Abstract
The Pseudomonas sp. P-1 strain, isolated from heavily petroleum hydrocarbon-contaminated soil, was investigated for its capability to degrade hydrocarbons and produce a biosurfactant. The strain degraded crude oil, fractions A5 and P3 of crude oil, and hexadecane (27, 39, 27 and 13 % of hydrocarbons added to culture medium were degraded, respectively) but had no ability to degrade phenanthrene. Additionally, the presence of gene-encoding enzymes responsible for the degradation of alkanes and naphthalene in the genome of the P-1 strain was reported. Positive results of blood agar and methylene blue agar tests, as well as the presence of gene rhl, involved in the biosynthesis of rhamnolipid, confirmed the ability of P-1 for synthesis of glycolipid biosurfactant. 1H and 13C nuclear magnetic resonance, Fourier transform infrared spectrum and mass spectrum analyses indicated that the extracted biosurfactant was affiliated with rhamnolipid. The results of this study indicate that the P-1 and/or biosurfactant produced by this strain have the potential to be used in bioremediation of hydrocarbon-contaminated soils.
Keywords: Hydrocarbon degradation, Biosurfactant production, Pseudomonas sp. P-1, Crude oil, Rhamnolipid, rhl gene
Introduction
Contamination of soil by petroleum hydrocarbons and their derivatives is a serious environmental problem all over the world. Among a variety of the remediation methods, bioaugmentation, involving the introduction of microorganisms into contaminated water or soil, is widely used for cleaning up environments polluted with organic compounds (Chang et al. 2011; Federici et al. 2012), co-contaminated with hydrocarbons and heavy metals (Alisi et al. 2009), or polluted with heavy metals (Płociniczak et al. 2013).
In bioaugmentation of petroleum-contaminated soil, hydrocarbon-degrading bacteria are applied to polluted environments in order to accelerate the degradation of toxic compounds (Margesin and Schinner 2001). One of the main problems that limit the effectiveness of this process is very low solubility and high hydrophobicity of oil pollutants. These compounds strongly bind to soil particles, and thereby they are poorly available for bacterial cells (Johnsen et al. 2005). Solution to this problem is the use of biosurfactants or biosurfactant-producing microorganisms. Biosurfactants are a structurally diverse group of surface-active substances produced by microorganisms that exhibit the ability to reduce surface and interfacial tension. They may enhance hydrocarbon bioremediation by two mechanisms. The first includes the increase the substrate availability for microorganisms, while the second involves interaction with the cell surface, which increases the hydrophobicity of the surface, allowing hydrophobic substrates to associate more easily with bacterial cells (Pacwa-Płociniczak et al. 2011).
The application of biosurfactants for large scale is limited by the high costs of their production. These costs may be reduced by the use of agro-industrial wastes to culture biosurfactant-producing bacteria (Płaza et al. 2011). Therefore, in our preliminary studies, the ability of P-1 to grow on various agro-industrial substrates has been tested. Among 21 tested media, molasses appeared to be the most suitable medium for culturing these bacteria (Pacwa-Płociniczak et al, Evaluation the usefulness of Pseudomonas aeruginosa P-1 strain for bioremediation of petroleum-polluted soils, Personal Communication).
The aims of the study were to (1) characterize the ability of the Pseudomonas sp. P-1 strain to degrade hydrocarbons and produce biosurfactants when grown on molasses, and (2) to characterize the structure of the biosurfactant produced by P-1.
Materials and methods
Bacterial strain—isolation and detection of biosurfactant production
Bacterial strain P-1 used in this study was isolated using an enrichment technique from petroleum-polluted soil taken from the area around of a 100-year-old oil refinery in Czechowice-Dziedzice, Upper Silesia, Poland, as described by Berry et al. (2006) and Płaza et al. (2006). Expedite methods were used to detect the ability of P-1 for biosurfactant production. Hemolytic activity was carried out as described by Carrillo et al. (1996). Strain P-1 was streaked onto blood agar plates containing 40 g of blood agar base (Becton Dickinson, Sparks, MD) and 50 ml of sheep blood (BIOMED-LUBLIN) per litre. Plates were incubated for 48 h at 28 °C. The plates were inspected for zones of clearing around the colonies, indicative of biosurfactant production. The synthesis of extracellular glycolipid biosurfactant was detected using the Siegmund and Wagner (1991) technique. A spot of culture grown on LB medium was placed on a methylene blue agar plate (0.2 g of cetyltrimethylammonium bromide (CTAB), 0.005 g of methylene blue, 20 g of glucose, 0.7 g of KH2PO4, 0.9 g of Na2HPO4, 2 g of NaNO3, 0.4 g of MgSO4 × 7H2O, 0.1 of CaCl2 × 2H2O, 20 g of agar and 2 ml of a trace elements solution containing 2 g of FeSO4 × 7H2O, 1.5 g of MnSO4 × H2O and 0.6 g of (NH4)6Mo7O24 × 4H2O per litre of deionized water). After 48 h of incubation at 28 °C, the plate was observed for the formation of a dark blue halo around the culture spot, indicating the formation of an insoluble ion pair between the anionic glycolipid biosurfactant and the cationic CTAB–methylene blue agar complex.
Identification of isolate
Strain P-1 was identified on the basis of 16S rRNA gene sequence analysis. For 16S rRNA gene amplification, the universal bacterial primers 8 F (5′ AGTTTGATCATCGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′) targeting fragment size 1,484 bp were used (Lonergan et al. 1996). The PCR was run with a mixture containing 1 μl of the DNA template, 0.2 μM of each primer, 10× reaction buffer (Fermentas), 1.5 mM of MgCl2 (Fermentas), 200 μM of dNTP and 1 U of DreamTaq DNA polymerase (Fermentas) in a C1000 Touch™ Thermal Cycler (BioRad). PCR amplification was performed at 94 °C for 5 min, 3 cycles at 94 °C for 45 s, 57 °C for 30 s, 72 °C for 120 s; 3 cycles at 94 °C for 45 s, 56 °C for 30 s, 72 °C for 120 s; 3 cycles at 94 °C for 45 s, 55 °C for 30 s, 72 °C for 120 s; 26 cycles at 94 °C for 45 s, 53 °C for 30 s, 72 °C for 120 s; and a final elongation cycle at 72 °C for 5 min. Gene sequencing was performed by using the Big Dye® Terminator Cycle Sequencing Kit (Applied Biosystem) and the AbiPrism®3100 Genetic Analyzer. The obtained sequences were compared to known 16S rRNA gene sequences using the BLAST server at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). DNA sequences were aligned using CLUSTAL W. Phylogenetic analyses were performed by the neighbour-joining (NJ) method testing the support for the phylogeny with a bootstrap analysis based on 1,000 replicates using the MEGA ver. 6.0.
Detection of genes encoding enzymes involved in rhamnolipid synthesis
The primers rhlABF 5′ CAG GCC GAT GAA GGG AAA TA 3′ and rhlABR 5′ AGG ACG ACG AGG TGG AAA TC 3′ (Kumar et al. 2008) targeting fragment size 777 bp were used to detect potential rhamnolipid synthesis by the P-1 strain. The PCR was run with a mixture containing 1 μl of the DNA template, 0.2 μM of each primer, 10× reaction buffer (Fermentas), 1.5 mM of MgCl2 (Fermentas), 200 μM of dNTP and 1 U of Taq DNA polymerase (Fermentas) in a C1000 Touch™ Thermal Cycler (BioRad). PCR amplification was performed at 95 °C for 5 min, and 30 cycles of 30 s at 95 °C, followed by annealing for 1 min at 50 °C and an extension step of 2 min at 72 °C and a final extension step of 10 min at 72 °C. The experiment included a control reaction mixture without added DNA.
Detection of gene encoding enzymes involved in hydrocarbon degradation
The primers alkBfd 5′ AAC TAC MTC GAR CAY TAC GG 3′ and alkBRd 5′ TGA MGA TGT GGT YRC TGT TCC 3′ (where M = AC, R = AG and Y = CT) (Powell et al. 2006) and nahAc-7 F 5′ ACT TGG TTC CGG AGT TGA TG 3′ and nahAc-7R 5′ CAG GTC AGC ATG CTG TTG TT 3′ (Park and Crowley 2006) targeting fragments 100 and 136 bp were used to detect alkane monooxygenase and naphthalene dioxygenase genes in the P-1 strain. The PCR was run with a mixture containing 1 μl of the DNA template, 0.2 μM of each primer, 10× reaction buffer (Fermentas), 1.5 mM of MgCl2 (Fermentas), 200 μM of dNTP and 1 U of DreamTaq DNA polymerase (Fermentas) in a C1000 Touch™ Thermal Cycler (BioRad). PCR amplification was performed at 95 °C for 5 min, and 35 cycles of 1 min at 94 °C followed by annealing for 30 s at 49 °C and an extension step of 45 s at 72 °C for the alkB gene and 37 cycles of 20 s at 94 °C followed by annealing for 15 s at 56 °C and an extension step of 15 s at 72 °C for the nahAc gene, and then a final extension step of 5 min at 72 °C. The experiment included a positive control with DNA isolated from the strain with high ability to degrade aromatic hydrocarbons (Arthrobacter sp. F1A strain, Markowicz and Piotrowska-Seget, Isolation and characterization of bacterial strains capable of degradation of phenanthrene, Personal Communication) for the nahAc gene and a control reaction mixture without added DNA for both genes.
Growth of isolates on hydrocarbons and determination of hydrocarbon concentration
Replicate flasks containing 100 ml of M9 minimal salt medium (Viramontes-Ramos et al. 2010) with 100 μl of crude oil, its fraction A5, P3 and hexadecane and 20 mg of phenanthrene were prepared. A5 and P3 are distillation fractions of crude oil containing components of diesel oils and light fuel oils and components of high fuel oils (raw material to mazout), respectively (Płaza et al. 2008). The flasks were inoculated with 1 ml of the P-1 strain suspended in sterile saline. Non-inoculated flasks were prepared as controls. The cultures were grown aerobically at 28 °C for 28 days with constant shaking (120 rpm). After that, the residual hydrocarbon concentrations were determined. Crude oil and its fraction A5 and P3 were extracted from bacterial culture using the liquid–liquid extraction technique with hexane as an extraction solvent. The extract was cleaned up on a column filled with Florisil sorbent and evaporated to a volume of 1 ml. Mineral oil (in the range C10–C40) was determined by gas chromatography coupled with a flame ionization detector (GC/FID), according to the accredited procedure PB-16 (2009). Hexadecane concentration was determined according to the method described by Wypych and Mańko (2002), in which HS-SPME-GC/MS was used. The analysis was carried out with a Star 3400 Cx gas chromatograph (equipped with a 63Ni Electron Capture Detector); it was coupled to a mass spectrometer, Saturn 3 and Autosampler 8200 Cx (Varian) with 10 ml autosampler vials. The chromatographic column with the phase DB624 and length 30 m × 0.32 mm ID (1.8 μm film thickness) was used. Helium was used as the carrier gas: purity 99.999 %, flow capacity of 1.0 ml min−1 (in a temperature of 35 °C). The gas chromatographic conditions were as follows: the oven temperature was held at 40 °C for 10 min, then increased to 250 °C at 10 °C min−1, and finally increased at 5 °C min−1 to 270 °C. The total analysis time was 45 min. The injector temperature was 250 °C. The MS operating conditions were the following: the mass range scanned was 30–250 amu at 1 s/scan, the temperature of the ion trap was 170 °C, multiplier voltage was 2,700 V and ionization energy was 70 eV (electron impact mode EI). The transfer line temperature was 250 °C. The temperature of the ECD detector (63Ni) was 300 °C. Three types of SPME fibre were used (Supelco, Bellefonte) in an autosampler set of the following stationary phases: 100 μm polydimethylosiloxane, 7 μm polydimethylosiloxane and 85 μm polyacrylate. The SPME fibre was conditioned prior to use in order to reduce bleeding by heating in a split/splitless injector with an open purger, in a helium stream: fibre 100 μm PDMS at a temperature of 250 °C (for 1 h), fibre 7 μm PDMS at a temperature of 320 °C (for 2 h) and fibre 85 μm polyacrylate at a temperature of 300 °C (for 2 h). Phenanthrene was extracted using the solid phase extraction (spe) technique with a mixture of octadecyl and amino phase as a sorbent and eluted with dichloromethane. Then, the eluate was evaporated to a volume of 1 ml. Phenanthrene was determined by high-performance liquid chromatography coupled with a fluorescence detector (HPLC/FLD), according to accredited procedure PB-06 (2010). For each measurement, three replicates were used.
Determination of rhamnolipid production and yield
The drop-collapse technique was carried out in the polystyrene lid of a 96-microwell plate (Biolog, Harward, CA, USA), as described by Płaza et al. (2006). One hundred microlitres of each supernatant was added to the wells of a 96-well microtitre plate lid, then 2 μl of crude oil was added to the surface of the culture supernatant, and the flattening of the oil drop indicating a positive result of the test was checked.
The oil-spreading technique was carried out according to Morikawa et al. (2000) and Youssef et al. (2004). Fifty millilitres of distilled water was added to Petri dishes, followed by the addition of 100 μl of crude oil to the surface of the water. Then, 10 μl of each culture supernatant was put onto the crude oil surface. The diameter of the clear zone on the oil surface was observed.
The emulsifying activity was determined using a modification of the method described by Cooper et al. (1987). To 2 ml of the bacterial cultures in a screw-cap tube, 3 ml of hydrocarbons (benzene, hexadecane, cyclohexane, diesel oil and A3 fraction of diesel oil) was added and vortexed at high speed for 2 min. The emulsion stability was determined after 24 h. The emulsification index (E24) was calculated as the percentage height of the emulsion layer to the total height of the liquid column.
Surface (ST) tension of the culture supernatants was determined with a Kruss Processor Tensiometer (model K12 Kruss, Germany) using the plate method. To increase the accuracy an average of triplicates was used for the study. All the assays were performed in triplicate with distilled water as the negative control and sodium dodecyl sulphate (SDS) as the positive control.
Rhamnolipid was quantified from the cell-free molasses culture as rhamnose (g l−1) using the phenol–sulphuric method (Dubois et al. 1956). One millilitre of the culture supernatant was mixed with 0.5 ml of 80 % phenol and 2.5 ml of concentrated sulphuric acid. After 10 min of incubation at room temperature, the absorbance was measured at 490 nm, and the rhamnose concentration was calculated using a standard curve prepared using different concentrations (0–4,000 mg l−1) of rhamnose. Rhamnolipid values were determined by multiplying rhamnose values by a coefficient of 3.4 obtained from the correlation (y = (0.0139x − 0.0058) × 0.68) of pure rhamnolipids/rhamnose (Benincasa et al. 2002). The experiment was carried out in triplicate.
Extraction and structural analysis of biosurfactant
To get a biosurfactant for chemical analysis, the P-1 strain was grown in a molasses medium on a rotatory shaker (120 rpm) at 28 °C for 96 h. Then, the culture was centrifuged at 12,000 rpm for 30 min at 4 °C to remove the cells, and the biosurfactant was recovered from the supernatant. Its pH was adjusted to 2.0 with 1 N HCl and left overnight at 4 °C. The cloudy supernatant was twice extracted with an equal volume of chloroform/methanol (2:1) solution in a separating funnel. The pooled organic phase was evaporated under vacuum (Buchi, Germany). Then, the obtained brownish oily residue was characterized with spectroscopic techniques.
Electrospray ionization (ESI) mass spectra of analysed product were recorded on the LC-MS system consisting of high-performance liquid chromatography (Ultimate 3000, Dionex) with a diode-array detector and a high-resolution micrOTOF-QII (Bruker Daltonics, Germany) time-of-flight mass spectrometer. The chromatographic separation was performed on a Kinetex C18, 2.6 μm, 100 × 4.6 mm column. An acetonitrile/water gradient was used, starting with 25 % acetonitrile for 5 min, followed by a ramp of 25–100 % acetonitrile for 30 min. The ESI mass spectrum in the negative ion mode was acquired using a capillary voltage of −3.5 kV, and desolvatation gas (nitrogen) was heated to 150 °C. Full scan data were obtained by scanning from range m/z 50 to 1,000. The analysed sample of brownish oily product before LC-MS analysis was dissolved in methanol/water (1:1, v/v) and filtered (0.22 μm).
Infrared absorption spectra were recorded on an FTIR Nicolet Nexus spectrophotometer using KBr discs prepared as follows: 2 mg of brownish oily biosurfactant was ground with 600 mg of KBr and pressed with 15,000 kg for 30 s to obtain translucent pellets. All measurements consisted of 20 scans with a spectral resolution 2 cm−1.
The assignments of the glycolipid signals were carried out using 1H-NMR and 13C-NMR spectra with Bruker Ultrashield 400 MHz. The NMR spectra were determined in deuterated DMSO (dimethyl sulphate-d6), using tetramethyl silane (TMS) as the internal standard.
Results
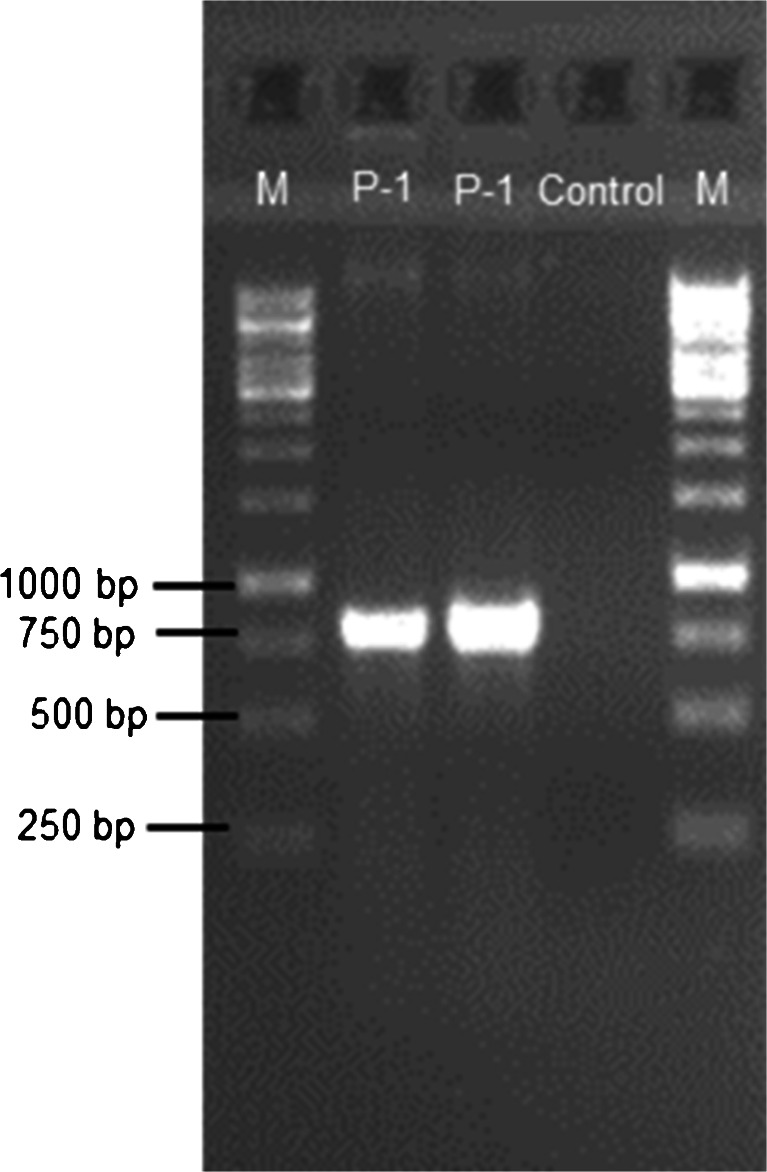
The application of an enrichment technique for the isolation of bacteria from petroleum-polluted soil from the area of 100-year-old oil refinery in Czechowice-Dziedzice (Poland) enabled us to find several bacterial strains with the ability to degrade hydrocarbons and produce biosurfactants. However, only one strain (P-1) demonstrated a positive reaction for both blood agar and methylene blue agar tests, suggesting the potential ability of the isolate to synthesize glycolipid biosurfactant. This ability was confirmed by the polymerase chain reaction of the rhl gene. When the primers rhlABF and rhlABR were used, a single DNA fragment of the expected size (777 bp) was amplified in the P-1 strain, and no amplification was observed in the control without DNA (Fig. 1).
Fig. 1.
Agarose gel electrophoresis of PCR products of rhamnosyl transferase gene; M—1 kb DNA ladder; P-1—Pseudomonas sp. P-1 strain; Control—sample without DNA (1 % agarose gel)
The strain was identified using 16S rDNA gene sequence analysis as Pseudomonas sp. and designed as the P-1 strain. The phylogenetic analysis showed that the 16S rDNA sequence of P-1 had 99 % sequence similarity with strains Pseudomonas aeruginosa DSM 50071 and P. aeruginosa PAO1 (Fig. 2).
Fig. 2.
Neighbour-joining phylogenetic tree of bacteria based on 16S rRNA gene sequence comparisons. Bootstrap values are indicated at the branches from 1,000 replications. GenBank accession numbers are given in parentheses
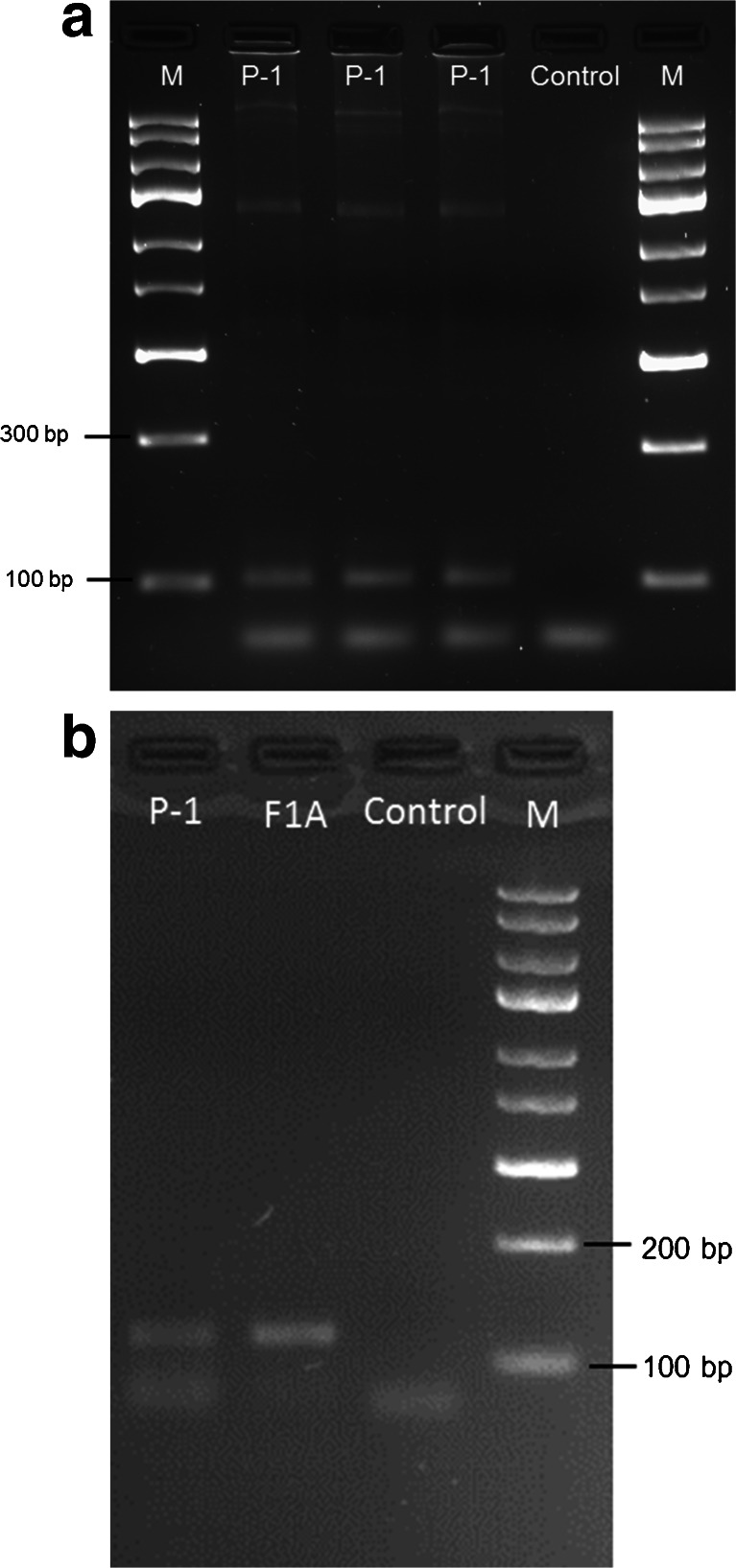
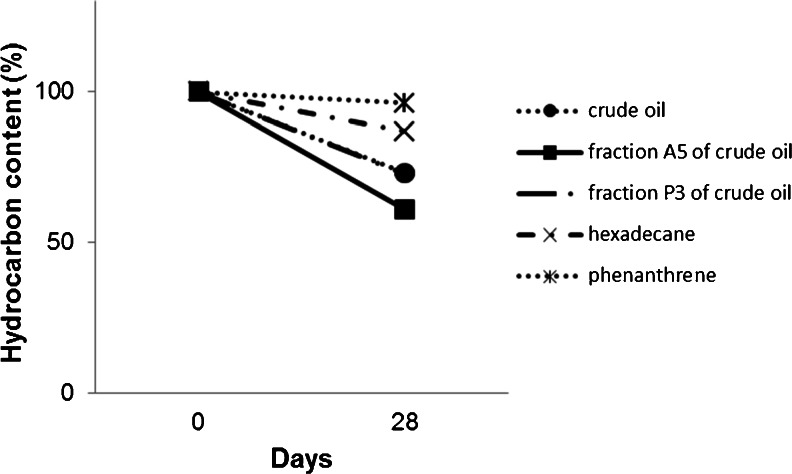
The use of the polymerase chain reaction and the sets of primers alkB and nahAc-7 confirmed the presence of genes involved in the degradation of alkanes and naphthalene in the genome of the P-1 strain. In the gel (Fig. 3a, b), both DNA products of the expected size, 100 and 136 bp, respectively, were observed. The ability of P-1 to degrade crude oil, fractions A5 and P3 of crude oil, hexadecane and phenanthrene was determined by measuring the amount of residual hydrocarbons in the culture medium after 28 days of culture. Among the analyzed hydrocarbons crude oil, fractions A5 and P3 of crude oil and hexadecane were degraded with various efficiency, and no degradation of hydrocarbon was observed in the medium supplemented with phenanthrene (96 % of phenanthrene remained in the culture) as a sole carbon source. The highest degradation efficiency was observed in the culture supplemented with fraction A5 of crude oil (39 % of hydrocarbon was degraded at the end of the experiment). In the media supplemented with crude oil and fraction P3 of crude oil, 27 % of hydrocarbons were degraded and only 13 % of hexadecane was degraded in the medium with this hydrocarbon as the only carbon source (Fig. 4).
Fig. 3.
Agarose gel electrophoresis of PCR products of a alkane monooxygenase gene; M—express DNA ladder; P-1—Pseudomonas sp. P-1 strain; Control—sample without DNA (3 % agarose gel); b naphthalene dioxygenase gene; M—100 bp DNA ladder; P-1—Pseudomonas sp. P-1 strain; F1A—reference strain with high ability to degrade aromatic hydrocarbons (Arthrobacter sp., Markowicz and Piotrowska-Seget, Isolation and characterization of bacterial strains capable of degradation of phenanthrene, Personal Communication); Control—sample without DNA (2 % agarose gel)
Fig. 4.
Degradation of hydrocarbons by P-1 strain
In order to check if P-1 produces biosurfactants on molasses, the surface-active properties of cell-free supernatant were determined (Table 1). The surface tension of the supernatant was reduced to 26.67 mN m−1, and the flat shape of the oil drop added to the surface of the culture supernatant, indicating a positive result in the drop-collapse method, was observed. Moreover, the clear zones generated in the oil-spreading test and the high emulsification activities of the strain P-1 were recorded. The concentration of the produced rhamnolipid was high and reached the value of 30.23 g l−1 (Table 1).
Table 1.
Surface-active properties of molasses culture and concentration of rhamnolipid produced by P-1
| Surface tension (mN × m−1 ) | Drop-collapse | Oil-spreading (mm) |
MBA | Emulsification index (%) | Rhamnolipid yield (g l−1) |
||
|---|---|---|---|---|---|---|---|
| C | D | H | |||||
| 26.67 ± 0.35 | + | 47.33 ± 1.15 | + | 68.8 ± 4.83 | 67.63 ± 2.26 | 70.1 ± 0.39 | 30.23 ± 2.97 |
± Standard deviation of three independent experiments
MBA methylene blue agar, C cyclohexane, D diesel oil, H hexadecane
The molecular composition of the biosurfactant produced by Pseudomonas sp. P-1 was evaluated by FT-IR. The obtained infrared spectrum disclosed a broad stretching peak at 3,370 cm−1, indicating the presence of –OH bonds (free hydroxyl groups of rhamnose rings). Absorption around 2,929 and 2,858 cm−1 is assigned to the symmetric stretch (–CH) of –CH2 and –CH3 groups of aliphatic chains. The absorption peak located at 1,734 cm−1 indicates the presence of ester carbonyl groups (–C = O bond) in biosurfactants. The ester carbonyl group from the bands at 1,128 cm−1 and C–O–C vibrations at 1,053 cm−1 (rhamnose rings) were also proved. Protein-related bands, the –C = O amide I (1,659 cm−1) and –NH/–C = O combination of the amide II bands (1,537 cm−1), were observed. It might be possible that the additional bands at 1,659 and 1,537 cm−1 resulted from polypeptide contamination resulting from cell debris co-precipitated with the biosurfactant during the extraction process. The absorption peak at 994 cm−1 may indicate the presence of polysaccharide or polysaccharide-like substances in the biosurfactants (Fig. 5)
Fig 5.
FT-IR spectrum of the crude biosurfactant produced by P-1 strain cultivated in molasses medium
The chemical structure of the rhamnolipids present in the analysed mixture of biosurfactants was confirmed by nuclear magnetic resonance spectroscopy. The obtained 1H NMR spectra strongly indicated that the brownish oily substance produced by P-1 was a mixture of glycolipids, and not another group of biosurfactants (Fig. 6a). In the 13C-NMR spectrum lipid, signals of CH2 from δ 21.9 to 31.1 and CH3 at δ 13.8, and ester and carboxylic signals at δ 171.5 and δ 176.95 were observed (Fig. 6b).
Fig. 6.
NMR spectra of crude biosurfactant produced by P-1 strain cultivated in molasses medium: a 1H NMR; b 13C NMR
Up to 11 rhamnolipid homologues were determined by LC-MS analysis in the mixture produced by the P-1 strain. Table 2 presents a list of RL congeners found in the analysed sample of crude biosurfactant. Negative ESI-MS spectra showed the main pseudo-molecular ions at m/z 677 and m/z 622, which corresponds to the deprotonated molecules [M-H]¯ of the dirhamnolipids Rha-Rha-C10-C12 or Rha-Rha-C12-C10, and Rha-Rha-C8-C10, respectively. Minor ions at m/z 649 (Rha-Rha- C10-C10), m/z 475 (Rha-C8-C10 or Rha-C10-C8) and m/z 503 (Rha-C10-C10) were observed in the analysed mixture of biosurfactants. RL with some unsaturated fatty acid, Rha-C10-C12:1/Rha-C12:1-C10 (m/z 529), Rha-Rha-C10-C12:1 (m/z 675) and Rha-C8:2 (m/z 301) were also present in the mixture. In general, the predominance of dirhamnolipids compared to monorhamnolipids was observed. The most frequent were di-rhamno-di-lipidic (four detected) and mono-rhamno-di-lipidic (three detected). The relative abundance of [M-H]− mono-rhamno-mono-lipidic, mono-rhamno-di-lipidic and di-rhamno-di-lipidic structures were 0.8, 31.8 and 67.4 %, respectively.
Table 2.
The chemical composition of rhamnolipid mixture produced by P. aeruginosa P-1 culture identified by LC-ESI-MS
| Rhamnolipid congener | Molecular formula | Molecular weight [g mol−1] | Pseudomolecular ion (m/z) | Ion fragments m/z | Retention time [min] | Relative abundance [%] |
|---|---|---|---|---|---|---|
| Rha-C8:2 | C14H22O7 | 302 | 301 | – | 9.2 | 0.8 |
| Rha-C8-C10/ Rha-C10-C8 * | C24H44O9 | 476 | 475 | 311; 169 | 18.5 | 14.1 |
| Rha- C10-C10 | C24H48O9 | 504 | 503 | 339; 333; 169;163;119;103 | 12.1 | 6.8 |
| Rha-C10-C12:1/ Rha-C12:1-C10 | C28H50O9 | 530 | 529 | 365; 333; 169; 163;119;103 | 16.1 | 10.9 |
| Rha-Rha-C8-C10 | C30H54O13 | 622 | 621 | 452; 311;169 | 16.9 | 21.3 |
| Rha-Rha-C10-C10 | C32H58O13 | 650 | 649 | 479; 339; 311; 246; 169 | 11.5 | 15.7 |
| Rha-Rha-C10-C12:1 | C34H60O13 | 676 | 675 | 479; 311; 195; 169; 119; 103 | 10.4 | 5.6 |
| Rha-Rha-C10-C12/ Rha-Rha-C12-C10 | C34H62O13 | 678 | 677 | 507; 479; 311; 205;197; 169 | 15.2 | 24.8 |
Negative ESI-MS, [M-H]− = pseudomolecular ions
Discussion
In the present study of the characteristics of the Pseudomonas sp. P-1 strain, a potential tool for the enhanced bioremediation of petroleum-contaminated soil was described. Members of the genus Pseudomonas, due to the presence of a complex enzymatic system, show a wide variety of metabolic and physiological properties. They are also the most predominant group of microorganisms that degrade xenobiotic compounds. The phylogenetic analysis of P-1 revealed that close relatives of this strain are strains belonging to P. aeruginosa. It has been reported that various bacteria from genus Pseudomonas, including P. aeruginosa strains, inhabit oil-contaminated soils and can break down more than 100 different organic compounds (Glick et al. 1994; Hong et al. 2005; Saikia et al. 2012). They are the best known bacteria capable of utilizing a number of aliphatic and aromatic hydrocarbons as carbon and energy sources (Das and Chandran 2011; Kadali et al. 2012; Puškárová et al. 2013). Strain Pseudomonas sp. P-1 was characterized by its high removal of hydrocarbons contained in crude oil and its fractions A5 and P3. It exhibited high ability to utilize light boiling fraction (A5) of crude oil and had similar potential to degrade crude oil and its high boiling fraction (P3). The reason of lower degradability of the P3 was its molecular size and physico-chemical parameters (especially water solubility). The ability of bacteria to degrade hydrocarbons from fraction A5 with higher efficiency as compared to fraction P3 was reported by Płaza et al. (2008). The capability of other Pseudomonas strains to remove crude oil, kerosene and lubricant oil (Silva et al. 2006), and crude, diesel and engine oils (Obayori et al. 2009) was reported earlier for P. aeruginosa AT18 and Pseudomonas sp. LP1 strains, respectively. P-1 also exhibited the ability to degrade hexadecane, but did not use naphthalene as a carbon source. This was surprising because the presence of genes responsible for naphthalene degradation in the genome of P-1 was proved by the PCR method. This indicates that P-1 was only able to utilize aliphatic hydrocarbons, and the naphthalene dioxygenase nahAc possessed by this strain gene was disabled. The ability of Pseudomonas strains to degrade only one type of hydrocarbon was also observed by Kumar et al. (2006), who found that IR1 was capable of utilizing two-, three- and four-ring PAHs but not hexadecane and octadecane.
Strains belong to the genus Pseudomonas are among the best known biosurfactant producers. Since biological surface-active compounds are considered to be very useful in improving the bioavailability of hydrocarbon pollutants in soil, the ability of biosurfactant production in combination with the capacity to degrade hydrocarbons makes pseudomonads one of the most useful tools in bioremediation of petroleum-polluted soils. The ability of P-1 to synthesize biosurfactants was tested on a blood agar plate. The obtained positive result for the hemolytic activity of P-1 was insufficient to confirm about biosurfactant production by this strain. Since it is known that pseudomonads possess, apart from biosurfactants, β-hemolysins, which also cause clearing of the blood agar, the application of additional tests is required. The blue agar plate method, specially developed for the detection of glycolipid production such as rhamnolipids by Pseudomonas sp., was proposed by Siegmund and Wagner (1991). A new approach for the determination of the potential ability of bacterial strains for biosurfactant production is the detection of genes coding enzymes involved in their biosynthesis (Satpute et al. 2010). The polymerase chain reaction screening method is now a widely used technique. This approach was used for the detection of Bacillus strains with the potential to produce lipopeptide biosurfactants (Hsieh et al. 2004; Tapi et al. 2010), and is becoming an increasingly employed approach in the search for biosurfactant-producing pseudomonads. The use of methylene blue indicator plates and PCR of rhamnosyl transferase gene (rhlAB) confirmed the production of anionic glycolipid biosurfactant (rhamnolipid) by strain P-1. The usefulness of the CTAB–methylene blue agar test for the screening of a bacterial rhamnolipid producer was reported by Gunther IV et al. (2005). Kumar et al. (2008) used both of these methods to indicate rhamnolipid synthesis by P. aeruginosa DHT2. Similarly, Shoeb et al. (2012) applied the detection of the rhlAB gene through PCR for establishing the ability of eight bacterial strains from the genus Pseudomonas to produce the rhamnolipid type of biosurfactant.
Interest in biosurfactant applications in many industries and environmental protection has recently significantly increased. However, the success of their production depends on the increase of yield, the development of economical engineering processes and the use of low cost effective renewable agro-industrial substrates for their production. The search for inexpensive raw materials is important in the overall economy of biosurfactant production since they account for 10–30 % of the total cost (Cameotra and Makkar 1998). The main problem in the use of raw substrates is the selection of suitable waste materials with the right balance of nutrients adequate for cell growth and biosurfactant accumulation. It is known that pseudomonads are able to grow on numerous low-cost raw substrates, but often they do not synthesize sufficient amounts of biosurfactant (Makkar et al. 2011). Of 21 low-cost raw materials, only molasses appeared to be good material for P-1 growth and rhamnolipid production with high efficiency (Pacwa-Płociniczak et al, Evaluation the usefulness of Pseudomonas aeruginosa P-1 strain for bioremediation of petroleum-polluted soils, Personal Communication). A few attempts to use agro-industrial waste substrates for biosurfactant production by bacteria from the genus Pseudomonas have been made. It has been demonstrated that rhamnolipids can be produced using distillery and curd whey waste (Dubey and Juwarkar 2001), waste soybean soapstock (Nitschke et al. 2005), glycerol (Silva et al. 2010) oil, or waste coconut frying oil (George and Jayachandran 2012). However, in most cases, the rhamnolipid yield using low-cost substrates does not exceed 10 g l−1. Optimization of rhamnolipid production using waste materials by changing carbon and nitrogen sources, the ratio of carbon to nitrogen source, pH and temperature conditions, studied by Xia et al. (2012), enhanced the rhamnolipid yield to 50.2 g l−1. Nevertheless, all the individual modifications of fermentation conditions make the biosurfactant synthesis more expensive, therefore the application of raw, unchanged materials as substrates for high biosurfactant synthesis is preferred. Here, in our study, we confirmed the usefulness of molasses as a material for the fermentative production of rhamnolipid by the P-1 strain. The obtained rhamnolipid synthesis can be considered as very high (30.23 g of rhamnolipid per litre of molasses medium) compared to other strains for which rhamnolipid yield did not exceed 0.4 g l−1 (Onbasli and Aslim 2009).
A practical measure of the utility of a biosurfactant solution or biosurfactant-producing strain is its ability to emulsify non-aqueous liquids (Ghurye et al. 1994).
The emulsification activity of the cell-free molasses was 68.8, 67.6 and 70.1 % with cyclohexane, diesel oil and hexadecane, respectively. In similar investigations, Pseudomonas biosurfactant producers were able to emulsify hydrocarbons with similar efficiency (Rahman et al. 2002; Kumar et al. 2008; Aparna et al. 2012). The obtained results suggested that the application of this cell-free molasses supernatant in the bioremediation process may enhance the availability of the hydrocarbons in soil.
The spectroscopic analysis of crude biosurfactant produced by P-1 cultivated in molasses medium confirmed that it was a mixture of glycolipids, and not another group of biosurfactants. This mixture consisted of different rhamnolipidic congeners (mono-rhamno- and di-rhamno-di-lipidic). The di-rhamno-di-lipidic congeners showed the highest relative abundance of [M-H]. It has been reported that the composition and predominance of a particular type of congener of rhamnolipids produced by pseudomonads often depend on the type of used carbon source (Déziel et al. 1999). Similar to our results, a higher proportion of dirhamnolipid compared to monorhamnolipid was obtained when Pseudomonas strains were cultivated using soybean oil (Rahman et al. 2002) and modified peptone glucose ammonium salt (PPGAS) containing 1 % of molasses (Aparna et al. 2012). However, the predomination of monorhamnolipids was observed when the Pseudomonas strain was grown in media composed of residues from soybean, corn, babassu, cottonseed and palm oil refineries (Nitschke et al. 2005).
Conclusions
The results of this study indicate that the Pseudomonas sp. P-1 strain has the ability to degrade various hydrocarbons (hexadecane, crude oil and fractions A5 and P3 of crude oil), and is efficient in rhamnolipid production and hydrocarbon emulsification. Our study confirmed that molasses can serve as an efficient and low-cost medium for bacterial culturing and synthesis of rhamnolipid by P-1. This finding has a practical advantage because the use of low-cost raw materials as a substrate for P-1 growth and biosurfactant production may result in wider application of this strain in bioremediation. The features of the P-1 strain make it promising agent for cleaning up environments contaminated with petroleum compounds. Because the potential of bacteria for bioremediation application is highly dependent on biotic and abiotic soil parameters, further studies to check its activity and ability to survive in hydrocarbon-contaminated soil will be carried out.
Acknowledgments
The paper was prepared in connection with the work done under the project no. N N523 418237 financed by the Polish Ministry of Science and Higher Education. The authors are grateful to the factories for kindly supplying the waste products. Author M.P.P. is a scholarship holder within the DoktoRIS project–scholarship program for the innovation of Silesia region supported by the European Community from the European Social Fund.
References
- Alisi C, Musella R, Tasso F, Ubaldi C, Manzo S, Cremisini C, Sprocati AR. Bioremediation of diesel oil in a co-contaminated soil by bioaugmentation with a microbial formula tailored with native strains selected for heavy metals resistance. Sci Total Environ. 2009;407:3024–3032. doi: 10.1016/j.scitotenv.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Aparna A, Srinikethan G, Smitha H. Production and characterization of biosurfactant produced by a novel Pseudomonas sp. 2B. Colloids Surf B. 2012;95:23–29. doi: 10.1016/j.colsurfb.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Benincasa M, Contiero J, Manresa A, Moraes IO. Rhamnolipid production by Pseudomonas aeruginosa LBI growing on soapstock as the sole carbon source. J Food Eng. 2002;54:283–288. doi: 10.1016/S0260-8774(01)00214-X. [DOI] [Google Scholar]
- Berry CJ, Story S, Altman DJ, Upchurch R, Whitman W, Singleton D, Płaza G Brigmon RL (2006). In: Clayton J, Lindner AS (eds) Remediation of hazardous waste in the subsurface. Bridging flask and field. American Chemical Society, Washington, pp 87-93
- Cameotra SS, Makkar RS. Synthesis of biosurfactants in extreme conditions. Appl Microbiol Biotechnol. 1998;50:520–529. doi: 10.1007/s002530051329. [DOI] [PubMed] [Google Scholar]
- Carrillo PG, Mardaraz C, Pitta-Alvarez SJ, Giulietti AM. Isolation and selection of biosurfactant-producing bacteria. World J Microbiol Biotechnol. 1996;12:82–84. doi: 10.1007/BF00327807. [DOI] [PubMed] [Google Scholar]
- Chang LK, Ibrahim D, Omar IC. A laboratory scale bioremediation of Tapis crude oil contaminated soil by bioaugmentation of Acinetobacter baumannii T30C. Afr J Microbiol Res. 2011;5:2609–2615. [Google Scholar]
- Cooper DG, Goldenberg BG. Surface-active agents from two Bacilllus species. Appl Environ Microbiol. 1987;53:224–229. doi: 10.1128/aem.53.2.224-229.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Chandran P. Microbiol degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. 2011;2011:1–13. doi: 10.4061/2011/941810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta. 1999;1440:244–252. doi: 10.1016/S1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- Dubey K, Juwarkar A. Distillery and curd whey wastes as viable alternative sources for biosurfactant production. World J Microbiol Biotechnol. 2001;17:61–69. doi: 10.1023/A:1016606509385. [DOI] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rubero P, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Federici E, Giubilei M, Santi G, Zanaroli G, Negroni A, Fava F, Petruccioli M, D’Annibale A. Bioaugmentation of a historically contaminated soil by polychlorinated biphenyls with Lentinus tigrinus. Microb Cell Factories. 2012;11:1–35. doi: 10.1186/1475-2859-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S, Jayachandran K. Production and characterization of rhamnolipid biosurfactant from waste frying coconut oil using a novel Pseudomonas aeruginosa D. J Appl Microbiol. 2012;114:373–383. doi: 10.1111/jam.12069. [DOI] [PubMed] [Google Scholar]
- Ghurye GL, Vipulanandan C, Wilson RC. A practical approach to biosurfactant production using non-aspetic fermentation of mixed cultures. Biotechnol Bioeng. 1994;44:661–666. doi: 10.1002/bit.260440514. [DOI] [PubMed] [Google Scholar]
- Glick BR, Pasternak JJ, Patten CL. Molecular biotechnology: principles and applications of recombinant DNA. Washington: ASM; 1994. [Google Scholar]
- Gunther NW, IV, Nuñez A, Fett W, Salaiman DKY. Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl Environ Microbiol. 2005;71:2288–2293. doi: 10.1128/AEM.71.5.2288-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Kim J, Choi OK, Cho KS, Ryu HW. Characterization of a diesel-degrading bacterium, Pseudomonas aeruginosa IU5, isolated from oil-contaminated soil in Korea. World J Microbiol Biotechnol. 2005;21:381–384. doi: 10.1007/s11274-004-3630-1. [DOI] [Google Scholar]
- Hsieh FC, Li MC, Lin TC, Kao SS. Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr Microbiol. 2004;49:186–191. doi: 10.1007/s00284-004-4314-7. [DOI] [PubMed] [Google Scholar]
- Johnsen AR, Wick LY, Harms H. Principles of microbial PAH-degradation in soil. Environ Pollut. 2005;133:71–84. doi: 10.1016/j.envpol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Kadali KK, Simons KL, Skuza PP, Moore RB, Ball AS. A complementary approach to identifying and assessing the remediation potential of hydrocarbonoclastic bacteria. J Microbiol Meth. 2012;88:348–355. doi: 10.1016/j.mimet.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Kumar M, Leon V, De Sisto Materano A, Ilzins OA, Galindo-Castro I, Fuenmayor SL. Polycyclic aromatic hydrocarbon degradation by biosurfactant-producing Pseudomonas sp. IR1. Z Naturforsch. 2006;61c:203–212. doi: 10.1515/znc-2006-3-409. [DOI] [PubMed] [Google Scholar]
- Kumar M, León V, De Sisto A, Ilzins OA, Luis L. Biosurfactant production and hydrocarbon-degradation by halotolerant and thermotolerant Pseudomonas sp. World J Microbiol Biotechnol. 2008;24:1047–1057. doi: 10.1007/s11274-007-9574-5. [DOI] [Google Scholar]
- Lonergan DJ, Jenter H, Coates JD, Philips EJ, Schmidt TM, Lovley DR. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkar RS, Cameotra SS, Banat IM. Advances in utilization of renewable substrates for biosurfactant production. AMB Express. 2011;1:1–19. doi: 10.1186/2191-0855-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R, Schinner F. Bioremediation (natural attenuation and biostimulation) of diesel-oil-contaminated soil in an alpine glacier skiing area. Appl Environ Microbiol. 2001;67:3127–3133. doi: 10.1128/AEM.67.7.3127-3133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Hirata Y, Imanaka T. A study on the structure–function relationship of the lipopeptide biosurfactants. Biochim Biophys Acta. 2000;1488:211–218. doi: 10.1016/S1388-1981(00)00124-4. [DOI] [PubMed] [Google Scholar]
- Nitschke M, Costa SGVAO, Haddad R, Gonçalves LAG, Eberlin ME, Contiero J. Oil wastes as unconventional substrates for rhamnolipid biosurfactant production by Pseudomonas aeruginosa LBI. Biotechnol Prog. 2005;21:1562–1566. doi: 10.1021/bp050198x. [DOI] [PubMed] [Google Scholar]
- Obayori OS, Ilori MO, Adebusoye SA, Oyetibo GO, Omotayo AE, Amund OO. Degradation of hydrocarbons and biosurfactant production by Pseudomonas sp. strain LP1. World J Microbiol Biotechnol. 2009;25:1615–1623. doi: 10.1007/s11274-009-0053-z. [DOI] [Google Scholar]
- Onbasli D, Aslim B. Biosurfactant production in sugar beet molasses by some Pseudomonas spp. J Environ Biol. 2009;30:161–163. [PubMed] [Google Scholar]
- Pacwa-Płociniczak M, Płaza GA, Piotrowska-Seget Z, Cameotra SS. Environmental applications of biosurfactants: recent advances. Int J Mol Sci. 2011;12:633–654. doi: 10.3390/ijms12010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Crowley E. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl Microbiol Biotechnol. 2006;72:1322–1329. doi: 10.1007/s00253-006-0423-5. [DOI] [PubMed] [Google Scholar]
- PB-06 (2010) Determination of polycyclic aromatic hydrocarbons (PAHs) in water by high pressure liquid chromatography (HPLC) (in Polish)
- PB-16 (2009) Determination of content of petroleum hydrocarbons—oil index in water and seawages by gas chromatography (in Polish)
- Płaza GA, Zjawiony I, Banat IM. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated and bioremediated soils. J Pet Sci Eng. 2006;50:71–77. doi: 10.1016/j.petrol.2005.10.005. [DOI] [Google Scholar]
- Płaza GA, Łukasik K, Wypych J, Nałęcz-Jawecki G, Berry C, Brigmon RL. Biodegradation of crude oil and distillation products by biosurfactants-producing bacteria. Pol J Environ Stud. 2008;17:87–94. [Google Scholar]
- Płaza GA, Pacwa-Płociniczak M, Piotrowska-Seget Z, Jangid K, Wilk KA. Agroindustrial wastes as unconventional substrates for growing of Bacillus strains and production of biosurfactant. Environ Prot Eng. 2011;37:63–71. [Google Scholar]
- Płociniczak T, Kukla M, Wątroba R, Piotrowska-Seget Z. The effect of soil bioaugmentation with strains of Pseudomonas on Cd, Zn and Cu uptake by Sinapis alba L. Chemosphere. 2013;91:1332–1337. doi: 10.1016/j.chemosphere.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Powell SM, Ferguson SH, Bowman JP, Snape I. Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol. 2006;52:523–532. doi: 10.1007/s00248-006-9131-z. [DOI] [PubMed] [Google Scholar]
- Puškárová A, Bučková M, Chovanová K, Harichová J, Karelová E, Godočiková J, Polek B, Ferianc P, Pangallo Diversity and PAH growth abilities of bacterial strains isolated from a contaminated soil in Slovakia. Biologia. 2013;4:587–591. doi: 10.2478/s11756-013-0193-3. [DOI] [Google Scholar]
- Rahman KSM, Rahman TJ, McClean S, Marchant R, Banat IM. Rhamnolipid biosurfactant production by strains of Pseudomonas aeruginosa using low-cost raw materials. Biotechnol Prog. 2002;18:1277–1281. doi: 10.1021/bp020071x. [DOI] [PubMed] [Google Scholar]
- Saikia RR, Deka S, Deka M, Banat IM. Isolation of biosurfactant-producing Pseudomonas aeruginosa RS29 from oil-contaminated soil and evaluation of different nitrogen sources in biosurfactant production. Ann Microbiol. 2012;62:753–763. doi: 10.1007/s13213-011-0315-5. [DOI] [Google Scholar]
- Satpute SK, Banat IM, Dhakephalkar PK, Banpurkar AG, Chopade BA. Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol Adv. 2010;28:436–450. doi: 10.1016/j.biotechadv.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Shoeb E, Badar U, Akhter J, Ansari FA, Waqar M, Ansari MA. Screening of surfactant producing bacterial strains isolated from soil samples of an automobile workshop. Karachi Univ J Sci. 2012;40:31–36. [Google Scholar]
- Siegmund I, Wagner F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol Tech. 1991;5:265–268. doi: 10.1007/BF02438660. [DOI] [Google Scholar]
- Silva RMP, Rodriguez AA, de Oca JMGM, Moreno DC. Biodegradation of crude oil by Pseudomonas aeruginosa AT18 strain. Tecnol Quimica. 2006;26:70–77. [Google Scholar]
- Silva SNRL, Farias CBB, Rufino RD, Luna JM, Sarubbo LA. Glycerol as substrate for the production of biosurfactant by Pseudomonas aeruginosa UCP0992. Colloids Surf B. 2010;79:174–183. doi: 10.1016/j.colsurfb.2010.03.050. [DOI] [PubMed] [Google Scholar]
- Tapi A, Chollet-Imbert M, Scherens B, Jacques P. New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl Microbiol Biotechnol. 2010;85:1521–1531. doi: 10.1007/s00253-009-2176-4. [DOI] [PubMed] [Google Scholar]
- Viramontes-Ramos S, Portillo-Ruiz MC, Ballinas-Casarrubias ML, Torres-Muñoz JV, Rivera-Chavira BE, Nevárez-Moorillón GV. Selection of biosurfactan/bioemulsifier-producing bacteria from hydrocarbon-contaminated soil. Braz J Microbiol. 2010;41:668–675. doi: 10.1590/S1517-83822010000300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypych J, Mańko T. Determination of volatile organic compounds (VOCs) in water and soil using solid phase microextraction. Chem Anal. 2002;47:507–530. [Google Scholar]
- Xia WJ, Luo Z, Dong HP, Yu L, Cui QF, Bi YQ. Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous Pseudomonas aeruginosa WJ-1 using waste vegetable oils. Appl Biochem Biotechnol. 2012;166:1148–1166. doi: 10.1007/s12010-011-9501-y. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Duncan KE, Nagle DP, Savager KN, Knapp RM, McInerney MJ. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004;56:339–347. doi: 10.1016/j.mimet.2003.11.001. [DOI] [PubMed] [Google Scholar]