Abstract
Oomycetes are filamentous organisms that cause notorious diseases, several of which have a high economic impact. Well known is Phytophthora infestans, the causal agent of potato late blight. Previously, in silico analyses of the genome and transcriptome of P. infestans resulted in the annotation of a large number of genes encoding proteins with an N-terminal signal peptide. This set is collectively referred to as the secretome and comprises proteins involved in, for example, cell wall growth and modification, proteolytic processes, and the promotion of successful invasion of plant cells. So far, proteomic profiling in oomycetes was primarily focused on subcellular, intracellular or cell wall fractions; the extracellular proteome has not been studied systematically. Here we present the first comprehensive characterization of the in vivo secretome and extracellular proteome of P. infestans. We have used mass spectrometry to analyze P. infestans proteins present in seven different growth media with mycelial cultures and this resulted in the consistent identification of over two hundred proteins. Gene ontology classification pinpointed proteins involved in cell wall modifications, pathogenesis, defense responses, and proteolytic processes. Moreover, we found members of the RXLR and CRN effector families as well as several proteins lacking an obvious signal peptide. The latter were confirmed to be bona fide extracellular proteins and this suggests that, similar to other organisms, oomycetes exploit non-conventional secretion mechanisms to transfer certain proteins to the extracellular environment.
Phytophthora infestans, the causal agent of tomato and potato late blight, is one of the most notorious plant pathogens in modern history. It was responsible for the Irish Potato Famine in the mid-19th century and recurrent outbreaks have been reported ever since. The Phytophthora genus comprises over hundred plant pathogenic species and belongs to the oomycetes, a lineage with filamentous organisms that morphologically resemble fungi but are more closely related to brown algae and diatoms (1, 2).
To facilitate growth, cell wall assembly, cell wall modification, and acquisition of nutrients, organisms require extracellular proteins. Prominent extracellular proteins are hydrolytic enzymes such as proteases, lipases, and glycosyl hydrolases, which digest complex substrates into small units that act as nutritional sources. Pathogen derived proteins facilitate host tissue degradation resulting in colonization or invasion, and they are considered to act as pathogenicity factors (3). Microbial pathogens also need an extensive set of proteins that play a role in host-pathogen interplay. For plant pathogens, these proteins are required during penetration and colonization of the plant tissue and are frequently referred to as effector proteins (3). The genomes of Phytophthora spp. encode hundreds of such putative effector proteins (4, 5). Two groups of effectors, apoplastic and cytosolic, are discerned dependent on the site of action. Among apoplastic effectors are protein inhibitors, secreted to counteract apoplastic host plant derived proteins, and hydrolytic enzymes such as proteases. Other apoplastic effectors interfere with the host membrane-cell wall integrity and can trigger host cell death (3, 6). Cytoplasmic effectors translocate into the plant cell, targeting various subcellular compartments where they modulate plant cell signaling, suppress immunity, and metabolic processes in the plant cytosol and nucleus for the pathogens benefit (7). In P. infestans these predicted host-translocated effectors encompass the RXLR (short for the four amino acids that form the motif, Arginine, Any, Leucine, and Arginine) and CRN (crinkling and necrosis inducing)1 effectors. These are large and complex protein families, with around 560 RXLRs and 200 CRNs members encoded in the genome (4). Apoplastic and cytosolic effector classes are mostly small modular proteins that contain an N-terminal signal peptide to facilitate secretion. Their C-terminal part comprises additional effector modules including host targeting signals, as is the case for RXLRs and CRNs, and a functional domain exerting its function (8). Both RXLR and CRN′s were originally identified as inducers of plant cell death and defense-related gene expression during in planta expression (3, 9) although not all CRNs promote infection (10). Effector genes frequently have distinct patterns of expression during various life stages and colonization of host plants (4).
Phytophthora research in the last decade benefitted largely from high-throughput bioinformatics tools. EST mining resulted in the identification of various putative extracellular proteins (9). With the elucidation of various Phytophthora and other oomycete genomes, a wealth of information was retrieved from genome sequences by in silico gene annotation (4, 5, 11–14). Genome mining resulted in the identification of many novel genes and a large repertoire of potential virulence factors (4, 5, 15, 16). In the P. infestans genome, a genome-wide inventory of genes encoding proteins with a signal peptide resulted in the initial identification of 2228 candidates, later refined to 1415 secretome proteins, many of which are potential pathogenicity factors (17). The in silico refinement was based on the archetypal secretion pathway, and, thus, it consisted in scoring for presence or absence of a signal peptide in combination with cellular compartment prediction and presence of transmembrane domains. There are several limitations in this in silico approach. Firstly, accurate gene annotation is essential. N-terminal inaccuracies result in signal peptide detection failures whereas other erroneous predictions can result in the misinterpretation of transmembrane domains or targeting sequences, which would lead to including or excluding them from the predicted secretome. Secondly, signal peptide sequences are extremely heterogeneous and weakly predicted ones were excluded. In addition, the term “secretome” is frequently misinterpreted as the extracellular proteome whereas it is limited to the collection of signal peptide containing proteins that are handled via the endoplasmic reticulum and Golgi apparatus before secretion (18). Many proteins identified in the plant cell apoplast belong to leaderless secretory proteins (LSP) (19, 20) and similar findings have been reported for fungi and animals (21, 22). Meanwhile, several unconventional protein secretion systems have been described including self-sustained protein translocation, ABC-transporter based secretion, exosome/autophagosome mediated secretion, and microvesicle shedding/blebbing (19, 22–26). It can be therefore anticipated that similar mechanisms exist in oomycetes.
One of the most powerful methods to evaluate the final outcome of gene expression is the identification of the resulting proteins using proteomics. This approach has frequently been applied in fungi to elucidate the proteome and secretome under various conditions including plant-pathogen interactions (27, 28). Using this technology, however, only limited information has been gained in Phytophthora. In P. palmivora three actin isoforms were identified by proteomics (29). Four enzymes, involved in amino acid biosynthesis were retrieved from P. infestans (30). Thirteen proteins with a life stage specific expression pattern were identified by 2D-gel electrophoresis, including CRN2 (31). For P. sojae and P. ramorum, a global proteomic approach was used to detect proteomic differences between life stages (32, 33), and a recent large-scale phosphoproteome analysis revealed the phosphorylation status of thousands of proteins and provided novel information on life stage specific phosphorylation events in P. infestans (34). Despite their importance, proteomic studies on Phytophthora extracellular proteins are even more limited. The identification of individual extracellular protein components in culture filtrates was described for elicitins (35–38), CBELs (39), and glucanase inhibitor proteins (GIPs; (40, 41)). Proteomic analysis of secreted proteins of P. infestans cultures grown on a synthetic medium resulted in the unambiguous identification of nine signal peptide containing proteins (9). Studies on cell wall located proteins of P. ramorum and P. infestans revealed the presence of effector proteins or pathogen-associated molecular pattern molecules, either as part of the incorporated or immobilized moiety. In addition, proteins both with and without predicted signal peptide were identified (42, 43). Despite these efforts, a more comprehensive overview of the in vivo extracellular proteome of P. infestans, or any other oomycete, is currently lacking.
Here we describe the in vivo repertoire of secreted and extracellular proteins from P. infestans. In an attempt to mimic various natural environments, mycelium was grown in liquid media varying in composition. We recently described that P. infestans secretes an enzyme with phospholipase D (PLD) activity (44). Here, this PLD activity was used as an extracellular marker to monitor the effect of media composition. We initially used (LC)-MS/MS to identify the proteins present in the extracellular medium based on the predicted secretome (17), but the search was further extended to identify additional proteins present in the medium that either do not have a signal peptide or were not predicted as secreted proteins. Our proteomics results did not only lead to the identification of many extracellular proteins that can now be considered either valid secretome proteins or LSP extracellular proteins, but it also led to the correction of many ORFs annotations in the P. infestans genome. This work provides, therefore, a comprehensive characterization of the in vivo secretome and extracellular proteome of P. infestans and it additionally supplies the data essential for future research.
EXPERIMENTAL PROCEDURES
Phytophthora infestans Culture Conditions and Sampling
P. infestans strains T30–4 and NL-88069 were routinely cultured at 18 °C in the dark on Rye agar medium supplemented with 2% sucrose (45). Mycelial plugs (Ø 0.5 cm) obtained from the edge of the growing colony were used to inoculate varying liquid cultures of 15 ml ranging from nutrient rich to nutrient poor media. The media used were V8 (nonclarified), V8 clarified (V8Cl), various dilutions (V81/2 and V81/4), Plich medium (PL, ± yeast), and Henniger medium (Hen) (46). The extracellular medium was harvested either after 10 days of sustained growth, or after overnight incubation with fresh medium, as described in previous studies (9, 38, 47). Medium was recovered or replaced by tilting of the Petri dish to such an angle that the mycelial mat remained undisturbed and the fluid congregated and could be retrieved by pipetting. Replacement of the growth medium involved rinsing the mycelial mat with growth medium. Upon collection of the samples the extracellular medium was immediately centrifuged for 2 min at 10,000 × g and the supernatant was collected and filtered through 0.2 μm filters. Viability staining revealed no significant damage of hyphae during the extracellular medium retrieval. Squeezing of mycelia was performed by folding mycelia into a stack and by pressing until all fluid was expelled. The mycelial mat was then refolded and allowed to rehydrate for ∼1 min. This process was repeated three times.
Phospholipid Analysis
Extracellular PLD activity was determined as described previously (44). Radio-labeling of P. infestans was performed by overnight incubation of mycelial plugs (grown in a 96 wells plate in V8), with 100 μCi carrier-free 32PO43− (GE Healthcare, Diegem, Belgium) in a volume of 200 μl. Upon addition of propanol (2% final concentration) the mycelial plugs were either incubated at room temperature or frozen in liquid nitrogen for 5 min and left to defrost for 15 min. Incubations were terminated by addition of 20 μl perchloric acid (50%, v/v) and the lipids isolated as described before (44). Phospholipids were separated using the alkaline solvent system (48). Radiolabeled phospholipids were visualized by phosphoimaging (Storm, Molecular Dynamics; Sunnyvale, CA, USA).
Protein Sample Preparation
All sample media were concentrated by ultrafiltration with a MWCO membrane (Vivaspin 15R, 2000 MW; Sartorius, Gottingen, Germany) and protein content was determined using the BCA Protein Quantification Kit (Thermo Fisher Scientific, San Jose, CA). 100 μg of sample were precipitated by the addition of six volumes of acetone (overnight, 4 °C). Precipitated proteins were then dissolved in 100 μl of 6 m Urea plus 200 mm NH4HCO3. Samples were reduced with dithiothreitol (15.5 μm, 1 h, 37 °C) and alkylated in the dark with iodoacetamide (3.1 μm, 30 min, 25 °C). The resulting protein extract was then diluted with 200 mm NH4HCO3 (dil. 1/6) and digested with 10 μg of trypsin (Promega, Madison, Wi; cat # V5113) (overnight, 37 °C). Finally, the peptide mix was acidified with formic acid and desalted with homemade Empore C18 column (3 m Inc.) prior to LC-MS/MS analysis (49).
Chromatographic and Mass Spectrometric Analysis
The peptide mixes were analyzed using a LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled to an EasyLC (Thermo Fisher Scientific (Proxeon), Odense, Denmark). Peptides were loaded directly onto the analytical column at a flow rate of 1.5–2 μl/min using a wash-volume of four times the injection volume, and were separated by reversed-phase chromatography using a 12-cm column with an inner diameter of 75 μm, packed with 5 μm C18 particles (Nikkyo Technos Co., Ltd. Japan). Chromatographic gradients started at 97% buffer A and 3% buffer B with a flow rate of 300 nl/min, and gradually increased to 93% buffer A and 7% buffer B in 1 min, and to 65% buffer A and 35% buffer B in 120 min. After each analysis, the column was washed for 10 min with 10% buffer A and 90% buffer B. Buffer A: 0.1% formic acid in water. Buffer B: 0.1% formic acid in acetonitrile.
The mass spectrometer was operated in positive ionization mode with nanospray voltage set at 2.2 kV and source temperature at 275 °C. Ultramark 1621 for the FT mass analyzer was used for external calibration prior the analyses. Internal calibration was performed using the background polysiloxane ion signal at m/z 445.1200. The instrument was operated in data dependent mode in which a survey scan was followed by the sequential fragmentation of the ten most intense precursors. Full MS scans were acquired with 2 microscans at resolution of 30,000, and a mass range of m/z 350–2000. Auto gain control (AGC) was set to 1e6, dynamic exclusion to 60 s, and charge state filtering was set to disqualify singly charged peptides. Normalized collision energy of 35% was used. Fragment ion spectra produced via high-energy collision dissociation (HCD) were acquired in the Orbitrap mass analyzer with a resolution of 7500. AGC was set to 5e4, isolation window to 2.0 m/z, and activation time to 0.1 ms. A maximum injection time of 100 ms was used during data acquisition. All data were acquired with Xcalibur software v2.1.
Data Analysis
The Proteome Discoverer software suite (v1.4.0.288 Thermo Fisher Scientific) and the Mascot search engine (v2.3.01, Matrix Science LTD, London, UK, (50)) were used for peptide identification. Data were initially searched against an in-house generated database based on the in silico predicted secretome of P. infestans as described by Raffaele et al. (17) (supplementary Table S1). A precursor ion mass tolerance of 7 ppm at the MS1 level was used, and up to three miscleavages for trypsin were allowed. The fragment ion mass tolerance was set to 20 mmu. Oxidation of methionine and N-terminal protein acetylation were defined as variable modification, whereas carbamidomethylation on cysteines was set as fixed modification. In all cases, false discovery rate (FDR) in peptide identification was evaluated by using a decoy database and it was set to a maximum of 1%. Identified proteins were grouped in protein groups using the algorithm implemented in Proteome Discoverer software suite (v1.4.0.288 Thermo Fisher Scientific), and only peptides uniquely mapping to a protein group were taken into consideration for the identification of protein groups and protein group members.
Unassigned spectra from the P. infestans secretome sample were searched against a spectral library created with all PSMs belonging to control media samples using Spectrast tool (51), and matching spectra were excluded from further analyses. The remaining unassigned spectra were analyzed with PEAKS v6.0 using a de novo sequencing strategy associated with database search (52). A subset of NCBInr including only P. infestans sequences (January 2012, nearly 37350 sequences) was used and FDR was set to a maximum of 5%. The identified proteins were annotated by comparison with other close organisms using Blast2GO (53). Newly identified proteins were manually curated and those related to secretion were added to the predicted secretome (17), which was used for a second database search (Mascot v2.4, Proteome Discoverer v1.4.0.288) using the same parameters of the initial database search. All used proteins (final versions used for analysis) are listed in supplementary Table S2. The acquired data in this study is publicly available in the ProteomeXchange repository with the accession number PXD000802.
Bioinformatic Analysis
Signal peptide prediction was performed using a combination of SignalP (version 3.0 and 4.1), TargetP (version 1.1), and TMHMM (version 2.0) with default settings (54–56). Proteins were considered secreted, if both the neural-network as well as the hidden-markov model in both SignalP versions identified a signal peptide. Moreover, to select a candidate as a secreted protein, we also required TargetP to predict the proteins to enter the secretory pathway. To prevent false positive assignments, we subsequently scanned the proteins for transmembrane domains using TMHMM. If no transmembrane or only a single transmembrane domain with significantly overlap (≥10 amino acid; start position of transmembrane domain within the first 35 amino acids) with the predicted signal peptide, was detected, we retained the protein. If more than a single transmembrane domain was predicted, the protein was discarded.
RESULTS
Phospholipase D Activity Release is Dependent on Media Composition
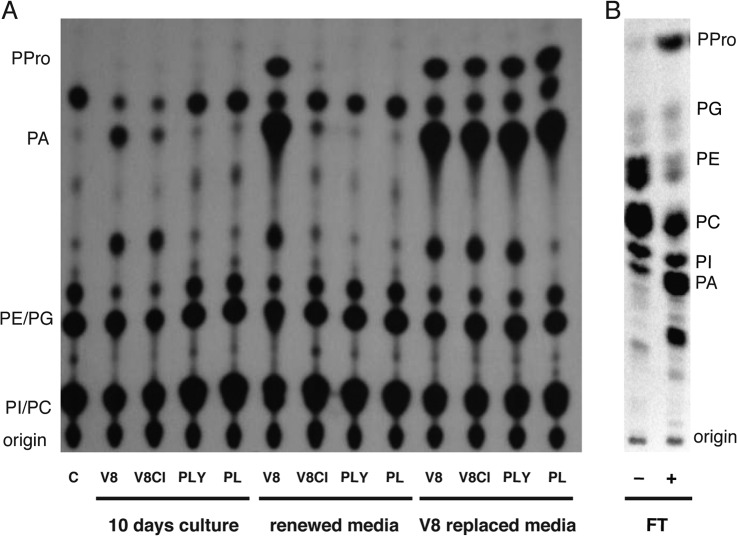
We previously showed that PLD activity was present in extracellular medium of P. infestans strains, as demonstrated by the production of phosphatidic acid (PA) and the PLD specific marker phosphatidylalcohol (PPro), when cultured on V8-agar or RS-agar plates flooded with V8 juice medium (44). P. infestans is capable of growing in various media ranging from vegetable based, nutritiously rich extracts (e.g. V8 juice or Rye sucrose; RS) to minimal media with poor nutritional content (e.g. Plich medium). It was deduced that PLD activity in the extracellular medium could act as an enzymatic marker to monitor P. infestans response to changing nutritional conditions. P. infestans strain NL-88069, was tested for extracellular PLD activity after 10 days of growth in V8 medium (V8), Clarified V8 (V8Cl), Plich with (PL+Y) and without yeast extract (PL-Y). Our results show that PLD activity correlated to the amount of nutritious elements present in the medium (Fig. 1A, 10 days culture). The highest activity, indicated by the production of PA and PPro, was detected in nutritious V8 medium whereas PL-Y extract lacked PLD activity. We anticipated that in all growth media, the mycelium is capable to secrete PLD activity but required a nutritional trigger. To test this hypothesis, we renewed the media 1 day before sampling. No PLD activity increase was detected under low nutritional conditions whereas PLD activity was detected in V8 based extracellular media. Surprisingly, the amount of PLD activity tremendously increased under renewed medium (Fig. 1A). This implies that renewal of V8 medium results in a quantitative release of active PLD enzyme. This point was strengthened by substituting all growth media with V8 juice 1 day before sampling which resulted in the detection of PLD activity for all used media at similar high levels (Fig. 1A, V8 replaced media). To discard the possibility that the detected PLD activity might be caused by hyphal rupture, we squeezed the mycelial mat repeatedly in the presence of sample medium, which resulted in only a minor increase in PLD activity. This increase was negligible when compared both to the activity release upon medium renewal and to the PLD activity obtained after snap-freezing the mycelial tissue (Fig. 1B). The latter treatment resulted in a major nonspecific breakdown of structural phospholipids such as phosphatidylcholine (PtdCho, −70%) and phosphatidylethanolamine (PtdEtn, −90%) (n = 3). Altogether, our results show that extracellular active PLD release is mediated by the growth media nutrient content.
Fig. 1.
Extracellular PLD activity depends on secretion rather than hyphal rupture and varies depending on the nutritional value of the growth media. A, Metabolically labeled phospholipids were isolated and vesicles generated as described before (44). Vesicles were incubated in the presence of 2% propanol with buffer only (C) or with extracellular medium derived from 10 days culture with indicated media, with overnight incubated renewed medium or overnight incubated with V8 medium. Lipids were visualized by phosphoimaging after extraction and separation by ethyl acetate TLC (44). The experiment was repeated twice with similar results. B, P. infestans mycelial plugs were metabolically labeled with 32P and left untreated, or snapfrozen and thawed (FT) for 15 min in the presence of 2% propanol. Phospholipids were extracted, separated by alkaline TLC (7474) and analyzed by phosphoimaging. The origin, phosphatidylinositol (PI), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidic acid (PA), and phosphatidylpropanol (PPro) are indicated. A representative experiment is shown.
Extracellular Proteome Covers a Significant Part of the In Silico Predicted Secretome
The medium-dependent PLD release suggests that the extracellular proteome composition is highly dependent on the environmental conditions and can be rapidly altered upon its modification or replacement. Therefore, to achieve optimal characterization of the secretome and extracellular proteome, we collected extracellular medium from P. infestans mycelium cultivated in various media with nutrient value differences. Extracellular medium was harvested in triplicates at 10 days after inoculation with a hyphal plug of strain T30–4. Proteins present both in the extracellular medium and in fresh growth media (controls) were analyzed per triplicate by LC-MS/MS (supplementary Table S3). Initially, we used the in silico secretome database described by Raffaele et al. (17) to corroborate the presence in extracellular media, and thus, validate these predicted secreted proteins as part of the secretome of P. infestans. During the data analysis, only unique peptides per protein group (defining a protein group as a set of undistinguishable proteins given the identified peptides), that were present in at least two replicates and absent in the control fresh medium were considered for protein identification. This analysis resulted in the identification of an initial set of 149 different protein groups that corresponded to 254 proteins. This made up for 18% of the original predicted secretome (17).
Based on the high number of not-annotated high-quality spectra present in the initial secretome analysis, we anticipated that there were many more proteins present in the extracellular medium that could be missed either because of genome annotation errors or to the lack of signal peptide (non-conventional secretion pathway). In order to identify additional extracellular proteins, we proceeded to re-annotate unassigned spectra from the initial database search using a de novo peptide sequencing strategy assisted with database search (Fig. 2A).
Fig. 2.
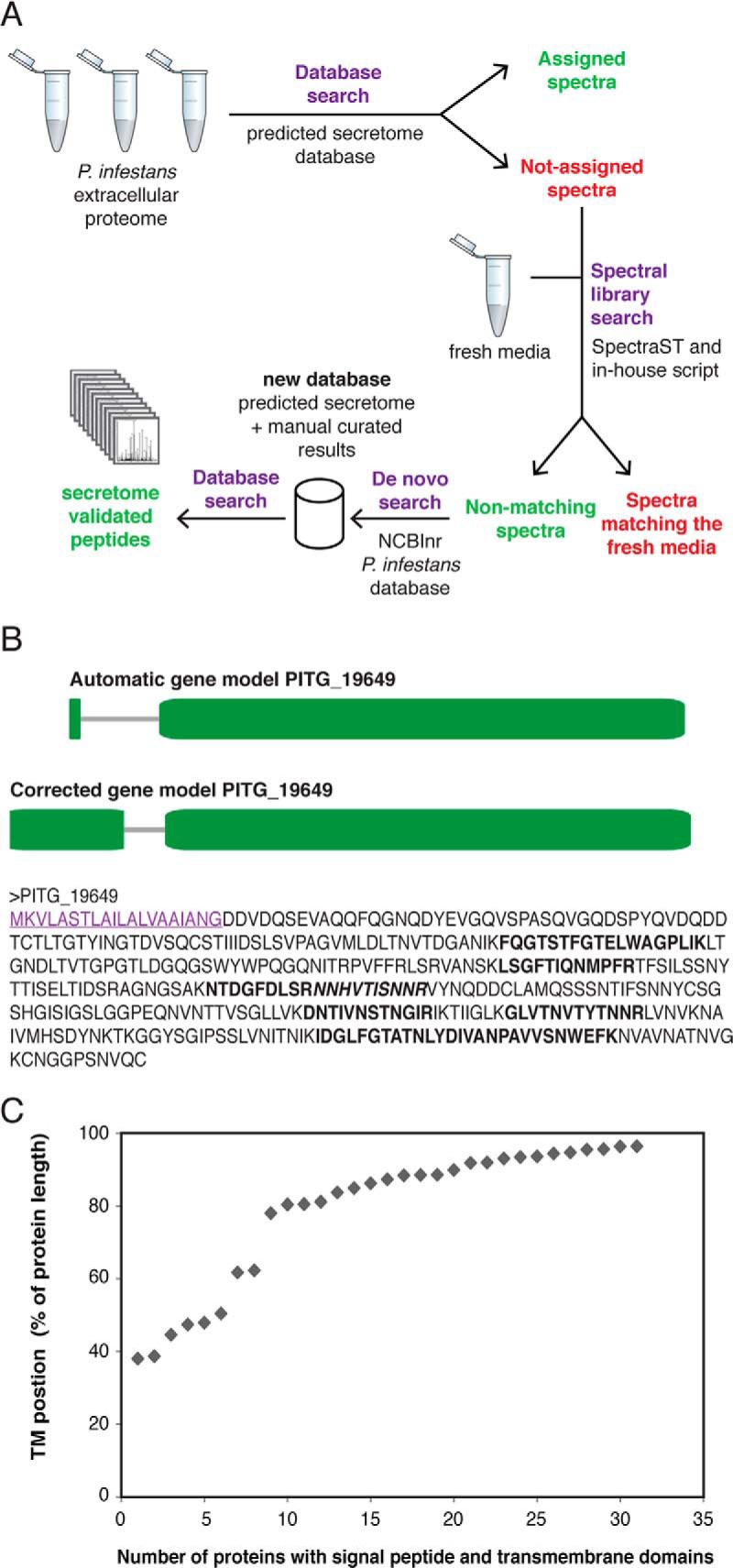
A, General mass spectrometric and data analysis workflow used in this study. B, Manual annotation of gene model PITG_19649 encoding an endopolygalacturonase. Top: automatic gene model (www.broadinstitute.org), numbers indicate protein length in AA; Middle: corrected gene model; Bottom: final protein prediction in AA. Signal peptide (SignalPv3.0: 1.000; SignalPv4.0: 0.862) is underlined. All amino acids retrieved in peptides by mass spec analysis are indicated in bold and adjacent peptides are differentiated by (non-)italics. C, Proteins identified from the workflow shown in A, were analyzed for the presence of a signal peptide (SignalP; version 3) and a single transmembrane domain (TMHMM). The transmembrane (TM) location is plotted as relative distance from the protein start for all 31 identified proteins.
The de novo proteome analysis assisted with a complete P. infestans database (NCBI, P. infestans) resulted in the identification of 1105 individual proteins based on unique peptides (supplementary Table S4), with the highest number of proteins obtained in P. infestans cultures grown in V8 medium (649 proteins) and the lowest amount of identified proteins in PL-Y medium (222 proteins). Among the identified proteins, there were 316 proteins containing a signal peptide of which 212 had already been predicted to be secreted proteins (17). Identified proteins by de novo sequencing were manually curated to search for additional evidences supporting potential secreted proteins. Putative secreted proteins were assessed based on automatic annotation by sequence homology with other phyla such as the oomycetes P. sojae and P. ramorum (53), and on the prediction of a signal peptide (54). For example, several peptides were identified for the C-terminal part of protein PITG_19649, which was not included in the initial secretome list (17), but which had been annotated as putative “endopolygalacturonase” based on the presence of a glycosyl hydrolase domain (4). This piece of evidence urged us to revise the 5′-end of the gene model resulting in a signal peptide bearing protein explaining its detection in the extracellular medium (Fig. 2B). Nearly 150 reference gene models were re-annotated in a similar manner when necessary (see supplemental Table S5). Whenever gene models were updated, the encoded proteins were re-assessed for the presence of signal peptides and compared with proteins encoded in close phyla. Forty-four proteins now encode a signal peptide that had not been considered in the reference annotation (17).
During the de novo analyses of the extracellular proteome we detected many transmembrane-containing proteins. Transmembrane containing proteins were previously identified from the oomycete cell walls (42, 43). In total, 31 proteins were identified by de novo proteome analysis that contained a signal peptide and a single transmembrane domain. The location of the transmembrane location was determined for each protein. This revealed that with only few exceptions all transmembrane domains were located in the C-terminal extreme of the protein (Fig. 2C).
Finally, a new database was built containing the in silico predicted secretome (17) and the manually curated proteins that were identified by de novo peptide sequencing (supplementary Table S2). A Mascot database search was performed on the new database, containing a total of 2253 entries from P. infestans, to validate the new peptide and protein identifications. Only unique peptides per protein group that were present in at least two replicates and absent in the control fresh medium (blank) were considered for protein identification. In total, 200 protein groups (283 proteins) corresponding to the secretome and extracellular proteome of P. infestans were identified, from which 201 proteins had already been predicted as secreted proteins (supplementary Tables S1, S6, and S7). Moreover, 227 proteins identified in this study contained a signal peptide, from which 14 proteins were not considered in the original list reported by Raffaele et al. (17) as they result from manual gene model corrections.
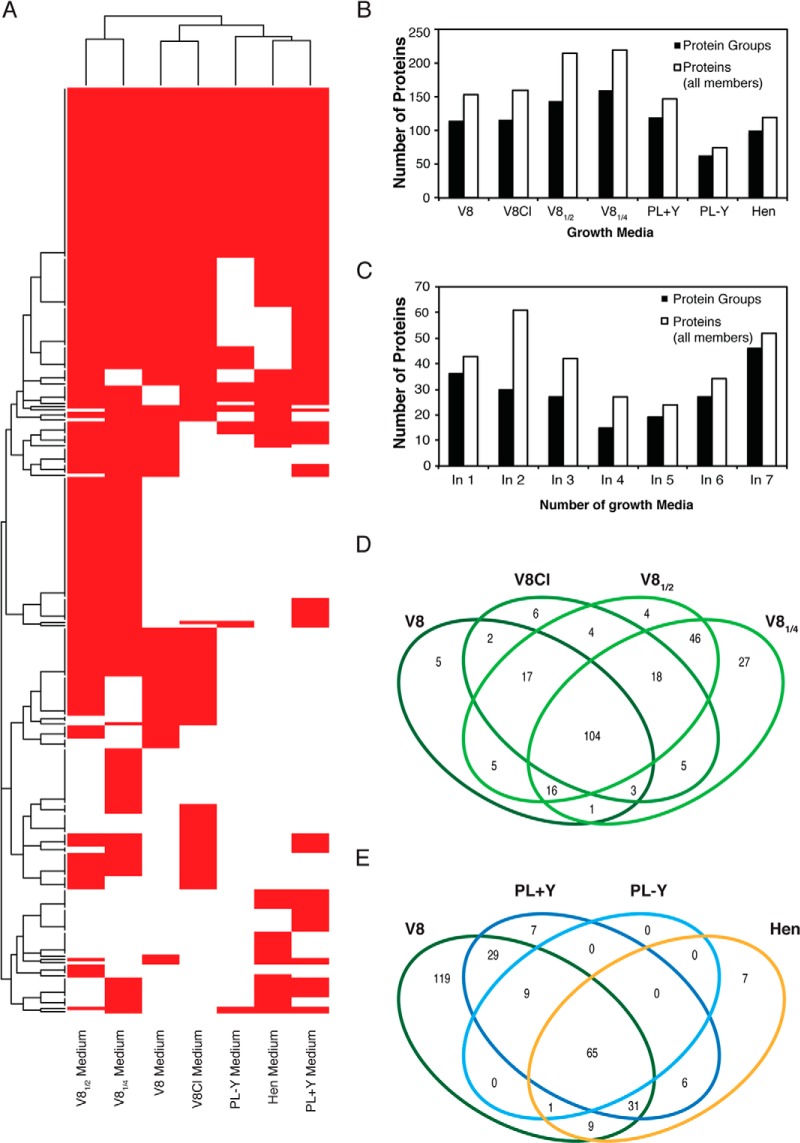
The number of identified proteins ranged from 63 protein groups (75 proteins) in the minimal PL-Y to 159 protein groups (220 proteins) in V81/4 medium thus correlating with the nutritional value of the media and their PLD activity. The complete distribution of hits per medium is shown in Fig. 3. About half of the identified extracellular proteins were detected in three or more growth media (Fig. 3C), and an important protein overlap was observed between cultures grown on the V8 family growth media and the rest of growth media, with almost no new proteins identified in the PL+Y, PL-Y and Hen media (Fig. 3D and 3E).
Fig. 3.
A, Heatmap representing the presence (red) and absence (white) of the identified proteins after a 10-day culture of P. infestans. B, Number of extracellular proteins and protein groups from P. infestans identified in different media. C, Number of extracellular identified protein groups and all protein group members (Proteins) and from P. infestans identified in at least n different media. D and E, Venn diagrams representing the overlap among all protein group members from P. infestans identified in different media. In all cases, P. infestans was cultured for 10 days, and protein identification was based on an in-house compiled database containing both the predicted secreted protein entries and the manually curated proteins identified by de novo strategy.
Incubation with Fresh Medium Results in Minor Changes in the In Vivo Secreted Proteome
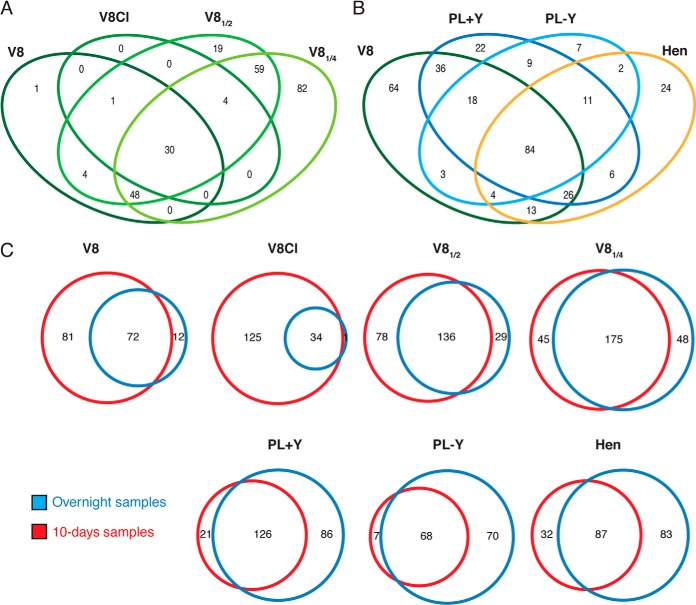
After identifying the proteins present in the secretome of the mycelial mat after 10 days of growth, we aimed to determine whether fresh media could trigger the release of new secreted proteins similarly to the observed induced release of active PLD. For this purpose, the 10-day mycelial mat was repeatedly washed and incubated overnight with fresh medium. Overnight extracellular media were processed as described above and subjected to LC-MS/MS analysis. Acquired data was analyzed using a Mascot database search with the same database and parameters as the ones used for the 10-day cultures (FDR ≤ 1%, peptide level). In total 330 proteins were detected of which 244 had previously been detected in the 10-day cultures. Because equivalent amounts per protein were retrieved per volume of extracellular medium, we conclude that there is a significant de novo protein secretion upon overnight incubation with fresh medium. However, the types and quantities of the proteins identified in these new samples were over 80% similar to the 10-day samples suggesting that the extracellular proteome composition does not respond majorly to the application of fresh medium (Fig. 4). The sequences of all the peptides identified by LC-MS/MS fragmentation are given in supplementary Table S8.
Fig. 4.
A, Venn diagrams representing the overlap among all protein group members from P. infestans identified in different media after an overnight culture. B, Venn diagrams comparing all the protein group members identified from P. infestans in different media after a 10-day culture and an overnight culture. In all cases, protein identification was based on an in-house compiled database containing both the predicted secreted protein entries and the manually curated proteins identified by de novo strategy.
DISCUSSION
The main goal of this study was to characterize the in vivo secretome and extracellular proteome of P. infestans during mycelial growth. Because purification of extracellular proteins from intracellular fluids of potato layers is technically challenging because of plant tissue collapse, we mimicked such natural hyphal growth conditions by using various media that differ in nutrient content. Fresh extracellular medium was also analyzed for protein composition but only a very limited number of proteins were identified, even in the full strength, non-clarified V8 media showing that original proteins coming from the fresh medium are highly diluted and degraded. The identification of secreted and extracellular proteins rendered a global P. infestans extracellular proteome that validates the existing predicted secretomes, which was highly based on in silico analysis.
Evaluation of PLD Activity in Different Media
Initially, we inferred that protein secretion activity is dependent on the growth conditions, as shown by the presence of the PLD activity in the different media. The release of extracellular PLD activity during an overnight incubation with fresh nutrient rich medium points to the rapid release of quantitative amounts of proteins suggesting the existence of highly regulated mechanisms. The mechanism that triggers protein secretion in Phytophthora remains to be determined but, as reported for fungi, nutrient sensing G protein-coupled receptors (GPCRs) could play a crucial role in this process (57). Indeed, the Phytophthora genomes harbor around 60 genes encoding putative GPCRs, among which are novel classes that might be involved in direct downstream signaling and play roles in chemotaxis as well as in developmental aspects (58, 59).
Initial Predicted Secretome and Definition of the Extracellular Proteome by De Novo Protein Identification
Once PLD activity was assessed, we used our experimentally generated dataset to search the in silico secretome database described by Raffaele et al. (17) and thus validate the predicted secreted proteins as part of the P. infestans secretome. A first analysis resulted in the identification of an initial set of 149 different protein groups that corresponded to 254 proteins, which represents the validation of around 18% of the original predicted secretome (17). However, because of the high number of high-quality spectra that were not annotated in this initial secretome analysis, we proceeded to re-annotate unassigned spectra from the initial database search. We performed a de novo peptide identification strategy assisted with database search in order to identify additional extracellular proteins that were not included in the predicted secretome.
Over one thousand individual protein groups were identified in these analyses and although several of the extracellular proteins identified were bearing a secretion signal peptide, many did not. Although hyphal lysis may occur, there are evidences that the presence of intracellular proteins in the extracellular medium—such as enolases, ribonucleases, and related proteins—might act as virulence factors and be involved in a variety of extracellular functions (22, 60, 61). Therefore, proteins identified by de novo sequencing were annotated by sequence homology with other oomycetes to search for additional evidence supporting their identification. Over a hundred reference gene models were re-annotated to confirm the presence of a signal peptide. Finally, the comparison of our de novo results, with previous studies in P. infestans, showed a considerable overlap among the identified proteins in the extracellular medium (9, 42), and similar results were obtained when comparing our dataset to the extracellular proteome of C. albicans (62).
Validation of the Predicted Secretome
To validate the in silico predicted secretome and control the false-discovery rate, a new database was build containing both the original in silico predicted secretome (17) and the manually curated proteins that were identified by de novo peptide sequencing (supplementary Table S2). In total, 200 protein groups (283 proteins; supplementary Table S7) corresponding to the secretome of P. infestans were identified, from which 201 proteins had already been predicted as secreted proteins in previous studies. Our study does not only validate a fraction of the in silico predicted secretome, but it greatly extends this validated subset (i.e. by over 40% as it adds 82 proteins to the 201 protein subset).
Among the proteins that were identified as part of the secretome of P. infestans are those that play a role in defense to oxidative stress such as catalases, peroxidases, and thioredoxin proteins (Table I). We could also identify five out of the seven berberine-like proteins encoded in the genome. These proteins that are involved in alkaloid biosynthesis and in the production of hydrogen peroxide through the oxidation of numerous metabolites (63), and they are thought to be important virulence factors induced during plant infection (17, 64). Alternatively, these proteins might also protect Phytophthora from plant counter defenses. Our dataset cannot shed light on the function of these proteins, but it clearly shows that berberine-like proteins are widely secreted by Phytophthora during hyphal growth.
Table I. Main protein families detected as part of the secretome of P. infestans.
| Protein families | Identifiers |
|---|---|
| Defense to oxidative stress | |
| Thioredoxines | PITG_10972, PITG_14917 |
| Catalases | PITG_05579 |
| Peroxidases | PITG_05579, PITG_18316, PITG_05254, PITG_04900 |
| berberine-like proteins | PITG_02928, PITG_02930, PITG_06585, PITG_06591, PITG_02935 |
| Pathogen associated proteins and cytosolic effectors | |
| RXLRs | PITG_19617, PITG_15972,PITG_08278, PITG_08943, PITG_20303, PITG_20301, PITG_04090, PITG_04085, PITG_20300, PITG_18683, PITG_14371, PITG_12706, PITG_16275, PITG_23131, PITG_23129, PITG_10654, PITG_15278, PITG_12710, PITG_09216, PITG_09218 |
| CRNs | PITG_14309, PITG_22536, PITG_17199, PITG_17185, PITG_05043, PITG_05049, PITG_18847, PITG_19377, PITG_19509, PITG_18503, PITG_18571, PITG_18497, PITG_19373 |
| Elicitins | PITG_12551, PITG_12561, PITG_21410, PITG_12562, PITG_16907, PITG_19604, PITG_01024 |
| NPPs (Necrosis inducing Phytophthora Proteins) | PITG_04208, PITG_16866, PITG_19938, PITG_22053, PITG_22734 |
| Trans-membrane domain-containing proteins and sheddases | |
| Single transmembrane proteins | PITG_06662, PITG_14156, PITG_04568, PITG_06481, PITG_07210, PITG_00646, PITG_19270, PITG_20079, PITG_07536, PITG_05561, PITG_10064, PITG_10543, PITG_09768, PITG_02058, PITG_07537, PITG_07523, PITG_06215, PITG_07521, PITG_00894, PITG_17411, PITG_06867, PITG_15746, PITG_11524, PITG_01397, PITG_01395, PITG_14917, PITG_09798, PITG_07774, PITG_11993, PITG_06170, PITG_09760 |
| Aspartic protease | PITG_09387 |
| Cysteine protease | PITG_02423, PITG_03020, PITG_12041, PITG_16276 |
| Metalloproteases | PITG_00056, PITG_09851 |
| SCP-like proteins | PITG_10409, PITG_10408, PITG_15746, PITG_11060, PITG_10410 |
Localized secretion of most effectors is known to take place at the haustorium, a specialized structure originating from the hyphae, which is not penetrating the plant cell but invigilates living host plant cells. In the fungus Magnaporthe oryzae, a differentiation in secretion systems was recently described. Apoplastic effectors are secreted from invasive hyphae whereas cytoplasmic effectors were delivered via the biotrophic interfacial complex (65). Although our data does not yet support such a distinguishing mechanism in Phytophthora, we detected a large quantity of predicted cytosolic effectors RXLRs (20 identified) and CRNs (13 identified) being released from hyphae in the absence of haustoria. In the case of CRNs, it remains challenging to identify the individual gene products because most peptides are shared among the different CRNs because of high conservation levels, only a single unique peptide was derived.
Moreover, we detected 11 out of the 40 elicitins encoded in the genome (4). Elicitins are oomycete specific proteins belonging to the pathogen associated molecular patterns (PAMPs) that trigger a hypersentive response in plants (66). Among those, INF1 was detected at high levels in all media and was thus the most prominently present elicitin together with INF2A-like, INF4, INF5A, and INF7 (38).
The analysis of the validated secretome also revealed a total of 31 proteins that contain a signal peptide in combination with a single transmembrane domain, which divides the mature proteins in an extracellular and a cytoplasmic domain. With few exceptions, nearly all single transmembrane domains were C-terminally located (Fig. 2C) and all detected peptides corresponded to the extracellular domain, upstream of the transmembrane region. Although unspecific protein shearing or degradation cannot be discarded, the presence of these proteins in the extracellular medium might also reflect ectodomain shedding. Shedding is the proteolysis of ectodomains of membrane proteins by a sheddase and it has been well established in various organisms in which both the extracellular and the cytosolic remnants might act as functional components (67–69). In fungi, membrane proteins, e.g. PraI, Msb2, have been described to contain a single transmembrane domain, which is shed by proteolytic cleavage by proteases, (68, 69). Therefore, the identification of transmembrane proteins as secreted proteins suggests the existence of a specific protein shedding activity in P. infestans. Although the concrete shedding enzymes in P. infestans remain to be identified, aspartic proteases, metalloproteases, cysteine proteases, and sperm-coating-like proteins could play a role in this process. Indeed, we identified one aspartic protease, two metalloproteases and four cysteine proteases among the secreted proteins in Phytophthora, as well as five SCP-like (Sperm-coating proteins) proteins (Table I). The SCP-like domain family is found in eukaryotes, prokaryotes and archaea and it includes mammalian cysteine-rich secretory proteins (CRISPS) involved in the reproductive system (70), and plant pathogenesis related proteins (PR-1s) (71–73). So far, no SCP-like protein of Phytophthora has directly been linked to pathogenesis or proteolytic activity although their identification among the secreted proteome could anticipate their role in protein shedding activity.
CONCLUSIONS
Living organisms interact with their environment by sensing cues and responding to them by different physiological adaptations. Our results suggest that P. infestans is capable to sample its environment for cues of nutrient composition, and while doing so, it surrounds itself with a set of extracellular proteins that prepares it for encountering and infection of the host plant. Here we provide a comprehensive characterization of the in vivo secretome and extracellular proteome of P. infestans and it additionally supplies the data files that will be essential for future research. Our proteomics results do not only lead to the identification of many extracellular proteins that can now be considered valid secretome proteins, but they also lead to the correction of many ORFs annotations in the P. infestans genome. The generated dataset demonstrates the validity and shortcoming of in silico analysis at the same time. In silico analysis obviously lacks sensitivity toward oomycete protein transport peculiarities, strengthened by the used cut-offs affecting the subcellular localization prediction. Strikingly, many proteins do not belong to the usual suspects such as those lacking a signal peptide, or those encoding valid transmembrane domain(s). Our approach considerably benefitted from manual gene model re-annotation, something unfeasible for full genome-based proteome analysis. However, it illustrates the potential scale of currently unconsidered proteins encoded in the genome.
Supplementary Material
Footnotes
Author contributions: H.J.M., C.C., F.G., and E.S. designed research; H.J.M., F.M.M., G.E., M.F.S., C.C., F.G., and E.S. performed research; H.J.M., F.M.M., G.E., M.F.S., C.C., F.G., and E.S. contributed new reagents or analytic tools; H.J.M., F.M.M., G.E., M.F.S., C.C., F.G., and E.S. analyzed data; H.J.M., F.G., and E.S. wrote the paper.
* This work has been supported by the PRIME-XS project, grant agreement number 262067, funded by the European Union 7th Framework Programme Programme (FP7/2007–2013). We thank NWO-STW for the VIDI grant (Nr. 10281) to H.J.G.M. The CRG/UPF Proteomics Unit is part of the Spanish Proteomics Platform “Plataforma de Recursos Biomoleculares y Bioinformáticos” (ProteoRed, Instituto de Salud Carlos III), and it is co-funded by the European Regional Development Fund (ERDF) in the framework of the Operational Programme of Catalonia 2007–2013. Objective 2 of regional competitiveness and employment. We also acknowledge support of the Spanish Ministry of Economy and Competitiveness, through the 'Centro de Excelencia Severo Ochoa 2013–2017' program (SEV-2012–0208).
 This article contains supplemental Tables S1 to S8.
This article contains supplemental Tables S1 to S8.
1 The abbreviations used are:
- CRN
- crinkling and necrosis producing
- PLO
- phospholipase D.
REFERENCES
- 1. Keeling P. J., Burger G., Durnford D. G., Lang B. F., Lee R. W., Pearlman R. E., Roger A. J., Gray M. W. (2005) The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676 [DOI] [PubMed] [Google Scholar]
- 2. Kroon L. P., Brouwer H., De Cock A. W., Govers F. (2012) The genus Phytophthora anno 2012. Phytopathology. 102, 348–364 [DOI] [PubMed] [Google Scholar]
- 3. Stassen J. H., van den Ackerveken G. (2011) How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14, 407–414 [DOI] [PubMed] [Google Scholar]
- 4. Haas B. J., Kamoun S., Zody M. C., Jiang R. H., Handsaker R. E., Cano L. M., Grabherr M., Kodira C. D., Raffaele S., Torto-Alalibo T., Bozkurt T. O., Ah-Fong A. M., Alvarado L., Anderson V. L., Armstrong M. R., Avrova A. O., Baxter L., Beynon J., Boevink P. C., Bollmann S. R., Bos J. I. B., Bulone V., Cai G., Cakir C., Carrington J. C., Chawner M., Conti L., Costanzo S., Ewan R., Fahlgren N., Fischbach M. l. A., Fugelstad J., Gilroy E. M., Gnerre S., Green P. J., Grenville-Briggs L. J., Griffith J. M., Grunwald N. J., Horn K., Horner N. R., Hu C. H., Huitema E., Jeong D. H., Jones A. M. E., Jones J. D. G., Jones R. W., Karlsson E. K., Kunjeti S. G., Lamour K., Liu Z., Ma L. J., Maclean D. J., Chibucos M. C., McDonald H., McWalters J., Meijer H. J. G., Morgan W., Morris P. F., Munro C. A., O'Neill K., Ospina-Giraldo M. D., Pinzon A., Pritchard L., Ramsahoye B., Ren Q., Restrepo S., Roy S., Sadanandom A., Savidor A., Schornack S., Schwartz D. C., Schumann U. D., Schwessinger B., Seyer L., Sharpe T., Silvar C., Song J., Studholme D. J., Sykes S., Thines M., van de Vondervoort P. J. I., Phuntumart V., Wawra S., Weide R., Win J., Young C., Zhou S., Fry W. E., Meyers B. C., van West P., Ristaino J. B., Govers F., Birch P. R. J., Whisson S. C., Judelson H. S., Nusbaum C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398 [DOI] [PubMed] [Google Scholar]
- 5. Tyler B. M., Tripathy S., Zhang X., Dehal P., Jiang R. H., Aerts A., Arredondo F. D., Baxter L., Bensasson D., Beynon J. L., Chapman J., Damasceno C. M., Dorrance A. E., Dou D., Dickerman A. W., Dubchak I., Garbelotto M., Gijzen M., Gordon S. G., Govers F., Grunwald N. J., Huang W., Ivors K. L., Jones R. W., Kamoun S., Krampis K., Lamour K. H., Lee M. K., McDonald W. H., Medina M., Meijer H. J. G., Nordberg E. K., Maclean D. J., Ospina-Giraldo M. D., Morris P. F., Phuntumart V., Putnam N. H., Rash S., Rose J. K. C., Sakihama Y., Salamov A. A., Savidor A., Scheuring C. F., Smith B. M., Sobral B. W. S., Terry A., Torto-Alalibo T. A., Win J., Xu Z., Zhang H., Grigoriev I. V., Rokhsar D. S., Boore J. L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 [DOI] [PubMed] [Google Scholar]
- 6. Kaschani F., Shabab M., Bozkurt T. O., Shindo T., Schornack S., Gu C., Ilyas M., Win J., Kamoun S., van der Hoorn R. A. L. (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Birch P. R. J., Armstrong M., Bos J., Boevink P., Gilroy E. M., Taylor R. M., Wawra S., Pritchard L., Conti L., Ewan R., Whisson S. C., van West P., Sadanandom A., Kamoun S. (2009) Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 60, 1133–1140 [DOI] [PubMed] [Google Scholar]
- 8. Schornack S., van Damme M., Bozkurt T. O., Cano L. M., Smoker M., Thines M., Gaulin E., Kamoun S., Huitema E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. U.S.A. 107, 17421–17426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torto T. A., Li S., Styer A., Huitema E., Testa A., Gow N. A., van West P., Kamoun S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stam R., Jupe J., Howden A. J. M., Morris J. A., Boevink P. C., Hedley P. E., Huitema E. (2013) Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One 8, e59517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baxter L., Tripathy S., Ishaque N., Boot N., Cabral A., Kemen E., Thines M., Ah-Fong A. M., Anderson R., Badejoko W., Bittner-Eddy P., Boore J. L., Chibucos M. C., Coates M., Dehal P., Delehaunty K., Dong S., Downton P., Dumas B., Fabro G., Fronick C., Fuerstenberg S. I., Fulton L., Gaulin E., Govers F., Hughes L., Humphray S., Jiang R. H. Y., Judelson H., Kamoun S., Kyung K., Meijer H. J. G., Minx P., Morris P., Nelson J., Phuntumart V., Qutob D., Rehmany A., Rougon A., Ryden P., Torto-Alalibo T., Studholme D., Wang Y., Win J., Wood J., Clifton S. W., Rogers J., V. d. A. G., Jones J. D. G., McDowell J. M., Beynon J., Tyler B. M. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330, 1549–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levesque C. A., Brouwer H., Cano L., Hamilton J. P., Holt C., Huitema E., Raffaele S., Robideau G. P., Thines M., Win J., Zerillo M. M., Beakes G. W., Boore J. L., Busam D., Dumas B., Ferriera S., Fuerstenberg S. I., Gachon C. M. M., Gaulin E., Govers F., Grenville-Briggs L., Horner N., Hostetler J., Jiang R. H. Y., Johnson J., Krajaejun T., Lin H., Meijer H. J. G., Moore B., Morris P., Phuntmart V., Puiu D., Shetty J., Stajich J. E., Tripathy S., Wawra S., van West P., Whitty B. R., Coutinho P. M., Henrissat B., Martin F., Thomas P. D., Tyler B. M., De Vries R. P., Kamoun S., Yandell M., Tisserat N., Buell C. R. (2010) Genome sequence of the necrotrophic plant pathogen, Pythium ultimum, reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang R. H. Y., de Bruijn I., Haas B. J., Belmonte R., Löbach L., Christie J., van den Ackerveken G., Bottin A., Bulone V., Díaz-Moreno S. M., Dumas B., Fan L., Gaulin E., Govers F., Grenville-Briggs L. J., Horner N. R., Levin J. Z., Mammella M., Meijer H. J. G., Morris P., Nusbaum C., Oome S., Phillips A. J., van Rooyen D., Rzeszutek E., Saraiva M., Secombes C. J., Seidl M. F., Snel B., Stassen J. H. M., Sykes S., Tripathy S., van den Berg H., Vega-Arreguin J. C., Wawra S., Young S. K., Zeng Q., Dieguez-Uribeondo J., Russ C., Tyler B. M., van West P. (2013) Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet. 9, e1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamour K., Mudge J., Gobena D., Hurtado-Gonzales O. P., Schmutz J., Kuo A., Miller N. A., Rice B. J., Raffaele S., Cano L., Bharti A. K., Donahoo R. S., Finley S. L., Huitema E., Hulvey J., Platt D., Salamov A., Savidor A., Sharma R., Stam R., Storey D., Thines M., Win J., Haas B. J., Dinwiddie D., Jenkins J., Knight J. R., Affourtit J., Han C. S., Chertkov O., Lindquist E. A., Detter C., Grigoriev I. V., Kamoun S., Kingsmore S. F. (2012) Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol. Plant-Microbe Interact. 25, 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meijer H. J., Govers F. (2006) Genomewide analysis of phospholipid signaling genes in Phytophthora spp.: novelties and a missing link. Mol. Plant-Microbe Interact. 19, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 16. Kay J., Meijer H. J. G., ten Have A., van Kan J. A. L. (2011) The aspartic proteinase family of three Phytophthora species. BMC Genomics 12, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raffaele S., Win J., Cano L. M., Kamoun S. (2010) Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rabouille C., Malhotra V., Nickel W. (2012) Diversity in unconventional protein secretion. J. Cell Sci. 125, 5251–5255 [DOI] [PubMed] [Google Scholar]
- 19. Agrawal G. K., Jwa N.-S., Lebrun M.-H., Job D., Rakwal R. (2010) Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10, 799–827 [DOI] [PubMed] [Google Scholar]
- 20. Regente M., Corti-Monzon G., Maldonado A. M., Pinedo M., Jorrin J., de la Canal L. (2009) Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 583, 3363–3366 [DOI] [PubMed] [Google Scholar]
- 21. Lum G., Min X. J. (2011) FunSecKB: the Fungal Secretome KnowledgeBase. Database 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oliveira D. L., Rizzo J., Joffe L. S., Godinho R. M., Rodrigues M. L. (2013) Where do they come from and where do they go: candidates for regulating extracellular vesicle formation in fungi. Int. J. Mol. Sci. 14, 9581–9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding Y., Wang J., Wang J., Stierhof Y. D., Robinson D. G., Jiang L. (2012) Unconventional protein secretion. Trends Plant Sci. 17, 606–615 [DOI] [PubMed] [Google Scholar]
- 24. Vallejo M. C., Matsuo A. L., Ganiko L., Medeiros L. C., Miranda K., Silva L. S., Freymuller-Haapalainen E., Sinigaglia-Coimbra R., Almeida I. C., Puccia R. (2011) The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-galactosyl epitopes. Eukaryot. Cell 10, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vallejo M. C., Nakayasu E. S., Matsuo A. L., Sobreira T. J., Longo L. V., Ganiko L., Almeida I. C., Puccia R. (2012) Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J. Proteome Res. 11, 1676–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albuquerque P. C., Nakayasu E. S., Rodrigues M. L., Frases S., Casadevall A., Zancope-Oliveira R. M., Almeida I. C., Nosanchuk J. D. (2008) Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 10, 1695–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cobos R., Barreiro C., Mateos R. M., Coque J. J. (2010) Cytoplasmic- and extracellular-proteome analysis of Diplodia seriata: a phytopathogenic fungus involved in grapevine decline. Proteome Sci. 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Espino J. J., Gutierrez-Sanchez G., Brito N., Shah P., Orlando R., Gonzalez C. (2010) The Botrytis cinerea early secretome. Proteomics 10, 3020–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shepherd S. J., van West P., Gow N. A. (2003) Proteomic analysis of asexual development of Phytophthora palmivora. Mycol. Res. 107, 395–400 [DOI] [PubMed] [Google Scholar]
- 30. Grenville-Briggs L. J., Avrova A. O., Bruce C. R., Williams A., Whisson S. C., Birch P. R., van West P. (2005) Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet. Biol. 42, 244–256 [DOI] [PubMed] [Google Scholar]
- 31. Ebstrup T., Saalbach G., Egsgaard H. (2005) A proteomics study of in vitro cyst germination and appressoria formation in Phytophthora infestans. Proteomics 5, 2839–2848 [DOI] [PubMed] [Google Scholar]
- 32. Savidor A., Donahoo R. S., Hurtado-Gonzales O., Land M. L., Shah M. B., Lamour K. H., McDonald W. H. (2008) Cross-species global proteomics reveals conserved and unique processes in Phytophthora sojae and Phytophthora ramorum. Mol. Cell. Proteomics 7, 1501–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Savidor A., Donahoo R. S., Hurtado-Gonzales O., VerBerkmoes N. C., Shah M. B., Lamour K. H., McDonald W. H. (2006) Expressed peptide tags: an additional layer of data for genome annotation. J. Proteome Res. 5, 3048–3058 [DOI] [PubMed] [Google Scholar]
- 34. Resjö S., Ali A., Meijer H. J. G., Seidl M. F., Snel B., Sandin M., Levander F., Govers F., Andreasson E. (2014) Quantitative label-free phosphoproteomics of six different life stages of the late blight pathogen Phytophthora infestans reveals abundant phosphorylation of members of the CRN effector family. J. Proteome Res. doi:10.1021/pr4009095 [DOI] [PubMed] [Google Scholar]
- 35. Huet J. C., Pernollet J. C. (1989) Amino acid sequence of cinnamomin, a new member of the elicitin family, and its comparison to cryptogein and capsicein. FEBS Lett. 257, 302–306 [DOI] [PubMed] [Google Scholar]
- 36. Huet J. C., Pernollet J. C. (1993) Sequences of acidic and basic elicitin isoforms secreted by Phytophthora megasperma megasperma. Phytochemistry. 33, 797–805 [DOI] [PubMed] [Google Scholar]
- 37. Kamoun S., van West P., de Jong A. J., de Groot K. E., Vleeshouwers V. G., Govers F. (1997) A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol. Plant-Microbe Interact. 10, 13–20 [DOI] [PubMed] [Google Scholar]
- 38. van West P., Kamoun S., van 't Klooster J. W., Govers F. (1999) Internuclear gene silencing in Phytophthora infestans. Mol. Cell. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 39. Mateos F. V., Rickauer M., Esquerre-Tugaye M. T. (1997) Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant-Microbe Interact. 10, 1045–1053 [DOI] [PubMed] [Google Scholar]
- 40. Rose J. K., Ham K. S., Darvill A. G., Albersheim P. (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell 14, 1329–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Damasceno C. M., Bishop J. G., Ripoll D. R., Win J., Kamoun S., Rose J. K. (2008) Structure of the glucanase inhibitor protein (GIP) family for Phytophthora species suggest co-evolution with plant endo-B-1,3-glucanases. Mol. Plant-Microbe Interact. 21, 820–830 [DOI] [PubMed] [Google Scholar]
- 42. Grenville-Briggs L. J., Avrova A. O., Hay R. J., Bruce C. R., Whisson S. C., van West P. (2010) Identification of appressorial and mycelial cell wall proteins and a survey of the membrane proteome of Phytophthora infestans. Fungal Biol. 114, 702–723 [DOI] [PubMed] [Google Scholar]
- 43. Meijer H. J.., Van de Vondervoort P. J., Yuan Yin Q. Y., De Koster C. G., Klis F. M., Govers F., de Groot P. W. (2006) Identification of cell wall-associated proteins from Phytophthora ramorum. Mol. Plant-Microbe Interact. 19, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 44. Meijer H. J. G., Hassen H. H., Govers F. (2011) Phytophthora infestans has a plethora of phospholipase D enzymes including a subclass that has extracellular activity. PLoS One 6, e17767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caten C. E., Jinks J. L. (1968) Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can. J. Bot. 46, 329–348 [Google Scholar]
- 46. Erwin D. C., Ribeiro O. K. (1996) Phytophthora diseases worldwide. American Phytopathological Society, St. Paul, MN [Google Scholar]
- 47. Gvozdeva E. L., Ievleva E. V., Gerasimova N. G., Ozeretskovskaya O. L., Valueva T. A. (2004) Exoproteinases of the oomycete Phytophthora infestans. Applied Biochemistry and Microbiology, 40, 165–169 [PubMed] [Google Scholar]
- 48. Munnik T., Musgrave A., Vrije d. T. (1994) Rapid turnover of polyphosphoinositides in carnation flower petals. Planta. 193, 89–98 [Google Scholar]
- 49. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 50. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 51. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., King N., Stein S. E., Aebersold R. (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 [DOI] [PubMed] [Google Scholar]
- 52. Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B. (2012) PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conesa A., Gotz S. (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 55. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 56. Sonnhammer E. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings / . International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology 6, 175–182 [PubMed] [Google Scholar]
- 57. Van Dijck P. (2009) Nutrient sensing G protein-coupled receptors: interesting targets for antifungals? Medical mycology : official publication of the International Society for Human and Animal Mycology. 47, 671–680 [DOI] [PubMed] [Google Scholar]
- 58. Hua C., Meijer H. J., de Keijzer J., Zhao W., Wang Y., Govers F. (2013) GK4, a G-protein-coupled receptor with a phosphatidylinositolphosphate kinase domain in Phytophthora infestans, is involved in sporangia development and virulence. Mol. Microbiol. 88, 352–370 [DOI] [PubMed] [Google Scholar]
- 59. Yang X., Zhao W., Hua C., Zheng X., Jing M., Li D., Govers F., Meijer H. J., Wang Y. (2013) Chemotaxis and oospore formation in Phytophthora sojae are controlled by G-protein-coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol. Microbiol. 88, 382–394 [DOI] [PubMed] [Google Scholar]
- 60. Rose J. K., Lee S. J. (2010) Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 153, 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Avilan L., Gualdron-Lopez M., Quinones W., Gonzalez-Gonzalez L., Hannaert V., Michels P. A., Concepcion J. L. (2011) Enolase: a key player in the metabolism and a probable virulence factor of trypanosomatid parasites-perspectives for its use as a therapeutic target. Enzyme Res. 2011:932549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chaffin W. L. (2008) Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72, 495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leferink N. G., Heuts D. P., Fraaije M. W., van Berkel W. J. (2008) The growing VAO flavoprotein family. Arch. Biochem. Biophys. 474, 292–301 [DOI] [PubMed] [Google Scholar]
- 64. Seidl M. F., Van den Ackerveken G., Govers F., Snel B. (2011) A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 155, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giraldo M. C., Dagdas Y. F., Gupta Y. K., Mentlak T. A., Yi M., Martinez-Rocha A. L., Saitoh H., Terauchi R., Talbot N. J., Valent B. (2013) Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4, doi:10.1038/ncomms2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huitema E., Vleeshouwers V. G., Cakir C., Kamoun S., Govers F. (2005) Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant-Microbe Interact. 18, 183–193 [DOI] [PubMed] [Google Scholar]
- 67. Parr-Sturgess C. A., Rushton D. J., Parkin E. T. (2010) Ectodomain shedding of the Notch ligand Jagged1 is mediated by ADAM17, but is not a lipid-raft-associated event. Biochem. J. 432, 283–294 [DOI] [PubMed] [Google Scholar]
- 68. Szafranski-Schneider E., Swidergall M., Cottier F., Tielker D., Román E., Pla J., Ernst J. F. (2012) Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 8, e1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Puri S., Kumar R., Chadha S., Tati S., Conti H. R., Hube B., Cullen P. J., Edgerton M. (2012) Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One 7, e46020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Koppers A. J., Reddy T., O'Bryan M. K. (2011) The role of cysteine-rich secretory proteins in male fertility. Asian J. Androl. 13, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Alexander D., Goodman R. M., Gut-Rella M., Glascock C., Weymann K., Friedrich L., Maddox D., Ahl-Goy P., Luntz T., Ward E., Ryals J. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. U.S.A. 90, 7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Niderman T., Genetet I., Bruyere T., Gees R., Stintzi A., Legrand M., Fritig B., Mosinger E. (1995) Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 108, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rauscher M., Adam A. L., Wirtz S., Guggenheim R., Mendgen K., Deising H. B. (1999) PR-1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broad bean. Plant J. 19, 625–633 [DOI] [PubMed] [Google Scholar]
- 74. Munnik T., de Vrije T., Irvine R. F., Musgrave A. (1996) Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271, 15708–15715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.