | |
| Q8KPP3 | Cytochrome b559 subunit α |
| Q54710 | Cytochrome b6-f complex subunit 4 |
| Q31NV8 | Apocytochrome f |
| Q54711 | Cytochrome b6 |
Next, we investigated different proteins from photosystems I and II and the ATP synthase complex. For photosystem I we found four proteins with significantly similar profiles (p value = 0.02). These showed increased abundance during the dark period. In photosystem II we found seven proteins with a significantly similar nocturnal trend (p value ≪ 0.001) and two proteins with similarly significant trends, with greater abundance during both dark and light phases (p value = 0.004). For the ATP synthase complex, we found that most of the components we identified had similar profiles (p value = 0.03). However, during the light period they differed slightly. Finally, for the ribosomal proteins, all the proteins from the small subunit showed significantly similar profiles (p value = 0.01), as did all proteins from the large subunit (p value = 10−4) (Fig. 3C). Some components of the two subunits also shared significant profiles (p value ≪ 0.001). Together, these observations show that many of the observed protein complexes or particular components of these complexes share abundance trends, hinting at the specific regulation of different subsets of proteins.
Focusing on Proteins Displaying Cyclic Abundances
The cluster analysis gave an indication of at least two clusters with possible cyclic trends and potential circadian-associated protein abundances (Fig. 3A). Cluster 2 showed little variability between each trend, whereas in cluster 1 both amplitude and variability were higher. To make a more precise assessment of the cyclic proteins, we performed a Pearson correlation and decided on the cyclic properties of the profiles based on the significant linear correlation between the data points from the first day and the second, corresponding to a p value less than 0.05, using IBM SPSS Statistics software.
The Pearson correlation analysis and a visual confirmation of the profiles resulted in the stringent definition of 77 well-defined cyclic abundance profiles. This group included several functionally uncharacterized or poorly annotated proteins. To facilitate the visualization of these profiles, we created a heatmap using R (Fig. 3D and Table II). This heatmap revealed 39 proteins featuring an abundance peak during the dark phase. In contrast, only 29 proteins exhibited a maximum abundance during the light phase. We also found proteins with certain ambiguities concerning the peak phase and the oscillation period (24 h or 12 h). Despite this, we considered those cases to correspond to cyclic proteins.
Table II. Proteins with cyclic abundance profiles. These protein profiles were first analyzed with respect to their significance using the interquartile range method, and then a Pearson correlation was performed comparing the first 24 h of the profiles with the second 24 h.
| Accession | Protein name |
|---|---|
| Q31LD0 | ATPase |
| Q8GAA6 | Queuine tRNA-ribosyltransferase |
| Q31P93 | Phosphomethylpyrimidine synthase |
| Q31NG2 | Metal dependent phosphohydrolase |
| Q31KC4 | Putative uncharacterized protein |
| Q31SA6 | Aminotransferase |
| Q31PT9 | Putative uncharacterized protein |
| Q31N33 | Putative uncharacterized protein |
| Q31QG0 | Transcriptional regulator, PadR family |
| Q31ME3 | RNA polymerase sigma factor rpoD2 |
| Q31KI8 | Heat shock protein Hsp20 |
| Q31RN5 | Putative uncharacterized protein |
| Q31KQ3 | Putative uncharacterized protein |
| Q31R35 | Putative uncharacterized protein |
| Q31LF7 | Elongation factor EF-G |
| Q31RH5 | UPF0102 protein Synpcc7942 0312 |
| Q31LE3 | Diguanylate cyclase with GAF sensor |
| Q31NA9 | Putative uncharacterized protein |
| Q31Q56 | Phosphoenolpyruvate synthase |
| Q31MS7 | Pyridine nucleotide transhydrogenase α subunit |
| Q31N29 | Group3 RNA polymerase sigma factor SigF |
| Q31P32 | Putative uncharacterized protein |
| Q31MS9 | NAD(P) transhydrogenase subunit β |
| Q31RI5 | Phospholipase D/transphosphatidylase |
| Q31MB4 | Phosphate starvation-induced protein |
| Q31MY3 | Putative uncharacterized protein |
| Q8GJN0 | Light-independent protochlorophyllide reductase subunit B |
| Q31P39 | Putative uncharacterized protein |
| Q31MA5 | Group3 RNA polymerase sigma factor SigF |
| Q31M40 | RNA polymerase sigma factor |
| Q55107 | Bicarbonate transport ATP-binding protein CmpC |
| Q31Q04 | Putative uncharacterized protein |
| Q31QH9 | Putative uncharacterized protein |
| Q31LM4 | Putative uncharacterized protein |
| Q31KV5 | 2-hydroxy-6-oxohepta-24-dienoate hydrolase |
| Q31RE3 | Probable peptidase |
| Q31KS7 | UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| Q31PT4 | Putative uncharacterized protein |
| Q31M89 | Putative uncharacterized protein |
| Q31SC6 | Putative uncharacterized protein |
| Q31PY8 | Putative uncharacterized protein |
| Q31PY3 | Response regulator receiver domain protein |
| Q31SC4 | Putative uncharacterized protein |
| Q31LP3 | Putative uncharacterized protein |
| Q31KP2 | Putative uncharacterized protein |
| Q31KE3 | Methionine aminopeptidase |
| Q31N58 | Imidazole glycerol phosphate synthase subunit HisH |
| P22879 | 60 kDa chaperonin |
| Q31LZ8 | LabA |
| Q31QF2 | 60 kDa chaperonin 1 |
| Q31Q35 | Allophycocyanin α chain-like |
| Q31MG6 | Carotene isomerase |
| Q31LB6 | Anthranilate phosphoribosyltransferase |
| Q31NW2 | DNA repair protein RadC |
| Q31R64 | Putative uncharacterized protein |
| Q31PC6 | Putative uncharacterized protein |
| Q31Q30 | Putative uncharacterized protein |
| Q31N79 | Putative uncharacterized protein |
| Q31K82 | Putative uncharacterized protein |
| Q31PZ4 | Phosphoesterase PHP-like |
| Q31NF7 | Diguanylate cyclase/phosphodiesterase |
| Q31KC3 | Two-component transcriptional regulator, winged helix family |
| P30331 | Metallothionein |
| Q31S88, Q31S89 | Pilin polypeptide PilA-like |
| Q31QW5 | Putative uncharacterized protein |
| Q31NF1 | Carbonate dehydratase |
| Q31MY2 | RNA polymerase sigma factor |
| Q31LV3 | Putative uncharacterized protein |
| Q31QQ2 | PDZ/DHR/GLGF |
| Q31RP4 | Possible high light inducible polypeptide HliC |
| Q31QC5 | C-terminal processing peptidase-2. Serine peptidase. MEROPS family S41A |
| Q31QX1 | C-terminal processing peptidase-2. Serine peptidase. MEROPS family S41A |
| Q31P82 | TPR repeat |
| Q31P37 | Rare lipoprotein A |
| Q31PR1 | Outer envelope membrane protein |
| Q8GAA4 | Putative uncharacterized protein sek0026 |
| Q31LP0 | RNA-binding region RNP-1 |
| Q31MS6 | Putative uncharacterized protein |
| Q31KD9 | General secretion pathway protein D |
The group of 29 proteins with greater abundance during the light period is reported to be involved in a broad variety of processes, including secretion, cell wall/membrane biogenesis, general stress, transcription, translation, protein turnover and folding, signal transduction, circadian rhythm, amino acid metabolism, and photosynthesis, with a small predominance of cell wall/membrane biogenesis. The group of proteins with their abundance peaking in the dark phase contained a variety of proteins involved in cell/wall biogenesis; chaperoning; signal transduction; transcription; translation; defense mechanisms; DNA repair; and different metabolisms such as coenzyme, vitamin, carbohydrate, amino acid, lipid, and energy-production related, including photosynthesis (light-capture related). Notably, the majority of the proteins that demonstrated greater abundance during the dark phase are involved in transcription. From these results one might suppose that the cyanobacterium has increased metabolism and maintenance during the night cycle relative to the day. Also, it is interesting that not all photosynthetic proteins showed cyclic profiles in our analysis, and those that did demonstrated a possibly diminished photosynthetic capacity during the day. The proteins ambiguous in their phase are part of secondary metabolite biosynthesis, transport and catabolism, inorganic ion transport, signal transduction, and transcription.
Interestingly, most of the protein abundance profiles did not have a sinusoidal peak shape. In fact, most of the profiles with maxima during the light period seemed to have a steeper increase in abundance followed by a slow decrease in abundance. Moreover, profiles with maxima during the dark period displayed exactly the opposite behavior. Consistently, the dark-to-light transition was marked by a steeper response than the LD transition, even though the sampling and LD transition were kept the same.
Among the proteins with abundance profiles over the complete period, we also found four proteins with abundance cycles shorter than 24 h. In fact, it has been shown by Westermark and Herzel (47) that components of the circadian clock can generate 12-h rhythms in the gene expression of animals. They demonstrated that 12-h genes have alternating peak heights, which is consistent with our observation at the protein level of pilin polypeptide PilA-like (signal transduction), carbonate dehydratase (nitrogen metabolism), 2-hydroxy-6-oxohepta-24-dienoate hydrolase (no specific function), and a putative uncharacterized protein. Recent reports describe rhythms shorter than 24 h, called ultradian rhythms, which have also been reported to occur in related cyanobacteria such as Cyanothece (48, 49) and Prochlorococcus (50).
Among all proteins with significantly changing abundances, we classified 14% as experiencing cyclic abundance. The percentage of proteins showing cyclic abundance was low relative to data reported on the transcript abundance. Transcript data for S. elongatus revealed that 30% to 60% of the mRNA levels exhibited cyclic profiles (21, 51). These somewhat conflicting results could indicate the existence of post-transcriptional and/or post-translational mechanisms regulating protein abundance, as discussed in the following section. However, it must be noted that differences in experimental conditions and in experimental and theoretical methods can influence the observed mRNA and protein abundance. The difference in methodology and the stringency of our data analysis might therefore account for some of the described differences.
In- and Out-of-phase Correlation between mRNA and Protein Levels of Cyclic Genes/Proteins
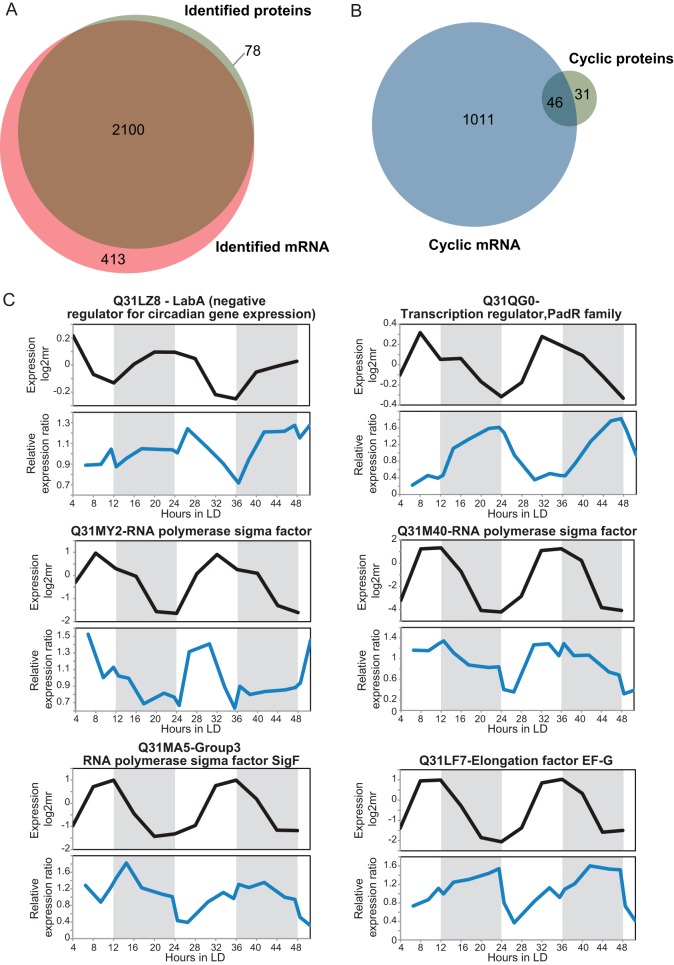
To compare circadian mRNA abundance profiles with the cyclic protein profiles, we used the published results from Ito et al. (21) on the quantitative profiles of S. elongatus transcripts during 48 h under continuous-light conditions. We took this dataset from Ito et al. because we noted that the majority of mRNA transcripts measured by them could also be quantified in our study at the protein level, and the taken time points matched best with our experimental setup (Fig. 4A). Of the identified transcripts, 800 were reported to be cyclic. However, when we performed a Pearson correlation analysis on the reported transcript profiles, we found that 1057 transcripts were cyclic. In a direct comparison of these cyclic mRNA to our 77 cyclic protein abundances, we found an overlap of 37 genes with the data reported by Ito et al. and an overlap of 46 genes with the data analyzed via Pearson correlation, uncovering an additional 13 cyclic genes in common with our cyclic proteins (Fig. 4B and supplemental Fig. S2). Moreover, an additional 27 proteins found to be cyclic in our study did not show cyclic behavior at the mRNA level. These results indicate a large discrepancy between the abundance at the transcript and at the protein level and clearly indicate post-transcriptional regulation.
Fig. 4.
Comparison of the proteome data with the mRNA data. A, comparison of the number of proteins identified in this work via quantitative LC-MS/MS with the number of transcripts identified by Ito et al. (21) via microarray. B, comparison of the number of proteins with cyclic abundance profiles with the number of transcripts with cyclic abundance profiles identified by Ito et al., but reanalyzed using Pearson correlation analysis, as described in “Experimental Procedures.” C, examples of the comparison of the abundance profiles of the proteins (blue lines) and transcripts (black lines) considered cyclic, showing in-phase or out-of-phase correlations between protein and transcript data. The dark period is represented by the gray area.
Next, we compared the cyclic protein profiles that overlapped with the cyclic mRNA data (46 proteins) in more detail. Here, we observed examples of proteins involved in several biological functions with a small prevalence for proteins related to DNA repair, transcription, and metabolism. The 31 cases in which only the protein abundances were cyclic showed equally diverse protein functions, with a small prevalence for signal transduction, transcription, and folding. Visual analysis of the peak phase of the mRNA profiles showed some ambiguity, similar to that observed for the cyclic analysis at the protein level. We observed two transcripts with 12-h cycles and abundance peaks within the subjective switch between day and night.
Interestingly, we noticed that the delays in the peak times between mRNA and proteins sometimes were not the same within a 48-h period. We performed a comparison of the peak times between our cyclic abundance profiles and the cyclic transcript profiles reported by Ito et al., which revealed delays between 0 and 22 h, hinting at in-phase and out-of-phase correlations between these two levels (Fig. 4C and supplemental Fig. S3). For 8 genes out of 46, there was a clear in-phase correlation between protein and mRNA profiles, and for 33 genes there was a clear out-of-phase correlation, with 15 being completely anti-phase.
Among the genes with delays shorter than 12 h, we mainly found proteins implicated in transcription, DNA repair, and metabolism. Genes with a longer delay were involved in cell wall biogenesis, transcription, general stress response, and coenzyme metabolism. Proteins related to transcription were observed to have time delays that differed from the transcript data (e.g. transcription factor PadR family and RNA polymerase sigma factor D5 versus RNA polymerase sigma factor D5/RNA polymerase sigma subunit C and RNA polymerase sigma factor F), indicating alternative means of post-transcriptional control of protein expression.
Our observations show a complex regulation of protein expression, which for several proteins occurs at the post-transcriptional level. For many proteins the observed cyclic behavior of mRNA abundance did not translate into the same behavior at the protein level. Moreover, among those gene products that did display cyclic behavior at both mRNA and protein levels, a large portion did not display similar abundance profiles and instead showed different levels of delay. These different delay times of actual translation of the mRNA into protein might be attributed to the necessity that the bacteria be able to quickly express certain proteins upon external signals.
DISCUSSION
Here we report one of the most complete proteomes to date, identifying evidence for 82% of the predicted genes at the protein level. Although the genome of the cyanobacterium S. elongatus is not as extensive as that in mammalian systems, this percentage is still impressive, especially when compared with the 60% to 70% reported for other, smaller genomes such as yeast (52).
We monitored the relative protein abundance of hundreds of proteins over two LD cycles with a total of 20 samples. We focused our analysis on the proteins showing a cycling abundance profile. 77 proteins were found to exhibit cycling abundance with a 24-h period, and these had quite distinct behavior. Firstly, the majority of cycling proteins were more abundant in the dark phase, which is unexpected for photosynthetic bacteria. We also observed many proteins within a single protein complex or pathway revealing similar abundance profiles, albeit not always cycling. Interestingly, a range of proteins was found to exhibit 12-h oscillations in abundance (e.g. carbonate dehydratase, Pilin polypeptide PilA-like, 2-hydroxy-6-oxohepta-24-dienoate hydrolase). Most remarkable was the observation of several cycling proteins whose cycle was clearly out of phase with the cycling behavior observed for the corresponding transcript. These data represent some of the clearest evidence for post-transcriptional regulation of many of the processes involved in the circadian rhythm and the clear need to analyze these processes at the protein level.
Although a few of the proteins we observed to be cyclic have been linked to the circadian rhythm in S. elongatus, most of these proteins have not yet been linked and originate from a variety of biological processes. Therefore, we believe our dataset provides several new starting points for further investigation of proteins and pathways involved in or regulated by circadian rhythms in cyanobacteria.
Supplementary Material
Acknowledgments
We acknowledge Anne Rediger, for her contribution to cell culturing and sample collection, and Stefanie Hertel, for helpful comments during the manuscript preparation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000510 and DOI 10.6019/PXD000510. This data deposition was supported by the PRIDE Team, EBI.
Footnotes
Author contributions: A.M.A., I.M.A., and A.J.H. designed research; A.C.G., H.P., and P.G. performed research; M.B. contributed new reagents or analytic tools; A.C.G., R.L., and B.v. analyzed data; A.C.G., A.M.A., I.M.A., and A.J.H. wrote the paper.
* This work was supported by The Netherlands Proteomics Centre and the Netherlands Organization for Scientific Research (NWO) with a VIDI grant to A.F.M.A. (723.012.102). Additionally, the PRIME-XS Project (Grant Agreement 262067), funded by the European Union Seventh Framework Program, is kindly acknowledged for financial support.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- LD
- light/dark
- TMT
- tandem mass tags
- SCX
- strong cation exchange
- MS/MS
- tandem mass spectrometry
- SWATH
- Sequential Windowed Acquisition of All Theoretical Spectra
- Pex
- period-extender (PadR family transcriptional regulator).
REFERENCES
- 1. Johnson C. H., Stewart P. L., Egli M. (2011) The cyanobacterial circadian system: from biophysics to bioevolution. Annu. Rev. Biophys. 40, 143–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson C. H., Golden S. S., Ishiura M., Kondo T. (1996) MicroReview circadian clocks in prokaryotes. Mol. Microbiol. 21, 5–11 [DOI] [PubMed] [Google Scholar]
- 3. Pittendrigh C. S. (1993) Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54 [DOI] [PubMed] [Google Scholar]
- 4. Dong G., Yang Q., Wang Q., Kim Y.-I., Wood T. L., Osteryoung K. W., van Oudenaarden A., Golden S. S. (2010) Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kondo T., Strayer C. A., Kulkarni R. D., Taylor W., Ishiura M., Golden S. S., Johnson C. H. (1993) Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 90, 5672–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andersson C. R., Tsinoremas N. F., Shelton J., Lebedeva N. V., Yarrow J., Min H., Golden S. S. (2000) Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305, 527–542 [DOI] [PubMed] [Google Scholar]
- 7. Holtman C. K., Chen Y., Sandoval P., Gonzales A., Nalty M. S., Thomas T. L., Youderian P., Golden S. S. (2005) High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 12, 103–115 [DOI] [PubMed] [Google Scholar]
- 8. Brettschneider C., Rose R. J., Hertel S., Axmann I. M., Heck A. J. R., Kollmann M. (2010) A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol. Syst. Biol. 6, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 [DOI] [PubMed] [Google Scholar]
- 10. Qin X., Byrne M., Xu Y., Mori T., Johnson C. H. (2010) Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 8, e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zwicker D., Lubensky D. K., ten Wolde P. R. (2010) Robust circadian clocks from coupled protein-modification and transcription-translation cycles. Proc. Natl. Acad. Sci. U.S.A. 107, 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hardin P. E., Hall J. C., Rosbash M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 [DOI] [PubMed] [Google Scholar]
- 13. Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., Iwasaki H. (2006) A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwasaki H., Williams S. B., Kitayama Y., Ishiura M., Golden S. S., Kondo T. (2000) A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233 [DOI] [PubMed] [Google Scholar]
- 15. Hanaoka M., Takai N., Hosokawa N., Fujiwara M., Akimoto Y., Kobori N., Iwasaki H., Kondo T., Tanaka K. (2012) RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J. Biol. Chem. 287, 26321–26327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taniguchi Y., Takai N., Katayama M., Kondo T., Oyama T. (2010) Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian KaiBC expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 3263–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmitz O., Katayama M., Williams S. B., Kondo T., Golden S. S. (2000) CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768 [DOI] [PubMed] [Google Scholar]
- 18. Mori T., Johnson C. H. (2001) Circadian programming in cyanobacteria. Semin. Cell Dev. Biol. 12, 271–278 [DOI] [PubMed] [Google Scholar]
- 19. Woelfle M. A., Xu Y., Qin X., Johnson C. H. (2007) Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 18819–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Tsinoremas N. F., Johnson C. H., Lebedeva N. V., Golden S. S., Ishiura M., Kondo T. (1995) Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 21. Ito H., Mutsuda M., Murayama Y., Tomita J., Hosokawa N., Terauchi K., Sugita C., Sugita M., Kondo T., Iwasaki H. (2009) Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 106, 14168–14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vijayan V., Jain I. H., O'Shea E. K. (2011) A high resolution map of a cyanobacterial transcriptome. Genome Biol. 12, R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wegener K. M., Singh A. K., Jacobs J. M., Elvitigala T., Welsh E. A., Keren N., Gritsenko M. A., Ghosh B. K., Camp D. G., Smith R. D., Pakrasi H. B. (2010) Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics 9, 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aryal U. K., Stöckel J., Welsh E. A., Gritsenko M. A., Nicora C. D., Koppenaal D. W., Smith R. D., Pakrasi H. B., Jacobs J. M. (2012) Dynamic proteome analysis of Cyanothece sp. ATCC 51142 under constant light. J. Proteome Res. 11, 609–619 [DOI] [PubMed] [Google Scholar]
- 25. Aryal U. K., Stöckel J., Krovvidi R. K., Gritsenko M. A., Monroe M. E., Moore R. J., Koppenaal D. W., Smith R. D., Pakrasi H. B., Jacobs J. M. (2011) Dynamic proteomic profiling of a unicellular cyanobacterium Cyanothece ATCC51142 across light-dark diurnal cycles. BMC Syst. Biol. 5, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stöckel J., Jacobs J. M., Elvitigala T. R., Liberton M., Welsh E. A., Polpitiya A. D., Gritsenko M. A., Nicora C. D., Koppenaal D. W., Smith R. D., Pakrasi H. B. (2011) Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PLoS One 6, e16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waldbauer J. R., Rodrigue S., Coleman M. L., Chisholm S. W. (2012) Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One 7, e43432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrios-Llerena M. E., Chong P. K., Gan C. S., Snijders A. P. L., Reardon K. F., Wright P. C. (2006) Shotgun proteomics of cyanobacteria—applications of experimental and data-mining techniques. Brief. Funct. Genomic. Proteomic. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 29. Rippka R., Deruelles J., Waterbury J., Herdman M., Stanier R. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 110, 1–61372487 [Google Scholar]
- 30. De Graaf E. L., Altelaar A. F. M., van Breukelen B., Mohammed S., Heck A. J. R. (2011) Improving SRM assay development: a global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res. 10, 4334–4341 [DOI] [PubMed] [Google Scholar]
- 31. Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van den Toorn H. W. P., Munõz J., Mohammed S., Raijmakers R., Heck A. J. R., van Breukelen B. (2010) RockerBox: analysis and filtering of massive proteomics search results. J. Proteome Res. 10, 1420–1424 [DOI] [PubMed] [Google Scholar]
- 33. Breitwieser F. P., Müller A., Dayon L., Köcher T., Hainard A., Pichler P., Schmidt-Erfurth U., Superti-Furga G., Sanchez J.-C., Mechtler K., Bennett K. L., Colinge J. (2011) General statistical modeling of data from protein relative expression isobaric tags. J. Proteome Res. 10, 2758–2766 [DOI] [PubMed] [Google Scholar]
- 34. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. R Development Core Team (2013) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 36. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 37. Saeed A., Bhagabati N., Braisted J., Liang W., Sharov V., Howe E., Li J., Thiagarajan M., White J., Quackenbush J. (2006) TM4 microarray software suite. Methods Enzymol. 411, 134–193 [DOI] [PubMed] [Google Scholar]
- 38. Peng M., Taouatas N., Cappadona S., van Breukelen B., Mohammed S., Scholten A., Heck A. J. R. (2012) Protease bias in absolute protein quantitation. Nat. Methods 9, 524–525 [DOI] [PubMed] [Google Scholar]
- 39. Ringrose J. H., van den Toorn H. W. P., Eitel M., Post H., Neerincx P., Schierwater B., Maarten Altelaar A. F., Heck A. J. R. (2013) Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat. Commun. 4, 1408. [DOI] [PubMed] [Google Scholar]
- 40. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kitayama Y., Iwasaki H., Nishiwaki T., Kondo T. (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takai N., Ikeuchi S., Manabe K., Kutsuna S. (2006) Expression of the circadian clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. J. Biol. Rhythms 21, 235–244 [DOI] [PubMed] [Google Scholar]
- 43. Kutsuna S., Kondo T., Ikegami H., Uzumaki T., Katayama M., Ishiura M. (2007) The circadian clock-related gene pex regulates a negative cis element in the kaia promoter region. J. Bacteriol. 189, 7690–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang D. W., Sherman B. T., Lempicki R. A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 45. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hansson J., Rafiee M. R., Reiland S., Polo J. M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. (2012) Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2, 1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westermark P., Herzel H. (2013) Mechanism for 12 hr rhythm generation by the circadian clock. Cell Rep. 3, 1228–1238 [DOI] [PubMed] [Google Scholar]
- 48. Cerveny J., Sinetova M. A., Valledor L., Sherman L. A., Nedbal L. (2013) Ultradian metabolic rhythm in the diazotrophic cyanobacterium Cyanothece sp. ATCC 51142. Proc. Natl. Acad. Sci. U.S.A. 110, 13210–13215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elvitigala T., Stöckel J., Ghosh B. K., Pakrasi H. B. (2009) Effect of continuous light on diurnal rhythms in Cyanothece sp. ATCC 51142. BMC Genomics 10, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shalapyonok A., Olson R. J., Shalapyonok L. S. (1998) Ultradian growth in Prochlorococcus spp. Appl. Environ. Microbiol. 64, 1066–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vijayan V., Zuzow R., O'Shea E. K. (2009) Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 22564–22568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gunaratne J., Schmidt A., Quandt A., Neo S. P., Saraç O. S., Gracia T., Loguercio S., Ahrné E., Xia R. L. H., Tan K. H., Lössner C., Bähler J., Beyer A., Blackstock W., Aebersold R. (2013) Extensive mass spectrometry-based analysis of the fission yeast proteome: the Schizosaccharomyces pombe PeptideAtlas. Mol. Cell. Proteomics 12, 1741–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.