Abstract
During the immune response, B cells undergo a programmed mutagenic cascade to promote increased affinity and expanded antibody function. The two processes, somatic hypermutation (SHM) and class switch recombination (CSR), are initiated by the protein activation-induced deaminase (AID), which converts cytosine to uracil in the immunoglobulin loci. The presence of uracil in DNA promotes DNA mutagenesis though a subset of DNA repair proteins. Two distinct mechanisms have been proposed to control uracil processing. The first is through base removal by uracil DNA glycosylase (UNG), and the second is through detection by the mismatch repair complex MSH2/6. In a study published in this issue of European Journal of Immunology, Dingler et al. [Eur. J. Immunol. 2014. 44: XXXX-XXXX] examine uracil processing in B cells in the absence of UNG and SMUG1 glycosylases. Similar to UNG, SMUG1 is an uracil glycosylase which can remove the uracil base. While Smug1−/− mice show no clear deficiency in SHM or CSR, Ung−/−Smug1−/− mice display exacerbated phenotypes, suggesting a back-up role for SMUG1 in antibody diversity. This new information expands the model of uracil processing in B cells and raises several interesting questions about the dynamic relationship between base excision repair and mismatch repair.
Keywords: class switch recombination, DNA repair, SMUG1, somatic hypermutation, UNG
To protect against the constant onslaught from pathogenic microorganisms, antibodies have evolved to detect and adapt to a large number of different molecules and antigens. During B-cell development, antibodies are first diversified by the process of V(D)J recombination of variable (V), diversity (D), and joining (J) gene segments to create a single variable exon for the heavy and light chains. This initial pool of different antibodies is further diversified after B-cell activation through the processes of somatic hypermutation (SHM) and class switch recombination (CSR). SHM is characterized by the introduction of nucleotide substitutions into the variable gene, which can alter the protein sequence of the antibody. Upon expression, the mutated antibody is then selected for increased affinity to antigen. In addition, nucleotide substitutions and DNA strand breaks occur in switch regions flanking the majority of constant gene exons in the heavy chain locus during CSR. The strand breaks are processed through recombination to bring downstream constant gene exons (Cγ, ε, or α) in close proximity to the variable exon. The change from Cµ expands antibody function as the IgG, IgE, and IgA antibodies interact with antigen and bind Fc receptors (Fcγ, ε, or α) found on immune effector cells to initiate specific immune responses. One fascinating feature of SHM and CSR is the finding that a single enzyme, activation-induced deaminase (AID), has been shown to initiate both processes [1, 2]. AID is a deaminase which functions to convert single-stranded cytosine residues into uracil (Figure 1A) [3, 4]. The mere presence of these uracil residues initiates a complex cascade of events which results in mutagenesis of immunoglobulin genes. While the mechanisms of uracil processing are still under investigation, seminal work from the laboratory of Michael Neuberger opened the floodgates for understanding and analyzing B-cell mutagenesis.
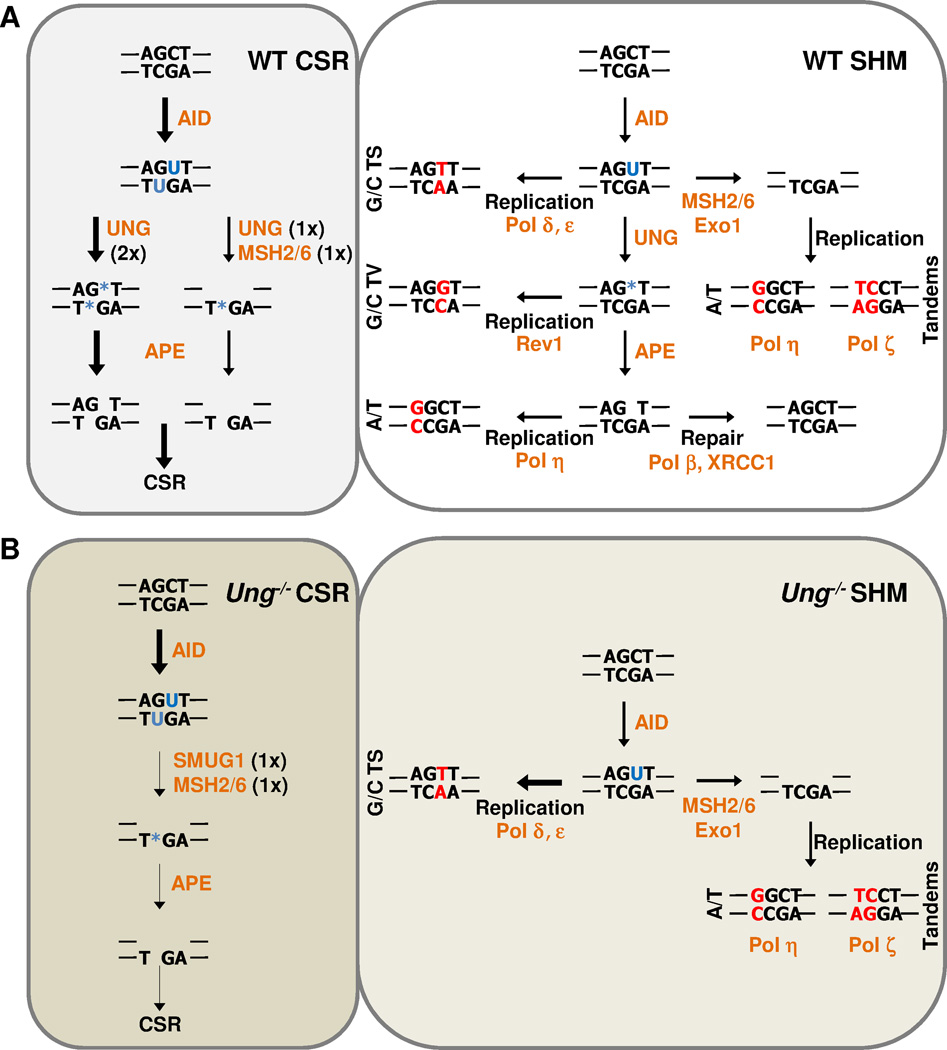
Figure 1. The Neuberger model.
The known mechanisms of uracil processing for CSR (left) and SHM (right) in (A) wild type or (B) Ung−/− mice are depicted. Arrow thickness represents the relative efficiency of the pathway. Blue nucleotides represent DNA lesion, (U – uracil, * - abasic site). Red nucleotides represent fixed mutations. Orange represents enzymes. 1×, creates one nick; 2×, creates two nicks, TS, transition; TV, transversion.
The Neuberger Model
The first definitive proof that SHM and CSR were regulated through mutagenic DNA repair came with the discovery that MSH2-deficient mice had a substantial loss in mutations at A/T residues, increased mutational hotspot focusing, and decreased CSR (reviewed in [5]). An essential member of the mismatch repair (MMR) pathway, MSH2 functions in detecting mismatches generated after DNA replication. Canonical MMR utilizes either the MSH2/6 heterodimer to recognize single nucleotide mismatches or the MSH2/3 heterodimer to recognize nucleotide insertions which create small loops of non-base paired nucleotides [6, 7]. Upon recognition of mismatches, additional proteins (MLH1, PMS2, EXO1) are recruited to the damage site to excise the DNA strand containing the mismatch, followed by accurate resynthesis by PCNA and Polymerase (Pol) δ. However, during SHM, germinal center B cells utilize ubiquitinated PCNA, which switches re-synthesis from Pol δ to highly error-prone Pols η and ζ, resulting in nucleotide substitutions [8–12]. An interesting proviso is that during SHM only a subset of the MMR proteins are involved. Just as with MSH2, both MSH6- and EXO1-deficient mice have decreased A/T substitutions, while MLH1- and PMS2-deficient mice display normal levels of A/T substitutions (reviewed in [13]). It is intriguing to speculate that during SHM, the inhibition or underutilization of MLH1 and PMS2 may inhibit the accurate repair of the uracils, resulting in increased mutagenesis.
While the MMR pathway defines the majority of A/T mutations during SHM, these mutations represent only 50% of total mutations that occur in wild-type mice. With the identification of AID as a DNA cytosine deaminase, Neuberger and colleagues proposed that SHM and CSR both originate from the processing of a U:G mismatch [4]. This model became dogma with the publication of data showing that either inhibition (in DT40 B cells) or deletion (in mouse Peyer’s patch B cells) of uracil-DNA glycosylase (UNG) alters mutation spectra (a 7-fold decrease in C/G transversions during SHM) and drastically inhibits CSR (~10-fold reduction in IgG1 ex vivo after stimulation of mouse B cells with lipopolysaccharide (LPS) and interleukin-4 (IL-4)) [14, 15]. During base excision repair (BER), UNG functions by recognizing and removing the uracil base from DNA (Figure 1A). The residual abasic site is then processed by an apurinic/apyrimidinic-endonuclease (Ape) to break the DNA backbone adjacent to the abasic site, creating a single-strand DNA nick (reviewed in [16]). The nick is then processed by Pol β and DNA ligase to remove the residual base, resynthesize the nick, and ligate the DNA. However, during SHM, the abasic site is used as a template for error-prone synthesis by the Rev1 protein to create C/G transversions (Figure 1A) [17]. While it is still unclear why Pol β cannot accurately repair all the abasic sites, recent evidence suggests that accurate BER occurs simultaneously with error-prone processing [18, 19]. In addition to altered SHM, Neuberger and colleagues also showed that in Ung−/− mouse B cells ex vivo CSR (via LPS and IL-4 stimulation) was reduced to near zero [15], solidifying the understanding that DNA breaks created by UNG-mediated uracil excision are the main driver of CSR. These findings from Msh2−/− and Ung−/− mice led to the model of uracil processing in activated B cells (Figure 1) [14, 15]. C/G mutations are generated by replication across from either uracil (C/G transitions) or abasic sites (C/G transversions) to fix mutations into the genome. A/T mutations are generated by MSH2/6 recognition of uracil and resynthesis of gapped DNA by error-prone polymerases. Thus ‘The Neuberger Model’ for SHM and CSR became the basis for further studies to fine-tune and expand the model.
Refining the model
While the model explains the primary mechanisms of SHM and CSR, a few questions still remain. Unlike MMR, which requires MSH2 under all conditions, uracil excision during BER can be accomplished by at least two major glycosylases: UNG and SMUG1. While UNG has distinct phenotypes during antibody diversification [15], the role of SMUG1 remains elusive. Overexpression of SMUG1 in wild type mice has been shown to decrease CSR and SHM [20], suggesting that SMUG1 may function in the accurate repair of uracil. Alternatively, when overexpressed in Ung−/−Msh2−/− mice, which are completely inhibited for CSR and A/T mutations [21], SMUG1 has also been shown to restore a modest amount of CSR and modest number of A/T mutations, suggesting a compensation for UNG deficiency [20]. However, these results are unable so far to definitively ascribe a role for SMUG1 in promoting or inhibiting mutagenesis during SHM and CSR.
In this issue of European Journal of Immunology, Dingler et al. [22] further analyze the effects of SMUG1 on antibody diversity. While Smug1−/− mice display no defects in CSR or SHM, mice with the simultaneous loss of SMUG1 and UNG begin to display distinct phenotypes. Similar to Ung−/−Msh2−/− mice [21], B cells from Ung−/−Smug1−/− mice have no ex vivo CSR to IgG1 after stimulation with LPS and IL4. Thus, in UNG-deficient short-term B-cell cultures, SMUG1 promotes MSH2-dependent switching at low levels [22]. These results establish a new hierarchy of uracil processing. The complete loss of CSR in Ung−/−Smug1−/− mice suggests that MSH2 requires a glycosylase for CSR under short-term conditions. This finding is intriguing, as it has been found that a low frequency of UNG-dependent G/C transversions are also MSH2 dependent, suggesting synergy between the two pathways [23]. This suggests that CSR proceeds through either a solitary UNG pathway with BER-dependent nicking on both strands, or a synergistic UNG/SMUG1-MSH2 pathway with BER creating one nick while MSH2 creates the second nick (Figure 1A). This model would fit with the finding that during SHM in mouse B cells, A/T mutations are strand biased (reviewed in [5]), suggesting that the majority of MSH2 function occurs in a strand-specific manner, which would decrease the probability of creating abundant double-strand DNA breaks. However, when given sufficient time in vivo, MSH2 can overcome the lack of both UNG and SMUG1 to generate nicks during CSR to generate serum IgG [22].
In terms of SHM, B cells from Ung−/−Smug1−/− mice display a decreased mutation frequency and a slight drop in A/T mutations [22]. While not as pronounced as the phenotype seen in Msh2−/−, Msh6−/−, Polh−/−, or PCNAK164R mice, the loss of both glycosylases seems to alter the spectra of SHM in Ung−/−Smug1−/− mice [22]. Interestingly, the ~14% drop in A/T mutations is similar to the level of residual A/T mutations seen in Msh2−/− mice. Can SMUG1 compensate for UNG in the UNG-Pol η pathway, as reported by Jacobs and colleagues [23]? However, the fact that Ung−/−Msh2−/− mice display only 2% A/T mutations in the presence of fully active SMUG1 suggests there is a more complicated mechanism controlling this subset of A/T mutations seen in Ung−/−Smug1−/− mice. One possibility is that MSH2/6 processing of SMUG1 excision sites is required to allow Pol η onto DNA, while it is not required for UNG sites. Taken together, the data presented by Dingler et al. [22] definitively demonstrate that SMUG1 can function in promoting CSR and SHM in the absence of UNG (Figure 1). However, a function for SMUG1 in wild type mice still remains elusive.
Remaining questions
With the creation and expansion of ‘The Neuberger Model’ [4], a significant problem still remains: why are deamination events not accurately repaired? In non-lymphoid cells, UNG protein levels are highest during early S-phase and show co-localization with replication foci to repair dUTP misincorporation [24]. However, it has been proposed that AID deamination events occur in G1 and that UNG processes these uracils in G1 [25, 26]. This leads to the possibility that UNG has different protein partners and/or has different post-translational modifications in different stages of the cell cycle (reviewed in [16]). Similar to UNG, MMR function is closely linked with DNA replication and recombination to repair nucleotide misincorporation [6, 7]. However, it is unclear whether cell cycle alters MSH2 function and/or protein partners. Another possibility is that AID generates a sufficient amount of uracils in the immunoglobulin loci to overwhelm the faithful repair capacity and thus forces mutagenesis. While initially proposed to function in accurate repair of uracils, the findings of Dingler et al. suggest that SMUG1 also functions in increasing mutagenesis in B cells [20, 22]. Thus more work is required to further explain the phenomenon of B-cell-mediated hijacking of DNA repair proteins.
Acknowledgements
This work was supported entirely by the Intramural Research program of the NIH, National Institute on Aging. We gratefully thank Lisa Russell and Kim Zanotti for insightful comments.
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, et al. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 5.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin Immunol. 2012;24:293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 7.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 9.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, et al. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saribasak H, Maul RW, Cao Z, Yang WW, Schenten D, Kracker S, Gearhart PJ. DNA polymerase zeta generates tandem mutations in immunoglobulin variable regions. J Exp Med. 2012;209:1075–1081. doi: 10.1084/jem.20112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saribasak H, Gearhart PJ. Does DNA repair occur during somatic hypermutation? Semin Immunol. 2012;24:287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 15.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 16.Krokan HE, Saetrom P, Aas PA, Pettersen HS, Kavli B, Slupphaug G. Error-free versus mutagenic processing of genomic uracil-Relevance to cancer. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 17.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saribasak H, Maul RW, Cao Z, McClure RL, Yang W, McNeill DR, Wilson DM, 3rd, et al. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med. 2011;208:2209–2216. doi: 10.1084/jem.20111135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Dingler FA, Kemmerich K, Neuberger MS, Rada C. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts A:T substitutions during somatic mutation. Eur J Immunol. 2014 doi: 10.1002/eji.201444482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krijger PH, Langerak P, van den Berk PC, Jacobs H. Dependence of nucleotide substitutions on Ung2, Msh2, and PCNA-Ub during somatic hypermutation. J Exp Med. 2009;206:2603–2611. doi: 10.1084/jem.20091707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharbeen G, Yee CW, Smith AL, Jolly CJ. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J Exp Med. 2012;209:965–974. doi: 10.1084/jem.20112379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]