Abstract
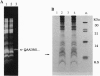
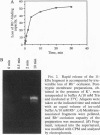
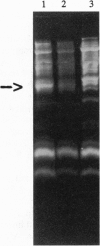
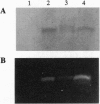
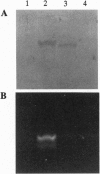
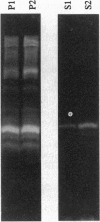
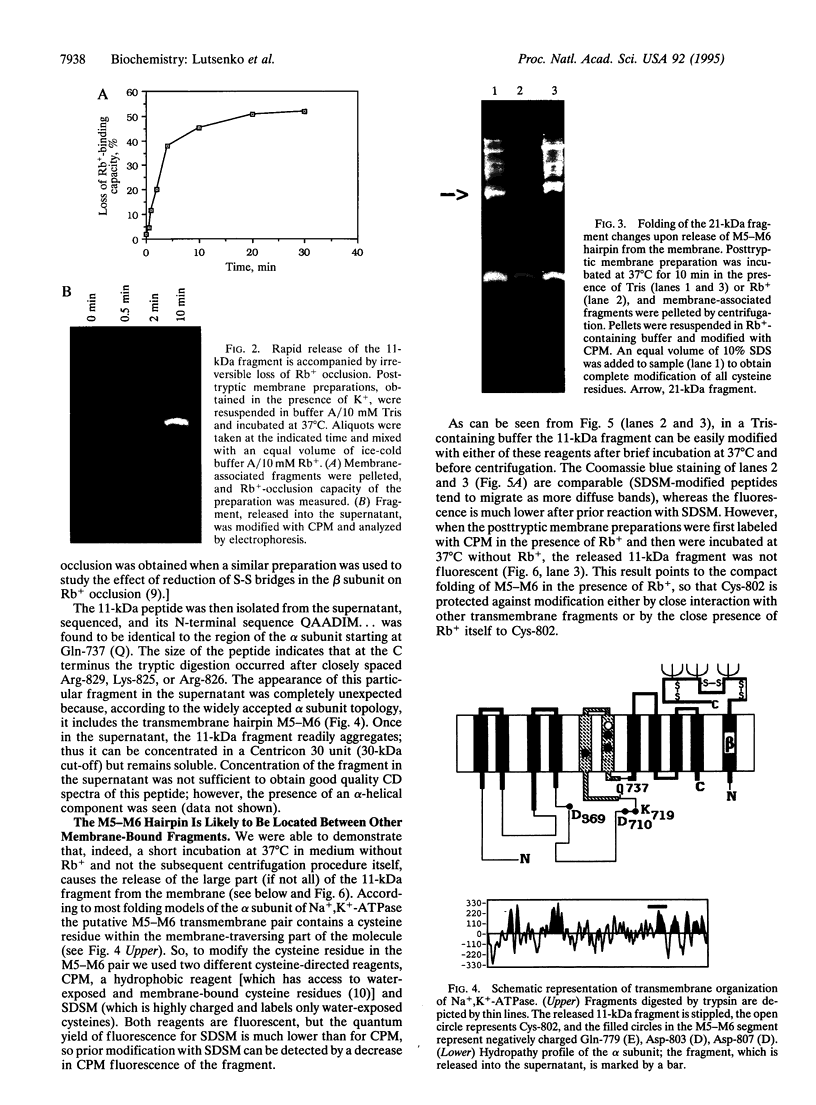
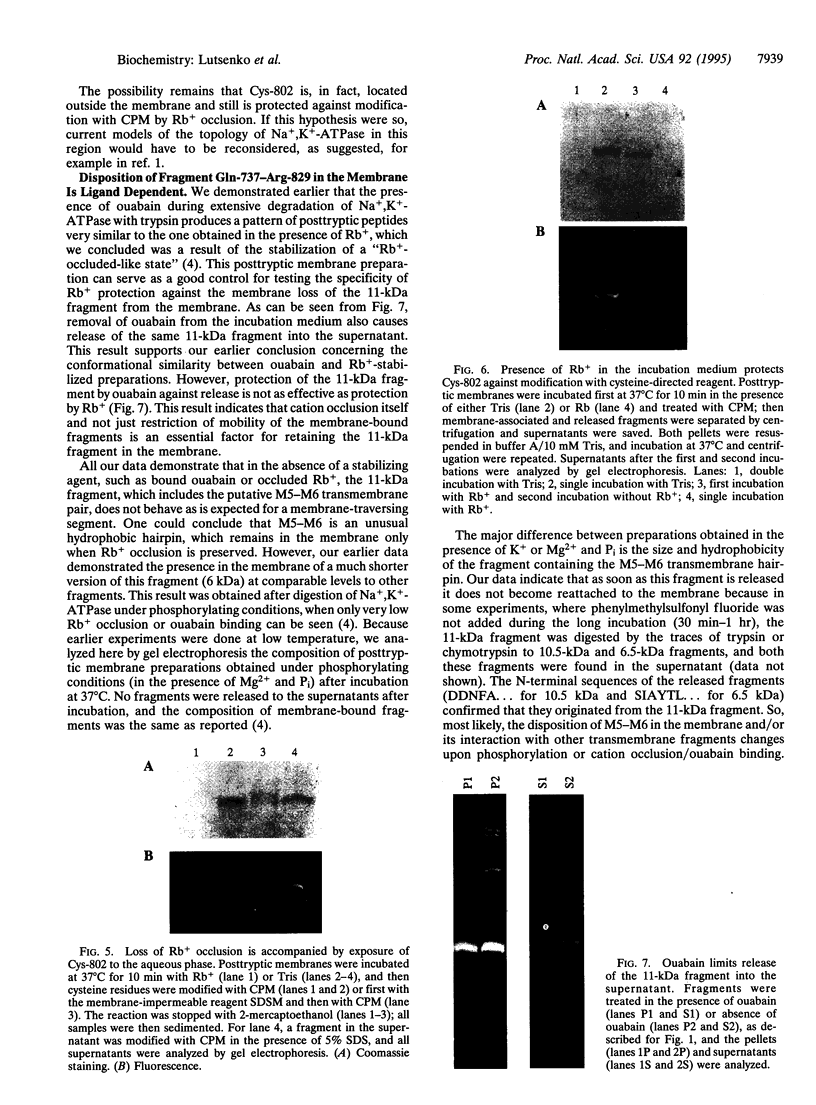
Extensive proteolytic digestion of Na+,K(+)-ATPase (EC 3.6.1.37) by trypsin produces a preparation where most of the extramembrane portions of the alpha subunit have been digested away and the beta subunit remains essentially intact. The fragment Gln-737-Arg-829 of the Na+,K(+)-ATPase alpha subunit, which includes the putative transmembrane hairpin M5-M6, is readily, selectively, and irreversibly released from the posttryptic membrane preparation after incubation at 37 degrees C for several minutes. Once released from the membrane, the fragment aggregates but remains water soluble. Occlusion of K+ or Rb+ specifically prevents release of the Gln-737-Arg-829 fragment into the supernatant. Labeling of the posttryptic membrane preparation with cysteine-directed reagents revealed that Cys-802 (which is thought to be located within the M6 segment) is protected against the modification by Rb+ while this fragment is in the membrane but can be readily modified upon release. Cation occlusion apparently alters the folding and/or disposition of the M5-M6 fragment in the membrane in a way that does not occur when the fragment migrates to the aqueous phase. The ligand-dependent disposition of the M5-M6 hairpin in the membrane along with recent labeling studies suggest a key role for this segment in cation pumping by Na+,K(+)-ATPase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argüello J. M., Kaplan J. H. Glutamate 779, an intramembrane carboxyl, is essential for monovalent cation binding by the Na,K-ATPase. J Biol Chem. 1994 Mar 4;269(9):6892–6899. [PubMed] [Google Scholar]
- Ayers F. C., Warner G. L., Smith K. L., Lawrence D. A. Fluorometric quantitation of cellular and nonprotein thiols. Anal Biochem. 1986 Apr;154(1):186–193. doi: 10.1016/0003-2697(86)90513-0. [DOI] [PubMed] [Google Scholar]
- Bamberg K., Sachs G. Topological analysis of H+,K(+)-ATPase using in vitro translation. J Biol Chem. 1994 Jun 17;269(24):16909–16919. [PubMed] [Google Scholar]
- Besancon M., Shin J. M., Mercier F., Munson K., Miller M., Hersey S., Sachs G. Membrane topology and omeprazole labeling of the gastric H+,K(+)-adenosinetriphosphatase. Biochemistry. 1993 Mar 9;32(9):2345–2355. doi: 10.1021/bi00060a028. [DOI] [PubMed] [Google Scholar]
- Canfield V. A., Levenson R. Transmembrane organization of the Na,K-ATPase determined by epitope addition. Biochemistry. 1993 Dec 21;32(50):13782–13786. doi: 10.1021/bi00213a005. [DOI] [PubMed] [Google Scholar]
- Capasso J. M., Hoving S., Tal D. M., Goldshleger R., Karlish S. J. Extensive digestion of Na+,K(+)-ATPase by specific and nonspecific proteases with preservation of cation occlusion sites. J Biol Chem. 1992 Jan 15;267(2):1150–1158. [PubMed] [Google Scholar]
- Esmann M., Karlish S. J., Sottrup-Jensen L., Marsh D. Structural integrity of the membrane domains in extensively trypsinized Na,K-ATPase from shark rectal glands. Biochemistry. 1994 Jul 5;33(26):8044–8050. doi: 10.1021/bi00192a008. [DOI] [PubMed] [Google Scholar]
- Jewell-Motz E. A., Lingrel J. B. Site-directed mutagenesis of the Na,K-ATPase: consequences of substitutions of negatively-charged amino acids localized in the transmembrane domains. Biochemistry. 1993 Dec 14;32(49):13523–13530. doi: 10.1021/bi00212a018. [DOI] [PubMed] [Google Scholar]
- Jorgensen P. L. Purification and characterization of (Na+, K+)-ATPase. V. Conformational changes in the enzyme Transitions between the Na-form and the K-form studied with tryptic digestion as a tool. Biochim Biophys Acta. 1975 Sep 2;401(3):399–415. doi: 10.1016/0005-2736(75)90239-4. [DOI] [PubMed] [Google Scholar]
- Jørgensen P. L., Andersen J. P. Structural basis for E1-E2 conformational transitions in Na,K-pump and Ca-pump proteins. J Membr Biol. 1988 Jul;103(2):95–120. doi: 10.1007/BF01870942. [DOI] [PubMed] [Google Scholar]
- Karlish S. J., Goldshleger R., Jørgensen P. L. Location of Asn831 of the alpha chain of Na/K-ATPase at the cytoplasmic surface. Implication for topological models. J Biol Chem. 1993 Feb 15;268(5):3471–3478. [PubMed] [Google Scholar]
- Karlish S. J., Goldshleger R., Stein W. D. A 19-kDa C-terminal tryptic fragment of the alpha chain of Na/K-ATPase is essential for occlusion and transport of cations. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4566–4570. doi: 10.1073/pnas.87.12.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingrel J. B., Kuntzweiler T. Na+,K(+)-ATPase. J Biol Chem. 1994 Aug 5;269(31):19659–19662. [PubMed] [Google Scholar]
- Lutsenko S., Kaplan J. H. An essential role for the extracellular domain of the Na,K-ATPase beta-subunit in cation occlusion. Biochemistry. 1993 Jul 6;32(26):6737–6743. doi: 10.1021/bi00077a029. [DOI] [PubMed] [Google Scholar]
- Lutsenko S., Kaplan J. H. Molecular events in close proximity to the membrane associated with the binding of ligands to the Na,K-ATPase. J Biol Chem. 1994 Feb 11;269(6):4555–4564. [PubMed] [Google Scholar]
- Modyanov N., Lutsenko S., Chertova E., Efremov R., Gulyaev D. Transmembrane organization of the Na+,K(+)-ATPase molecule. Acta Physiol Scand Suppl. 1992;607:49–58. [PubMed] [Google Scholar]
- Ohta T., Nagano K., Yoshida M. The active site structure of Na+/K+-transporting ATPase: location of the 5'-(p-fluorosulfonyl)benzoyladenosine binding site and soluble peptides released by trypsin. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2071–2075. doi: 10.1073/pnas.83.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E., David P., Shainskaya A., Tal D. M., Karlish S. J. Effects of competitive sodium-like antagonists on Na,K-ATPase suggest that cation occlusion from the cytoplasmic surface occurs in two steps. J Biol Chem. 1993 Aug 15;268(23):16929–16937. [PubMed] [Google Scholar]
- Ovchinnikov YuA, Dzhandzhugazyan K. N., Lutsenko S. V., Mustayev A. A., Modyanov N. N., Dzhandzugazyan KN=Dzhadzhugazyan K. N. Affinity modification of E1-form of Na+, K+-ATPase revealed Asp-710 in the catalytic site. FEBS Lett. 1987 Jun 8;217(1):111–116. doi: 10.1016/0014-5793(87)81253-x. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shani M., Goldschleger R., Karlish S. J. Rb+ occlusion in renal (Na+ + K+)-ATPase characterized with a simple manual assay. Biochim Biophys Acta. 1987 Nov 2;904(1):13–21. doi: 10.1016/0005-2736(87)90081-2. [DOI] [PubMed] [Google Scholar]
- Shin J. M., Besancon M., Simon A., Sachs G. The site of action of pantoprazole in the gastric H+/K(+)-ATPase. Biochim Biophys Acta. 1993 Jun 5;1148(2):223–233. doi: 10.1016/0005-2736(93)90133-k. [DOI] [PubMed] [Google Scholar]
- Sweadner K. J., Arystarkhova E. Constraints on models for the folding of the Na,K-ATPase. Ann N Y Acad Sci. 1992 Nov 30;671:217–227. doi: 10.1111/j.1749-6632.1992.tb43798.x. [DOI] [PubMed] [Google Scholar]
- Yoon K. L., Guidotti G. Studies on the membrane topology of the (Na,K)-ATPase. J Biol Chem. 1994 Nov 11;269(45):28249–28258. [PubMed] [Google Scholar]