Abstract
Manganese (Mn)-induced neurodegenerative toxicity has been associated with a distorted iron (Fe) metabolism at both systemic and cellular levels. In the current study, we examined whether the oxidation states of Mn produced differential effects on certain mitochondrial [Fe-S] containing enzymes in vitro. When mitochondrial aconitase, which possesses a [4Fe-4S] cluster, was incubated with either Mn(II) or Mn(III), both Mn species inhibited the activities of aconitase. However, the IC10 (concentration to cause a 10% enzyme inhibition) for Mn(III) was ninefold lower than that for Mn(II). Following exposure of mitochondrial fractions with Mn(II) or Mn(III), there was a significant inhibition by either Mn species in activities of Complex I whose active site contains five to eight [Fe-S] clusters. The dose–time response curves reveal that Mn(III) was more effective in blocking Complex I activity than Mn(II). Northern blotting was used to examine the expression of mRNAs encoding transferrin receptor (TfR), which is regulated by cytosolic aconitase. Treatment of cultured PC12 cells with Mn(II) and Mn(III) at 100 μM for 3 days resulted in 21 and 58% increases, respectively, in the expression of TfR mRNA. Further studies on cell growth dynamics after exposure to 25–50 μM Mn in culture media demonstrated that the cell numbers were much reduced in Mn(III)-treated groups compared to Mn(II)-treated groups, suggesting that Mn(III) is more effective than Mn(II) in cell killing. In cells exposed to Mn(II) and Mn(III), mitochondrial DNA (mtDNA) was significantly decreased by 24 and 16%, respectively. In contrast, rotenone and MPP+ did not seem to alter mtDNA levels. These in vitro results suggest that Mn(III) species appears to be more cytotoxic than Mn(II) species, possibly due to higher oxidative reactivity and closer radius resemblance to Fe.
Keywords: manganese, iron, aconitase, Complex I, speciation, transferrin receptor, PC12 cells, mitochondria, mitochondrial DNA, cytotoxicity, Fe-S cluster
Chronic manganese (Mn) intoxication in humans causes permanent neurodegenerative damage in the nigrostriatal region, resulting in a syndrome similar to Parkinson’s disease (PD; Cook et al., 1974; Mena et al., 1967). While the etiology of idiopathic Parkinson’s disease (IPD) remains unclear, high levels of total iron (Fe), decreased ferritin, oxidative stress, and abnormal mitochondrial Complex I activity have been repeatedly observed in the substantia nigra of IPD patients (Dexter et al., 1991; Jenner et al., 1992; Sofic et al., 1991; Ye et al., 1996). A recent population study has also established that serum Fe concentrations are significantly reduced in IPD patients compared with controls, suggesting a compartment shift in Fe from blood to tissues, including brain (Logroscino et al., 1997). The role of Fe in etiopathology of IPD has been extensively reviewed by Jenner et al. (1992) and Youdim et al. (1993).
Previous studies from this laboratory have established that Mn-induced neurotoxicities appear to be associated with its interaction with Fe at systemic and cellular levels (Zheng et al., 1998, 1999; Zheng and Zhao, 2001). Following Mn exposure, there is a predominant influx of Fe from the blood into the cerebrospinal fluid (CSF) and from extracellular matrix to intracellular space (Zheng et al., 1999; Zheng and Zhao, 2001). From a chemical point of view, Mn resembles Fe in several ways, such as having similar ionic radii, carrying similar 2+ and 3+ valent charges under physiological conditions, and possessing a similar binding affinity for the carrier protein transferrin (Aschner et al., 1999). Because of these similarities, we propose that Mn may not only interact with the Fe regulatory mechanism, but also act directly on certain enzymes whose active centers comprise [Fe-S] clusters, such as aconitase, NADH-ubiquinone reductase (Complex I), and succinic dehydrogenase (SDH, Complex II).
Aconitase, an enzyme present in mitochondria (and cytoplasm) is responsible for the conversion of citrate to isocitrate in the tricarboxylic acid cycle. It contains a [4F3-4S] cubane cluster, which serves as the enzymatic binding site for citrate (Henson and Cleland, 1967; Kennedy et al., 1983). Cytosolic aconitase, also known as iron-regulatory protein-1 (IRP-1 or ACO1), regulates intracellular Fe metabolism via its binding and unbinding to the mRNAs containing stem–loop structures, also referred to as iron-responsive elements (IRE). When cellular Fe levels are insufficient, IRP-1 assumes a [3Fe-4S] configuration, loses its enzymatic activity, and is transformed into an mRNA-binding protein. The net result of this RNA: protein interaction is an increase in cellular Fe uptake (Kennedy et al., 1983). Results from our previous studies have shown that Mn exposure, either in vitro or in vivo, inhibits the enzymatic activity of aconitase (Zheng et al., 1998, 1999). Since the coordination chemistry of Mn closely resembles that of Fe, we suspect that Mn may insert itself into the fourth Fe site of aconitase. While suppressing the enzyme’s catalytic function, it may increase the protein’s ability to bind to mRNA, which favors the expression of transferrin receptor (TfR) and restrains the translation of ferritin. An increased cellular level of Fe is expected to facilitate Fe-mediated cytotoxicity (Zheng et al., 1998; Zheng and Zhao, 2001; Zheng, 2001).
Complex I, the first enzyme in the mitochondrial respiratory chain, also contains [Fe-S] clusters in its active sites (Ohnishi, 1998). The functional unit of Complex I is thought to contain 13 to 14 conserved subunits, among which at least two [2Fe-2S] and four [4Fe-4S] clusters have been identified (Ohnishi, 1998). These clusters are ligated to 4 cysteinyl sulfurs from the polypeptide chain of the apoprotein and are responsible for electron transfer from NADH to acceptor ubiquinone. Deficiency in Complex I activity in specific brain regions has been associated with IPD. For example, a significant reduction in the activity of Complex I has been found postmortem in the substantia nigra, platelets, and skeletal muscle of patients with IPD (Haas et al., 1995; Mizuno et al., 1998; Parker et al., 1989; Schapira et al., 1990). Animal studies using model compounds such as 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine to induce Parkinson’s lesions also confirm the correlation between a defected Complex I and PD-type neuropathological damages (Burns et al., 1983; Langston et al., 1983; Nicklas et al., 1985; Ramsay et al., 1989). A recent study further demonstrates that chronic exposure to rotenone, a Complex I inhibitor, can cause highly selective nigrostriatal dopaminergic degeneration that is associated with certain PD syndromes in rats (Betarbet et al., 2000).
Upon entering the cells, Mn mainly accumulates in mitochondria (Liccione and Maines, 1988). The mitochondria take up Mn(II) presumably via a Ca(II) uniporter (Chance, 1965; Gavin and Gunter, 1996). The slow efflux of Mn by mitochondria accounts for the excess accumulation of Mn ions in this subcellular organelle (Gavin and Gunter, 1996). Since certain critical enzymes in the mitochondrial respiratory chain contain [Fe-S] structures, it seems likely that one of the Mn species in the mitochondria, either in the form of Mn(II) or Mn(III), may compete with Fe in the [Fe-S] cluster, leading to a disruption in energy production and the subsequent mitochondrial dysfunction.
Mn can assume multiple oxidation valence states under physiological conditions, primarily in the form of Mn(II) and Mn(III). Different oxidation states result in Mn species with distinct ionic size and reactivity, rendering one particular species more favorable in replacement of Fe in [Fe-S] centers or in oxidation/reduction reactions. For the past decade, research on Mn toxicities has focused on the use of Mn(II) (mainly as MnCl2) due to its high aqueous solubility and easy handling. However, the question as to what oxidation state of Mn in the body is responsible for Mn-induced cytotoxicity has not yet been explored.
This study was designed to test the hypothesis that exposure to different oxidative Mn species may induce differential cytotoxicities. We used a dopaminergic neuronal-derived PC12 cell line to compare the inhibitory effect of Mn(II) and Mn(III) on the activities of mitochondrial aconitase and Complex I. We also sought to investigate the effects of Mn(II) and Mn(III) on the expression of TfR mRNAs, which is regulated by IRP-1, and to study the cytotoxic effect of Mn(II) and Mn(III) on the growth dynamics of cultured cells. Since the levels of mitochondrial DNA (mtDNA) are sensitive to free radical-initiated oxidative stress (Wallace, 1992), we further examined whether exposure to Mn(II) or Mn(III) altered the levels of mtDNA.
MATERIALS AND METHODS
Chemicals
Chemicals were obtained from the following sources: manganese chloride (MnCl2 · 4H2O), NADH β-form, 2,3-dimethoxy-5-methyl-6-[3-methyl-2-butenyl]-1,4-benzoquinone (ubiquinone-5; CoQ1), antimycin A, potassium cyanide, phosphatidylcholine, Hepes, EGTA, sodium pyruvate, penicillin–stryptomycin, rotenone, cis-aconitic acid, ferrous ammonium sulfate [Fe(NH4)2(SO4)2 · 6H2O], L-citric acid, isocitric acid, triethanolamine–HCl, Triton X-100, sucrose, β-nicotinamide adenine dinucleotide phosphate (β-NADP), isocitrate dehydrogenase, purified porcine aconitase (30 units/g), XbaI, PvuII, and RNase A from Sigma Chemical Co. (St. Louis, MO); manganese acetate dihydrate (C6H9MnO6 · H2O) from Fluka Chemical Corp (Milwaukee, WI); 1-methyl-4-phenylpyridine (MPP+) from Research Biochemicals International (Natick, MA); NH4H2PO4 from Aldrich Chemical Co. (Milwaukee, WI); Dulbecco’s modified essential medium (DMEM), RPMI 1640 growth medium, fetal bovine serum (FBS), and antibiotic–antimycin from Gibco (Grand Island, NY); and RNA Zol B assay kit from Tel-Test “B” (Friendswood, TX). All reagents were of analytical grade, HPLC grade, or the best available pharmaceutical grade.
Preparation of Mn solutions
Mn(II) solution as MnCl2 was prepared by directly dissolving MnCl2 salts in distilled, deionized water at a concentration of 100 mM as the stock solution. The working solutions were diluted from the stock on the day of use. The stock was prepared on a weekly basis and stored at room temperature.
Mn(III) solutions were prepared freshly at the day of experimentation using various approaches. For enzymatic studies, Mn(III) acetate dihydrate was first dissolved in 100% ethanol to a concentration of 50 mM. The solution was then filtered through a sterile filter (25-mm 0.2-μm nylon membrane) attached to a Becton Dickinson 3-ml syringe. An aliquot of this solution (no more than 10 μl) was added directly into a total of 500 μl of the reaction mixture for the aconitase activity assay to produce final concentrations of 25–1000 μM. In the Complex I study, an aliquot of this solution (no more than 10 μl) was added into a total of 1 ml of the reaction mixture for final concentrations of 25–500 μM. Both enzymatic activities were not affected by the presence of small volumes of ethanol.
For the cell culture study, Mn(III) acetate dihydrate was dissolved in 100% ethanol as a 50-mM stock. After filtration, an aliquot (10–20 μl) of the stock was mixed with an equal volume (10–20 μl) of DMSO. The mixed Mn(III) solution was than added into the culture medium to the required Mn concentration, ranging from 25 to 200 μM (with <0.1% ethanol and <0.1% DMSO). The solvents used did not affect the cell growth of PC12 cells. Mn concentrations in the reaction mixture and in culture medium were verified by atomic absorption spectrophotometry.
Preparation of mitochondrial fraction from rat brain
The mitochondria were isolated according to the method described by Whitfield et al. (1981). Rats were euthanized by cervical dislocation. The whole brains were removed and rinsed in saline and homogenized in a homogenizing buffer containing 0.25 M sucrose, 10 mM Hepes (pH 8.0), and 2 mM EGTA on ice. The homogenate (10 ml) was centrifuged at 1100g, 4°C for 10 min to remove residual nuclei or unbroken cells. Mitochondrial fractions were separated from the supernatant solution by centrifugation at 14,000g, 4°C for 10 min. The pellet was washed three times with 10 ml of homogenizing buffer, centrifuged twice at 8,000g for 10 min, and finally at 14,000g for 10 min. The final mitochondrial fraction was resuspended in 0.5 ml of homogenizing buffer and stored at −70°C until use. Prior to experimentation, the mitochondrial fraction underwent freeze–thaw treatment three times and was sonicated by 10 pulses at a level of 3.5 and output of 20% in a Sonifer sonicator (VWR) on ice.
Cell cultures and growth dynamics study
PC12 cells were purchased from American Type Culture Collection (ATCC) (Manassas, VA). The cells were cultured in 150-cm2 flasks in a growth medium consisting of RPMI supplemented with 10% horse serum, 5% FBS, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 0.2% D-glucose, 10 mM Hepes, and 1 mM sodium pyruvate. All experiments were carried out with cells suspended in the culture media, rather than grown on collagen-coated tissue culture dishes. The culture medium was changed every 2 days. During the initial cell plating, the total cell numbers were managed in the range of 2.5 to 4.6 × 105 cells in 5 ml of growth medium.
The cells were exposed to Mn in culture media containing either Mn(II) predissolved in water or Mn(III) predissolved in equal volumes of ethanol and DMSO. Occasionally, Mn(II) predissolved in ethanol and DMSO was tested in the cell culture study. At the end of exposure, the cultures were mixed by repeatedly pipetting with a 10-ml Costar Stripette. A aliquot (1 ml) of cell suspension was removed and placed in an Eppendorf tube. A single-cell suspension was established by triturating the cells through a 22-G needle attached onto a syringe. A 100-μl aliquot of cell suspension was then mixed with 100 μl of 0.4% trypan blue solution (w/v). Following cell staining for 5 min, an aliquot (10 μl) of stained cell suspension was pipetted onto a hemocytometer and counted for cell number. Fresh culture medium (2 ml) was then added into the original culture for the duration of experiment.
Protein concentrations were determined by a Bio-Rad assay kit (Bio-Rad Laboratories, Richmond, CA) using bovine serum albumin as standard.
Determination of aconitase activity
The activity of aconitase was assayed by determining the rate of formation of the intermediate product, cis-aconitate, from the substrate DL-isocitrate as described previously (Zheng et al., 1998). Purified mitochondrial aconitase (5 μg protein) was pretreated with various concentrations of either Mn(II) or Mn(III) followed by preactivation with 25 μM Fe(II) (as ferrous ammonium sulfate) in a buffer consisting of 40 mM Hepes (pH 8.5) and 10 mM cysteine in a total volume of 50 μl at 37°C for 5 min. The enzymatic reaction was initiated by adding 20 μl of pretreated samples into a total of 1 ml of the reaction buffer containing 20 mM triethanolamine–HCl (pH 7.5) and 1.0 mM DL-isocitrate. The changes of absorbance at 240 nm were recorded for 10 min by a Perkin–Elmer Lambda-11 spectrophotometer and used for the calculation of the enzyme activity. The aconitase activity was expressed as the formation of cis-aconitate (increases in absorbance)/mg protein/min. Mn(III) was dissolved in ethanol. All samples were run in triplicate.
Determination of Complex I activity
The activity of Complex I was determined by a modified method of Hatefi (1978). The rate of oxidation of NADH by Complex I was estimated by measurement of the rate of consumption of NADH. The reaction mixture (in a total 1 ml of volume) contained 10 mM Tris–HCl (pH 8.0), 50 mM KCl, 2 μg/ml antimycin A (to block Complex III), 2 mM KCN (to block Complex IV), 0.1 mg/ml phophatidylcholine, 40 μg protein of mitochondrial fractions, 0.1 mM NADH, and 10 μl of various concentrations of either Mn(II) or Mn(III). The reaction was initiated by adding 4 μl 10 mM CoQ1 into the above reaction mixture and incubating at 37°C for 10 min. The decline of NADH absorbance was then monitored at 340 nm. The rate of conversion of NADH to NAD+ was used to calculate Complex I activity and expressed as the conversion of μmol NADH/mg protein/min.
It was noted that while the rate of enzymatic reaction reached the maximum in a potassium phosphate buffer, a visible precipitate was observed when Mn concentrations exceeded 1 mM in that solution. We then tried a Hepes buffer, which prevented the metal precipitation; however, it led to a reduced Vmax. In contrast, Tris buffer prevented the metal precipitation, while allowing a better reaction rate. We also noticed that that the combination of CoQ1 as the electron acceptor with phospholipids greatly improved the assay linearity and sensitivity. Mn(III) in ethanol was used in the experiments.
Northern blot of TfR mRNA
Cultured PC12 cells were incubated with either 100 μM Mn(II) or Mn(III) (predissolved in ethanol/DMSO) in culture medium for 3 days. At the end of exposure, the cells were washed with PBS and harvested for Northern blot analysis of TfR mRNA. Total RNA was extracted from the control and Mn-treated PC12 cells using the RNA Zol B kit following the instructions of the manufacturer. RNA was normalized by loading equal amounts of total RNA as determined by absorbance at 260 nm. Total RNA (10 μg) was electrophoresed on 1% formaldehyde–agarose gels, followed by soaking in 0.05 mM NaOH for 20 min and then 20× saline sodium citrate (SSC) for 40 min. After the RNA was transferred to nylon filter in 20× SSC overnight, the filter was UV irradiated, prehybridized in 0.5 M phosphate buffer (pH 7.2) containing 7% SDS and 1% BSA at 65°C for 15 min, and then hybridized at 65°C overnight in the same solution containing 2 × 107 cpm/ml of random-primed 32P-labeled TfR cDNA probe. The probe was a 507-bp fragment of the 5′-end of a full-length 3.4-kb rat TfR cDNA, subcloned to pTZ19U-RTR, which was a gift of Dr. Griswold at Washington State University (Roberts and Griswold, 1990). The filters were washed three times for 15 min each using 2× SSC with 0.1% SDS at 65°C, followed by 1× SSC with 0.1% SDS at 25°C once for 15 min. The filters were then autoradiographed with Kodak Biomax MR film (Eastman Kodak, Rochester, NY) in an intensifying screen at −70°C for 7–14 days.
To quantitate TfR mRNA, the filters were further hybridized with random-primed 32P-labeled β-actin RNA and autoradiographed for 6–8 h. The X-ray images of the Northern blots were scanned by an Epson Expression 636 scanner into a Macintosh Power PC G3 computer, followed by analysis of the density of mRNA bands corresponding to TfR and β-actin using NIH Image 1.57 software package. The densities of TfR mRNA in control and Mn-treated groups were normalized to those of β-actin mRNA in each corresponding lane.
Southern blot of mtDNA
Southern blot of mtDNA was performed using the method described by Miyako et al. (1997). The cells in Mn-treated and control groups were harvested and the total DNA was extracted with DNAzol reagent (Life Technologies, Inc.) according to the manufacturer’s instructions. The extracted DNA was digested with RNase A and PvuII and solubilized in 0.1 × TE (1 mM Tris–HCl (pH 7.5) and 0.1 mM EDTA). The concentrations of DNA were determined by measuring the absorbance at a wavelength of 260 nm. An aliquot (2–5 μg) of DNA was electrophoresed in 0.8% agarose gel and transferred onto a MSI nylon transfer membrane (Fisher Scientific). The mitochondrial gene and 18S rRNA gene (as an internal standard) were hybridized with the probes of 0.8-kbp XbaI fragment of mitochondrial DNA (nt 7441–8260) and 1.5-kbp XbaI fragment of 18S rRNA gene (nt 450–1955), respectively. Both probes were labeled with [α-32P]dCTP using a Megaprime DNA labeling kit (Amersham). The membranes were then autoradiographed and the densities of bands corresponding to mtDNA and 18S rRNA were quantified as described above.
Statistics
Statistical analyses of the differences between groups were performed by using two-way ANOVA. The differences between two means were considered significant if p values were equal or less than 0.05.
RESULTS
Solubilization of Mn(III) Salt
To solubilize Mn(III) species, we have tested several commonly used solvent systems in bioassays, including Emulphor, polyethylene glycol, Tween series, rat plasma, DMSO, ethanol, and the combination of these solvents (Table 1). Among the tested solvents, we found that absolute ethanol proved to be the best to solubilize Mn(III) as Mn(III) acetate dihydrate. The concentration of this solution can be made to as high as 100 mM. At 50 mM and even lower concentrations, all salts went into the solution without visible residuals. When small aliquots of this solution were added into enzyme reaction mixture, no visible precipitation was observed.
TABLE 1.
Solubility of Mn(III) as Mn Acetate Dihydrate in Various Solvents
| Vehicle | Concentration | Solubility |
|---|---|---|
| 100% Ethanol | <50 mM 100 mM |
+ +/− |
| DMSO | 1 mM | − |
| E mulphor EL-620p | 1 mM | − |
| Polyethylene glycol (MW400) | 1 mM | − |
| 2% Tween 80 | 1 mM | − |
| Rat plasma | 1 mM | +/− |
| 100% Ethanol + DMSO (1:1, v/v) | 25 mM | + |
| 100% Ethanol + DMSO (1:1, v/v) + culture medium | 500 μM | + |
Note. The solubility was judged by visible precipitation of Mn acetate dihydrate following addition to the solvents. +, completely dissolved: +/−, partially dissolved; −, not dissolved.
For the cell culture study, direct addition of Mn(III)–ethanol solution to the culture medium resulted in a granule-type of precipitation. Thus, tests were made by premixing Mn(III)–ethanol with rat plasma or DMSO. The experiments with rat plasma did not improve solubility. However, mixing Mn(III)–ethanol with DMSO at the ratio of 1:1, followed by addition this mixture to the culture medium, prevented the precipitation and proved to be useful in cell culture studies. This recipe was then used in subsequent experiments.
Inhibition of Aconitase Activity by Mn(II) and Mn(III)
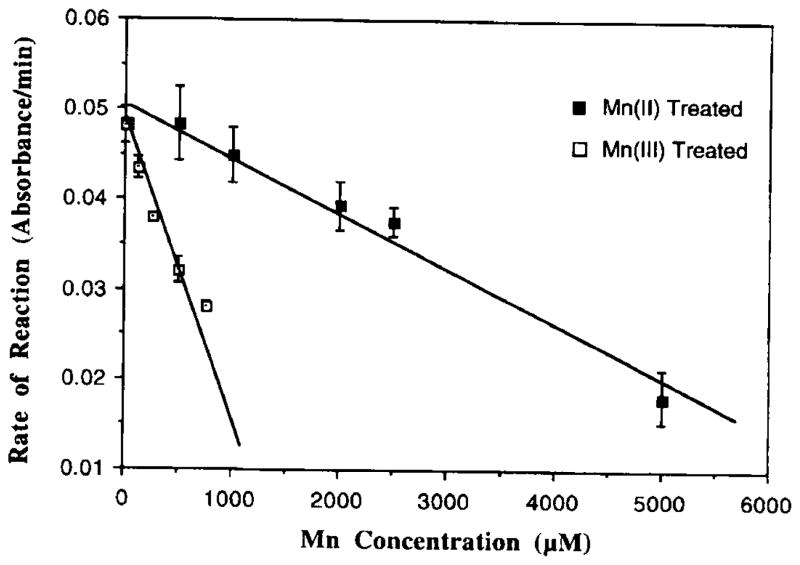
When mitochondrial aconitase was incubated with Mn(II), a minimum concentration of 1 mM of Mn(II) was required to initiate a notable inhibitory effect (6.6% inhibition) (Fig. 1). Increasing concentrations of Mn(II) in the reaction mixture decreased the enzymatic activity. At 5 mM, the effect of Mn(II) on the aconitase reached the maximum, which was about 36% inhibition of the aconitase activity (Fig. 1). Experiments with Mn(II) dissolved in ethanol at 1 mM produced the result (6.4% inhibition) that was comparable to those seen in experiments with Mn(II) in water.
FIG. 1.
Inhibition by Mn(II) or Mn(III) of mitochondrial aconitase activity in vitro. Mitochondrial aconitase (5 μg protein) was pretreated with various concentrations of Mn for 5 min followed by Fe (50 μM) activation for another 10 min. The rate of formation of aconitase from DL-isocitrate was monitored at 240 nm. Values represent means ± SD (n = 3).
In comparison, the minimum concentration required for Mn(III) to inhibit the enzyme was 125 μM. At 700 μM of Mn(III), there was a 43% inhibition compared to controls (Fig. 1). When the dose–response curves were compared between Mn(II) and Mn(III) groups, Mn(III)-treated groups showed a steeper slope than that of Mn(II) group (Fig. 1). The IC10, defined as the Mn concentration to cause 10% inhibition of aconitase activity and estimated from the linear regression of dose–response curves, was 124 μM for Mn(III), while it was 1164 μM for Mn(II), about 10-fold higher than that of Mn(III).
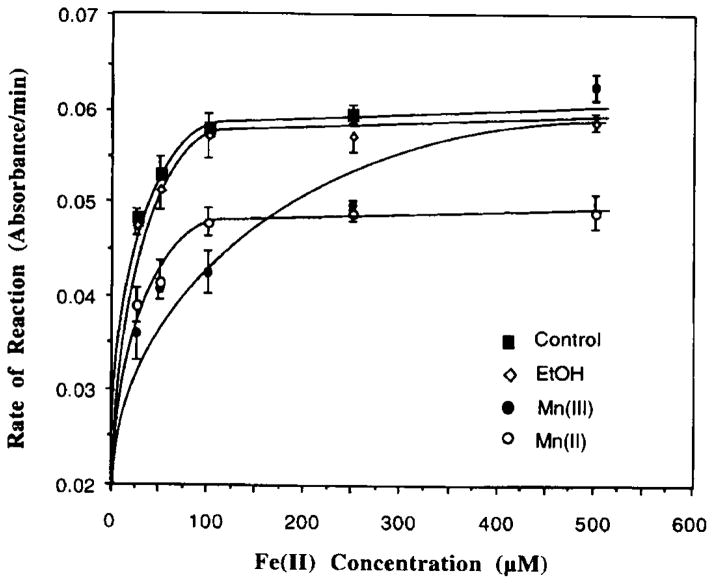
The rate of the aconitase-catalyzed reaction also varied as a function of Fe concentrations present in the reaction mixture (Fig. 2). Addition of excess amounts of Fe into Mn(II)- or Mn(III)-pretreated enzyme fractions restored the aconitase activity (Fig. 2); however, the reaction rate did not return to its full potential until 500 μM of Fe(II) was added into the reaction mixture of the Mn(III) group. Notably also, to restore the same reaction rate, Mn(II) preincubated samples required a higher concentration of Fe(II) than those treated with Mn(III).
FIG. 2.
Reversal of the Mn-induced inhibition of aconitase by Fe. Mitochondrial aconitase (5 μg) was pretreated with or without 500 μM Mn(III) or 2 mM Mn(II) for 5 min followed by Fe activation at various concentration for 5 min. Values represent means ± SD (n = 3).
In the same reaction system, addition of ethanol, a solvent for Mn(III) acetate, did not interfere with the aconitase activity.
Inhibition of Mitochondrial Complex I Activity by Mn(II) and Mn(III)
Following exposure of mitochondrial fractions to Mn(II) or Mn(III) at concentrations ranging from 50 to 500 μM for 20 min, Complex I activity was significantly inhibited by both Mn species (Fig. 3A). The inhibitory effects were Mn-concentration dependent. At 500 μM, Mn(III) and Mn(II) caused 76% (p < 0.01) and 60% (p < 0.01) inhibition of the Complex I activity, respectively. Mn(II) dissolved in ethanol at the same concentration resulted in the same degree of inhibition (data not shown). There were also statistically significant differences between Mn(II)- and Mn(III)-treated groups at 300 μM (p < 0.05) and 500 μM (p < 0.01) of Mn concentrations in Complex I inhibition. Thus, Mn(III) species appeared to be more effective than that of Mn(II) in inhibition of Complex I activity.
FIG. 3.
Inhibition by Mn(II) or Mn(III) on Complex I activity of rat brain mitochondria. (A) Dose–response relationship of Mn actions. Mitochondrial fractions were incubated with 50–500 μM Mn(II) (as MnCl2) or Mn(III) (as C6H9MnO6) for 20 min. (B) Time course of Mn actions. Mitochondrial fractions were incubated with 300 μM Mn(II) or Mn(III) for 5, 10, 20, and 30 min. The rate of conversion of NADH to NAD+ was monitored at 340 nm. Data represent means ± SD of duplicate determinations (n = 3–4). *p < 0.05. **p < 0.01 compared to control values; *p < 0.05, **p < 0.01 for Mn(III)- compared to Mn(II)-treated groups at the same concentration.
The time-course studies indicated a progressive inhibition of this enzyme by both Mn species. Two-way ANOVA revealed a statistically significant difference in time curves between Mn-treated groups and controls (p < 0.01 for both comparisons) (Fig. 3B). Mn(III) appeared to be more effective than Mn(II) at later times; however, the differences between Mn(III)- and Mn(II)-treated groups did not reach statistical significance (p > 0.05) (Fig. 3B).
Steady-State TfR mRNA Levels as Affected by Mn(II) and Mn(III) Treatment
One of the functions of cytosolic aconitase is to bind to the IRE in mRNAs encoding TfR or other proteins. An enhanced binding affinity of IRP-1 to 5′-IRE-containing mRNA can result in an up-regulation of these mRNAs. Since Mn may replace Fe in IRP-1, we examined the effect of Mn on the expression of TfR mRNA in cultured PC12 cells.
Following Mn treatment at 100 μM for 3 days, the expression of TfR mRNA in both Mn(II) and Mn(III) groups was evidently increased on the autoradiograph by Northern blot (Fig. 4). Quantitative analysis of the optical density of the bands corresponding to TfR mRNA, which was normalized to β-actin mRNA in the same lane, revealed that Mn(II) treatment enhanced the expression of TfR mRNA in PC12 cells by 21% (p < 0.05) (Table 2). Exposure to Mn(III) greatly enhanced the expression of TfR mRNA by 58% of controls (Fig. 4, Table 2).
FIG. 4.

Manganese exposure promotes the expression of TfR mRNA in cultured PC12 cells. The cells were incubated with 100 μM of either Mn(II) or Mn(III) for 3 days. There was 10 μg of total RNA loaded on each lane. Lane 1. control cells with normal culture medium without Mn(II); lane 2. control cells with medium containing DMSO without Mn(III); lane 3, Mn(II)-treated cells; and lane 4, Mn(III)-treated cells.
TABLE 2.
Effect of Mn(II) and Mn(III) on the Expression of TfR mRNA in Cultured PC12 Cells
| Control | Mn-treated | % increase | |
|---|---|---|---|
| Mn(II) study | 0.836 ± 0.216 | 1.012 ± 0.151* | 21.0 |
| Mn(III) study | 0.695 ± 0.091 | 1.101 ± 0.194** | 58.3 |
Note. PC12 cells were treated with either Mn(II) or Mn(III) at 100 μM for 3 days. The expression of TfR mRNA and β-actin mRNA (internal standard) were determined by Northern blot. The optical density of bands for TfR mRNA was normalized to the abundance of β-actin mRNA in the same lane, and data represent the ratio of TfR/β-actin abundance. The culture media in the Mn(III) study contained DMSO in both control and Mn(III)-treated groups. Values are means ± SD, n = 5–6.
p < 0.05 compared to control.
p < 0.01 compared to the control.
Cytotoxic Effects of Mn(II) and Mn(III) on the Growth of PC12 Cells
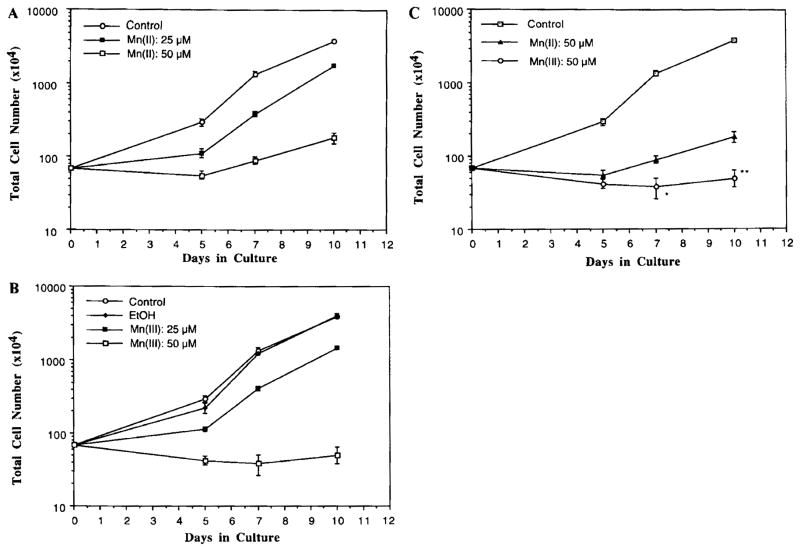
The cytotoxic effect of both Mn(II) and Mn(III) on the cell growth of neuronal typical PC12 cells was investigated at Mn concentrations of 25 and 50 μM in the culture medium. In comparison to controls, Mn(III) treatment prevented cell growth after 5 days of incubation at 25 μM (Fig. 5A). Under the microscope, the cells were visibly swollen, and there was evidence of cytoplasmic granules (data not shown). Treatment of cells with Mn(III) at 50 μM for 10 days resulted in a 99% reduction of the cell growth (Fig. 5A). Two-way ANOVA revealed an overall significant reduction in total cell numbers after Mn(III) treatment.
FIG. 5.
Manganese exposure hindered the cell growth of neuronal PC12 cells. The cells were exposed to 25 or 50 μM of either Mn(II) or Mn(III) at day 0. The viable cells were counted at times indicated. (A) Cell growth curves following Mn(II) treatment. (B) Cell growth curves following Mn(III) treatment (C) Comparison of cell growth curves between Mn(II) and Mn(III) groups. Data represent means ± SD (n = 3). *p < 0.05, **p < 0.01 between Mn(II) and Mn(III) groups.
Mn(II) treatment at both concentrations significantly reduced the total cell numbers by two-way ANOVA (Fig. 5B). However, the abnormal cytology, such as the cytoplasmic granules, did not become apparent until the cells were incubated with Mn(II) at concentrations higher than 500 μM.
When the comparison was made between Mn(II) and Mn(III) groups, there was a statistically significant difference between the two groups in cell growth curves by two-way ANOVA (Fig. 5C). By day 10, the cell numbers in the Mn(III) group were about 60% that of Mn(II) group. Thus, Mn(III) species appeared to be more cytotoxic than Mn(II) species.
The possible interference from the solvent on the Mn(II) groups was also investigated. Mn(II) was predissolved in ethanol/DMSO in a volume equivalent to Mn(III) preparation, followed by addition to the culture medium. After exposure of the cells at 50 μM Mn(II) for 5 days, Mn(II) predissolved in ethanol/DMSO produced the same degree of inhibition (74%) as did Mn(II) in water (75%). Thus, the solvent system seemed unlikely to affect the outcomes.
Effects of Mn(II) and Mn(III) on Mitochondrial DNA Contents
Since Mn extensively accumulates in mitochondria, we sought to test the hypothesis that failure in energy production and ensuing oxidative stress in mitochondria may affect mtDNA. The effect of Mn on mtDNA was compared to those of rotenone and MPP+, two well-defined Complex I inhibitors. Following treatment of PC12 cells with 100 μM of either Mn(II) or Mn(III) in culture media for 3 days, the densities of 16-kbp bands corresponding to the mtDNA were reduced by 24 and 16% in Mn(II) and Mn(III) treated groups, respectively (Fig. 6, Table 3). Mn(II) species seemed likely to cause more severe reduction of mtDNA than Mn(III); however, the difference between Mn(II)- and Mn(III)-treated groups was not statistically significant.
FIG. 6.

Decrease in mtDNA levels following Mn exposure. PC12 cells were incubated with 100 μM of either Mn(II) or Mn(III), 0.1 μM rotenone, or 25 μM MPP+ for 3 days. There was 2–5 μg of DNA loaded on each lane. Lane 1, control cells with normal culture medium; lane 2, Mn(II)-treated cells; lane 3, Mn(III)-treated; lane 4, rotenone-treated cells; and lane 5, MPP+-treated cells.
TABLE 3.
Effect of Mn, Rotenone, and MPP+ on the Levels of mtDNA in Cultured PC12 Cells
| mtDNA/18S rRNA | Decrease (%) | |
|---|---|---|
| Control | 1.019 ± 0.049 | 100.00 |
| Mn(II) (100 μM) | 0.773 ± 0.126 | −24.16* |
| Mn(III) (100 μM) | 0.859 ± 0.026 | −15.74* |
| Rotenone (0.5 μM) | 0.994 ± 0.018 | −2.94 |
| MPP+ (25 μM) | 0.977 ± 0.010 | −4.23 |
Note. Following incubation PC12 cells with Mn(II). Mn(III), rotenone, or MPP+ for 3 days, an aliquot (2–5 μg) of total DNA was used for Southern blot analysis. The optic density of bands for mtDNA was normalized to the abundance of 18S rRNA (internal standard) in the same lane, and data represent the ratio of mtDNA/18S rRNA abundance. Values are means ± SD, n = 3–4.
p < 0.05 compared to the control.
Treatment with rotenone at 0.1 μM for 3 days did not alter mtDNA levels nor did the exposure to MPP+ at 25 μM for the same duration (Fig. 6).
DISCUSSION
Mn ions can assume multiple oxidative valence states under physiological conditions ranging from Mn(II), Mn(III), to Mn(IV). Much of the literature covering Mn toxicity has been built on experiments using Mn(II), for its salts are quite water soluble. While Mn(III) exists naturally as salts of acetate, phosphate, or trioxide, none of these salts are readily water soluble. In this study, we have tested seven commonly used solvent systems and concluded that a combination of ethanol with DMSO proved to be an ideal vehicle for the in vitro study of Mn(III) (as Mn acetate dihydrate).
The results from the current study provide strong evidence that the valance state of Mn determines the degree to which Mn exerts cytotoxicities. Mn(III) species was more potent than Mn(II) in inhibiting mitochondrial [Fe-S]-containing enzymes such as aconitase and Complex I, in elevating the level of TfR mRNA that is regulated by [Fe-S]-containing cytosolic aconitase, and in retarding the growth and differentiation of cultured dopaminergic PC12 cells. These data support our hypothesis that a higher oxidation state of Mn, i.e., Mn(III), is more cytotoxic than Mn(II) in alteration of cell functions.
Our previous studies have shown that exposure to Mn(II), either in vitro or in vivo, leads to a significant alteration in aconitase activity (Zheng et al., 1998). The current results further establish that, to achieve the minimum magnitude of inhibition, i.e., at IC10, the required Mn(II) concentration was nearly 10 times higher than that of Mn(III) exposure. In other words, Mn(III) is more effective than Mn(II) in inhibition of aconitase. Two possible mechanisms might explain the extraordinary activity of Mn(III) species.
First, the higher oxidation species of Mn may replace Fe in the catalytic site of aconitase more readily than the lower ones. The ionic size of a metal generally depends on its oxidation state, with the ionic radii decreasing as the oxidation state increases. Two oxidation states of Fe possess a radius of 65 and 78 ppm for Fe(III) and Fe(II), respectively, while for Mn, the radii are 65 and 83 ppm for Mn(III) and Mn(II), respectively (Nieboer and Fletcher, 1996). Given that the Mn(III) species has an ionic size identical to Fe(III), it seems likely that the higher oxidation state of Mn may optimally fit into the geometric space of aconitase, substitute for Fe in the [Fe-S] cluster, and in so doing inhibit enzymatic reactions. In the case of Mn(II) exposure, however, the oxidation of Mn(II) to Mn(III), which is a rather slow, oxygen-dependent process, may turn into a critical rate-limiting step. Hence, the Mn(II)-elicited toxicity may become apparent only in the presence of a high concentration of Mn(II) species.
Second, the higher oxidation state of Mn may be more reactive than lower oxidation species in influencing the interchange of aconitase between oxidation and reduction states. Under normal conditions, a reductive switch of the [3Fe-4S] cluster to the [4Fe-4S] cluster is required for aconitase activity (Beinert and Kennedy, 1993; Robbins and Stout, 1989). The substrate (citrate) binds only to the added fourth Fe, thereby being converted to isocitrate. By oxidatively converting the 4Fe to the 3Fe cluster, the enzyme loses its catalytic function and is then transformed to an mRNA-binding protein. The latter form favors protein binding to the mRNA carrying IRE stem–loops (e.g., TfR mRNA). Taking into account the higher oxidative activity of Mn(III), we postulate that Mn(III), as a better oxidizing agent than Mn(II), may catalyze the conversion of the active site of aconitase from a [4Fe-4S] state to a [3Fe-4S] state. Correspondingly, Mn(III) itself may be reduced to Mn(II) during this oxidation/reduction reaction. The proposed interaction between Mn and [Fe-S] clusters in aconitase is shown below.

The results from our TfR mRNA assay also show that Mn(III) increased the levels of TfR mRNA nearly three times as much as Mn(II) in cultured PC12 cells. The enhanced expression of TfR mRNA could be a direct result of Mn interaction with IRP-1, a cytosolic aconitase with a [4Fe-4S] cluster (Zheng et al., 1998, 1999). It should be noted that only the [3Fe-4S] configuration of IRP-1 is capable of binding to TfR mRNA. Thus, our current results further substantiate our hypothesis that Mn(III), as a reactive species of Mn toxicity, may catalyze the change of aconitase to the oxidative state.
Unlike aconitase, there is no known interstructural exchange among six [Fe-S] clusters in Complex I. Complex I possesses at least two [2Fe-2S] and four [4Fe-4S] clusters in its 13–16 subunits (Ohnishi, 1998). No reports, however, indicate the presence of a labile Fe atom among these clusters. Thus, it is not entirely clear whether Mn(III) may be able to catalyze the oxidative reaction so as to deprive one of the irons from [Fe-S] clusters in Complex I. However, it is possible that Mn(III), owing to its similar ionic radius to Fe(III), may directly substitute for Fe in the catalytic active site, leading to the enzyme inhibition.
The copy numbers of mtDNA have been shown to be sensitive to oxidative stress and have been used as an indicator for the functional integrity of mitochondria (Melov et al., 1999; Tabrizi and Schapira, 1999). Initiation of mtDNA transcription is mainly regulated in the D-loop region, which is reportedly more susceptible to oxidative stress than other regions. The high level of oxidative stress in mitochondria, resulting from excessive Fe accumulation, altered Complex I activity, and arrested energy production after Mn exposure, would be expected to reduce the level of mtDNA. This seems to be true based on the results of this study. Both Mn species significantly lowered the levels of mtDNA by Southern blot analysis. However, it is somewhat puzzling that Mn(II) appeared to be more effective than Mn(III) on mtDNA, although the differences did not reach statistical significance. Currently, we do not have any explanation for this difference. However, it is noteworthy that treatment of PC12 cells with either rotenone or MPP+, two well-known Complex I inhibitors, did not greatly affect mtDNA levels in the current study. It appears that the altered Complex I activity might not necessarily cause the reduction of mtDNA.
Based on the results from the current assays, it is not surprising to see that a much more severe cell growth retardation occurred in Mn(III)-treated groups than in Mn(II) groups. This is primarily due to a more potent toxic effect of Mn(III) than Mn(II) in inhibition of mitochondrial [Fe-S] cluster enzymes, i.e., aconitase and Complex I, which ultimately led to a dysfunctional mitochondrial respiration. A greater effect of Mn(III) than Mn(II) on the expression of TfR mRNA could also promote more cellular uptake of Fe from the extracellular matrix in Mn(III)-treated cultures, bringing about Fe-initiated oxidative stress. The higher cytotoxicity of Mn(III) than Mn(II) has also been reported by other groups (Ali et al., 1995; Archibald and Tyree, 1987; Donaldson et al., 1982). These investigators have reported that the trivalent or higher valence states of Mn may produce the reactive oxidative species, promote the oxidation of catecholamines, and enhance lipid peroxidation. Taken together, we conclude that Mn(III) species, once entering the body or being converted from Mn(II), are more cytotoxic than Mn(II) species. Given the role that Mn(III) can play in initiating cytotoxicity, particularly in the brain, more research should be conducted to determine the mechanism by which Mn(III) is produced, including the site of its biotransformation and the enzyme systems that may catalyze the oxidation/reduction of Mn species.
Acknowledgments
We are greatly indebted to Dr. Michael Griswold at Washington State University for providing transferrin receptor cDNA at our request. This research was supported in part by National Institute of Environmental Health Sciences Grants RO1-ES08146 and P30-ES09089.
References
- Ali SF, Duhart HD, Newport GD, Lipe GW, Slikker W. Manganese-induced reactive oxygen species: Comparison between Mn2+ and Mn3+ Neurodegeneration. 1995;4:329–334. doi: 10.1016/1055-8330(95)90023-3. [DOI] [PubMed] [Google Scholar]
- Archibald FS, Tyree C. Manganese poisoning and the attack of trivalent manganese upon catecholamines. Arch Biochem Biophys. 1987;256:638–650. doi: 10.1016/0003-9861(87)90621-7. [DOI] [PubMed] [Google Scholar]
- Aschner M, Vrana KE, Zheng W. Manganese uptake and distribution in the central nervous system (CNS) Neurotoxicology. 1999;20:173–180. [PubMed] [Google Scholar]
- Beinert H, Kennedy MC. Aconitase, a two-faced protein: Enzyme and iron regulatory factor. FASEB J. 1993;7:1442–1449. doi: 10.1096/fasebj.7.15.8262329. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Burns RS, Chiueh CC, Markey SP, Ebert MH, Jacobowitz DM, Kopin IJ. A primate model of parkinsonism: Selective destruction of dopaminergic neurons in the pars compacta of substantia nigra by N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Proc Natl Acad Sci USA. 1983;80:4546–4550. doi: 10.1073/pnas.80.14.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B. The energy-linked reaction of calcium with mitochondria. J Biol Chem. 1965;240:2729–2748. [PubMed] [Google Scholar]
- Cook DG, Fahn S, Brait KA. Chronic manganese intoxication. Arch Neurol. 1974;30:59–64. doi: 10.1001/archneur.1974.00490310061010. [DOI] [PubMed] [Google Scholar]
- Dexter DT, Carayon A, Javoy-Agid F, Agid Y, Wells FR, Daniel SE, Lees AJ, Jenner P, Marsden CD. Alterations in the levels of iron, ferritin and other trace metals in Parkinson’s disease and other neurodegenerative diseases affecting the basal ganglia. Brain. 1991;114:1953–1975. doi: 10.1093/brain/114.4.1953. [DOI] [PubMed] [Google Scholar]
- Donaldson J, McGregor D, LaBella F. Manganese neurotoxicity: A model for free radical mediated neurodegeneration? Can J Physiol Pharmacol. 1982;60:1398–1405. doi: 10.1139/y82-208. [DOI] [PubMed] [Google Scholar]
- Gavin CE, Gunter TE. Manganese dynamics in brain mitochondria. In: Yasui M, Strong MJ, Ota K, Verity MA, editors. Mineral and Metal Neurotoxicology. CRC Press; New York: 1996. pp. 305–310. [Google Scholar]
- Haas RH, Nasirian F, Nakano K, Ward D, Pay M, Hill R, Shults CW. Low platelet mitochondrial complex 1 and complex II/III activity in early untreated Parkinson’s disease. Ann Neurol. 1995;37:714–722. doi: 10.1002/ana.410370604. [DOI] [PubMed] [Google Scholar]
- Hatefi Y. Preparation and properties of NADH-ubiquinone oxidoreductase (complex I) Methods Enzymol. 1978;53:11–14. doi: 10.1016/s0076-6879(78)53006-1. [DOI] [PubMed] [Google Scholar]
- Henson CP, Cleland WW. Purification and kinetic studies of beef liver cytoplasmic aconitase. J Biol Chem. 1967;242:3833–3838. [PubMed] [Google Scholar]
- Jenner P, Schapira AH, Marsden CD. New insights into the cause of Parkinson’s disease. Neurology. 1992;42:2241–2250. doi: 10.1212/wnl.42.12.2241. [DOI] [PubMed] [Google Scholar]
- Kennedy MC, Emptage MH, Dreyer JL, Beinert H. The role of iron in the activation–inactivation of aconitase. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- Liccione J, Maines M. Selective vulnerability of glutathione metablism and cellular defense mechanisms in rat striatum to manganese. J Pharmacol Exp Ther. 1988;247:156–161. [PubMed] [Google Scholar]
- Logroscino G, Marder K, Graziano JH, Freyer G, Slavkovich V, Lolacono N, Cote L, Mayeux R. Altered systemic iron metabolism in Parkinson’s disease. Neurology. 1997;49:714–717. doi: 10.1212/wnl.49.3.714. [DOI] [PubMed] [Google Scholar]
- Melov S, Coskun PE, Wallace DC. Mouse models of mitochondrial disease, oxidative stress, and senescence. Mutat Res. 1999;434:233–242. doi: 10.1016/s0921-8777(99)00031-2. [DOI] [PubMed] [Google Scholar]
- Mena I, Marin O, Fuenzalida S, Cotzias GC. Chronic manganese poisoning: Clinical picture and manganese turnover. Neurology. 1967;17:128–136. doi: 10.1212/wnl.17.2.128. [DOI] [PubMed] [Google Scholar]
- Miyako K, Kai Y, Irie T, Takeshige K, Kang D. The content of intracellular mitochondrial DNA is decreased by 1-methyl-4-phenylpyridinium ion (MPP+) J Biol Chem. 1997;272:9605–9608. doi: 10.1074/jbc.272.15.9605. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Hattori N, Matsumine H. Neurochemical and neurogenetic correlates of Parkinson’s disease. J Neurochem. 1998;71:893–902. doi: 10.1046/j.1471-4159.1998.71030893.x. [DOI] [PubMed] [Google Scholar]
- Nicklas WJ, Vyas I, Heikkila E. Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenylpyridine, a metabolite of the neurotoxin 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 1985;36:2503–2508. doi: 10.1016/0024-3205(85)90146-8. [DOI] [PubMed] [Google Scholar]
- Nieboer E, Fletcher GG. Determinants of reactivity in metal toxicology. In: Chang LW, editor. Toxicology of Metals. CRC Press; New York: 1996. pp. 113–132. [Google Scholar]
- Ohnishi T. Iron-sulfur clusters/semiquinones in complex I. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- Parker WD, Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- Ramsay RR, Youngster SK, Nicklas WJ, Mckeown KA, Jin YZ, Heikkila RE, Singer TP. Structural dependence of the inhibition of mitochondria respiration and of NADH oxidase by 1-methyl-4-phenylpyridinium (MPP+) analogs and their energized accumulation by mitochondria. Proc Natl Acad Sci USA. 1989;86:9168–9172. doi: 10.1073/pnas.86.23.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins AH, Stout CD. Structure of activated aconitase: Formation of the [4Fe-4S] cluster in the crystal. Proc Natl Acad Sci USA. 1989;86:3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KP, Griswold MD. Characterization of rat transferrin receptor cDNA: The regulation of transferrin receptor mRNA in testes and in Sertoli cells in culture. Mol Endocrinol. 1990;4:531–542. doi: 10.1210/mend-4-4-531. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Sofic E, Paulus W, Jellinger K, Riederer P, Youdim MB. Selective increase of iron in substantia nigra zona compacta of parkinsonian brains. J Neurochem. 1991;56:978–982. doi: 10.1111/j.1471-4159.1991.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Tabrizi SJ, Schapira AH. Secondary abnormalities of mitochondrial DNA associated with neurodegeneration. Biochem Soc Symp. 1999;66:99–110. doi: 10.1042/bss0660099. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Disease of the mitochondrial DNA. Annu Rev Biochem. 1992;61:978–982. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Whitfield CD, Bostedor R, Goodrum D, Haak M, Chu EHY. Hamster cell mutants unable to grow on galactose and exhibiting an overlapping complementation pattern are defective in the electron transport chain. J Biol Chem. 1981;256:6651–6656. [PubMed] [Google Scholar]
- Ye FQ, Allen PS, Martin WR. Basal ganglia iron content in Parkinson’s disease measured with magnetic resonance. Mov Dis. 1996;11:243–249. doi: 10.1002/mds.870110305. [DOI] [PubMed] [Google Scholar]
- Youdim MB, Ben-Shachar D, Riederer P. The possible role of iron in the etiopathology of Parkinson’s disease. Mov Dis. 1993;8:1–12. doi: 10.1002/mds.870080102. [DOI] [PubMed] [Google Scholar]
- Zheng W. Toxicology of choroid plexus: Special reference to metal-induced neurotoxicities. Microsc Res Tech. 2001;52:89–103. doi: 10.1002/1097-0029(20010101)52:1<89::AID-JEMT11>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Ren S, Graziano JH. Manganese inhibits mitochondrial aconitase: A mechanism of manganese neurotoxicity. Brain Res. 1998;799:334–342. doi: 10.1016/s0006-8993(98)00481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao Q. Cellular overload of iron following manganese exposure in cultured neuronal, but not neuroglial cells. Brain Res. 2001;897:175–179. doi: 10.1016/s0006-8993(01)02049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao Q, Slavkovich V, Aschner M, Graziano JH. Alteration of iron homeostasis following chronic exposure to manganese in rats. Brain Res. 1999;833:125–132. doi: 10.1016/s0006-8993(99)01558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]