Abstract
Background
The high prevalence of tobacco use in some developing nations, including Bangladesh, poses several public health challenges for these populations. Smoking behavior is determined by both genetic and environmental factors; however, the genetic determinants of smoking behavior have not been previously examined in a Bangladeshi or South Asian population. We performed a genome-wide association study (GWAS) of tobacco smoking behavior among a population-based sample of 5,354 (2,035 ever smokers and 3,319 never smokers) men and women in Bangladesh.
Methods
Genome-wide association analyses were conducted for smoking initiation (ever versus never smokers), smoking quantity (cigarettes per day), age of smoking initiation, and smoking cessation (former versus current smokers). Sex-stratified associations were performed for smoking initiation.
Results
We observed associations for smoking initiation in the SLC39A11 region at 17q21.31 (rs2567519, P = 1.33 × 10−7) among males and in the SLCO3A1 region at 15q26 (rs12912184, P = 9.32 × 10−8) among females.
Conclusions
These findings suggest possible underlying mechanisms related to solute carrier transporter genes, which transport neurotransmitters, nutrients, heavy metals, and other substrates into cells, for smoking initiation in a South Asian population in a sex-specific pattern. Genetic markers could have potential translational implications for the prevention or treatment of tobacco use and addiction in South Asian populations and warrant further exploration.
Keywords: angladesh, environment and public health, genome-wide association study, smoking initiation, tobacco smoke
INTRODUCTION
Tobacco smoke is a known human carcinogen and has been implicated in epidemiologic studies to be associated with increased risk of several cancers [1]. While the prevalence of tobacco smoking has begun to decline in many developed nations, it has been continuing to rise in developing nations, including Bangladesh and other South Asian nations [2]. There are several public health challenges related to the high prevalence of tobacco use in these populations. Tobacco smokers represent a sizable portion of the population who are at increased risk of future morbidity and mortality. Tobacco consumption in Bangladesh also generates a sizable burden on the health of the population through household expenditures on tobacco as opposed to other basic resources such as food, housing, health or education [2 3]. Therefore, tobacco control is a public health priority in Bangladesh, which could be better aided with an understanding of the various determinants of smoking behavior in this population.
Smoking behavior is a complex phenotype determined by a combination of environmental and genetic determinants [4]. However, an understanding of the genetic basis of smoking behavior remains limited, especially for developing country populations [5]. To date, several genome-wide association studies (GWAS) of smoking behavior have been conducted [6–14], with several chromosomal regions observed to be associated with various phenotypes of smoking behavior. However, these association studies have been conducted primarily in populations of European ancestry in developed nations, with only a few exceptions [12–14]. No previous study has comprehensively assessed genetic determinants of smoking behavior in a Bangladeshi or other South Asian population that comprise nearly a quarter of the world’s population.
In this analysis, we conducted a GWAS to evaluate genetic determinants of smoking behavior in a Bangladeshi population. We evaluated genome-wide associations of single nucleotide polymorphisms (SNPs) with smoking initiation (ever versus never smokers), smoking quantity (cigarettes per day), age of smoking initiation, and smoking cessation (former versus current smokers).
SUBJECTS AND METHODS
Study population
Individuals included in this study were enrolled in two population-based longitudinal studies: the Health Effects of Arsenic Longitudinal Study (HEALS) or the Bangladesh Vitamin E and Selenium Trial (BEST). HEALS, described previously in detail [15], is a cohort study established to investigate health outcomes associated with chronic arsenic exposure from groundwater in a population sample of adults in Araihazar, Bangladesh. Eligibility criteria for participation included being married (to minimize loss to follow-up), aged between 18 and 75 years, and resident in the study area for at least 5 years. A total of 20,033 (11,746 during 2000–2002 and 8,287 during 2006–2008) men and women were enrolled into the HEALS cohort. Trained study physicians, blinded to participants’ exposure to arsenic, conducted in-person interviews and clinical evaluations, and collected urine and blood samples from participants in their homes using structured protocols. BEST is a 2×2 factorial randomized chemoprevention trial evaluating the long-term effects of vitamin E and selenium supplementation on non-melanoma skin cancer risk [16]. BEST participants are residents of Araihazar (the same geographic area as HEALS participants), Matlab, and surrounding areas. Eligibility criteria included aged between 25 and 65 years, permanent residence in the study area, manifest arsenical skin lesions, and no prior cancer history. During 2006–2009, a total of 7,000 individuals were enrolled into the study. BEST uses many of the same study protocols as HEALS, including recruitment, interview data (including smoking and covariate data) and biospecimen collection and processing.
There were 2,035 ever smokers and 3,319 never smokers eligible for these analyses from the combined HEALS and BEST cohorts, with available GWAS data. The study protocols were approved by the relevant Institutional Review Boards in the US (The University of Chicago and Columbia University), and Bangladesh (Bangladesh Medical Research Council and ICDDR,B). Informed consent was obtained from all participants prior to the baseline interview of the original studies.
Smoking phenotypes
For the purposes of these analyses, we considered four smoking phenotypes: smoking initiation (ever versus never smokers), smoking quantity (cigarettes per day), age of smoking initiation, and smoking cessation (former versus current smokers). Self-reported smoking status was ascertained through the baseline interview administered by a trained study physician using a structured questionnaire [15 17]. See Supplementary Information for additional details regarding the smoking phenotype questionnaire ascertainment. Individuals reported smoking status as never, former, or current cigarette smoker at baseline. In these analyses, we considered two binary phenotypes: smoking initiation (ever versus never smoker) and smoking cessation (current versus former smoker). Since the prevalence of smoking among Bangladeshi women is low due to cultural norms [18], analyses for smoking initiation were conducted separately for males and females. We also considered two continuous phenotypes: self-reported age of smoking initiation and average number of cigarettes per day. These variables were evaluated as continuous phenotypes among ever smokers. The distributions of the smoking behavior phenotypes, stratified by sex, are shown in Table 1.
Table 1.
Characteristics of study sample by sex
| Characteristic | Males (n=2591) N (%) |
Females (n=2763) N (%) |
|---|---|---|
| Baseline age, years | ||
| 18–34 | 659 (25.4) | 1164 (42.1) |
| 35–45 | 898 (34.7) | 973 (35.2) |
| 46–70 | 1034 (39.9) | 626 (22.7) |
| Baseline smoking status | ||
| Never | 754 (29.1) | 2565 (92.8) |
| Former | 366 (14.1) | 87 (3.1) |
| Current | 1471 (56.8) | 111 (4.1) |
| Smoking quantity (among ever smokers) | ||
| Mean CPD ± SD | 12.2 ± 8.3 | 4.4 ± 4.2 |
| 1–5 | 923 (50.3) | 166 (84.3) |
| 6–9 | 223 (12.2) | 19 (9.6) |
| 11–20 | 486 (26.5) | 8 (4.1) |
| 21–30 | 170 (9.3) | 4 (2.0) |
| ≥31 | 32 (1.7) | 0 (0) |
| Age of smoking initiation (among ever smokers) | ||
| Mean age of smoking initiation ± SD | 19.1 ± 6.7 | 21.8 ± 10.3 |
| ≤18 | 1014 (55.3) | 89 (45.2) |
| 19–30 | 731 (39.9) | 74 (37.6) |
| ≥31 | 88 (4.8) | 34 (17.3) |
Abbreviations: cigarettes per day, CPD; standard deviation, SD.
Genotyping
For BEST and HEALS (2006–2008 cohort), DNA extraction was carried out from the whole blood using the QIAamp 96 DNA Blood Kit from Qiagen (Valencia, USA). For HEALS (2000–2002 cohort), DNA was extracted from clot blood using the Flexigene DNA kit from Qiagen (Valencia, USA). Any DNA sample with a concentration <40 ng/µL, and/or 260/280 ratio outside the range of <1.6 to ≥2.1 (measured by Nanodrop 1000), and/or fragmented DNA <2 Kb (assessed by smearing in Agilent BioAnalyzer) was excluded.
Genotyping was performed using the Illumina HumanCytoSNP-12 BeadChip utilizing 250 ng DNA according to the manufacturer’s protocol. There were 5,499 DNA samples genotyped. We excluded samples with very poor call rates (<97%; n=12); individuals with gender mismatches (n=79); and duplicate samples (n=54). No individuals had outlying autosomal heterozygosity or inbreeding values. This QC resulted in 5,354 individuals with high-quality genotype data included in these analyses.
Among 299,140 genotyped SNPs, we implemented the following quality control (QC) exclusion criteria for SNPs using PLINK[19]: (i) SNPs without rs numbers; (ii) SNP call rate <95%; (iii) monomorphic SNPs; (iv) Hardy-Weinberg P<1 × 10−10. This resulted in 257,768 SNPs. Imputation was performed using MaCH on the basis of the HapMap 3 Gujarati Indians in Houston (GIH) population (Build 36). We also implemented the following QC exclusion criteria for SNPs post-imputation: (i) MAF <0.01 and (ii) SNP imputation score <0.3. Both genotyped and imputed SNPs were included in these analyses, which yielded 1,211,988 million SNPs after QC procedures.
Gene expression
RNA was extracted from mononuclear cells preserved in RLT buffer, stored at −80°C, using RNeasy Micro Kit from Qiagen (Valencia, USA). The concentration and quality of RNA was checked on Nanodrop 1000. cRNA synthesis was done from 250 ng of RNA using Illumina TotalPrep 96 RNA Amplification kit. Gene expression was measured using the Illumina HumanHT-12-v4 BeadChip utilizing 750 ng of cRNA according to the manufacturer’s protocol. The chip contains a total of 47,231 probes covering 31,335 genes. We restricted our analyses to specific probes for expression quantitative trait loci (eQTL) analyses, which yielded 31,583 probes. Quantile normalized expression values were log2 transformed and adjusted for batch variability using ComBat software [20]. Gene expression data was available for 1,799 individuals (808 females and 991 males) included in these analyses.
Statistical models
Population structure due to relatedness and population stratification was previously examined in this study sample and has been described [21]; we found very little evidence of population stratification in this sample. Efficient Mixed-Model Association eXpedited (EMMAX) [22] software was used to assess associations for smoking phenotypes using genotyped and imputed SNP data. A linear mixed model regression incorporating the estimated relatedness matrix among individuals for cryptic relatedness (rather than principle components) was used for each SNP, adjusting for sex, age, age×age, and genotyping batch in overall analyses, with SNPs on the X chromosome coded as (0, 1) to indicate the number of minor alleles for males. Sex-specific analyses were adjusted for age, age×age, and genotyping batch. Continuous smoking phenotypes (age of smoking initiation and cigarettes per day) were log transformed to approximate a normal distribution. We considered SNPs to be genome-wide significant if the significance exceeded P <5 × 10−8 [23]. Regional association plots were generated using LocusZoom [24]. The Versatile Gene-based Association Study (VEGAS) approach was used to conduct gene-based tests, by summing the association signal from all the SNPs within a gene and correcting the sum for linkage disequilibrium (LD) using HapMap2 CHB+JPT to generate a test chi-squared statistic [25]. The eQTL analyses were conducted to evaluate associations between the top variant genotypes with gene expression levels genome-wide. Additive linear models for each gene expression probe, stratified by sex and adjusting for age and smoking status, were run using the Matrix eQTL package implemented in R software [26].
RESULTS
The characteristics of the study sample are shown in Table 1. The prevalence of smoking was much higher in males (ever smokers, 70.9%) as compared to females (ever smokers, 7.2%). The average number of cigarettes smoked per day among ever smokers was 11.5 ± 8.3 overall, with males skewed toward a larger quantity smoked compared to females. The average age of smoking initiation among ever smokers was 19.3 ± 7.2 years, with males slightly skewed toward a younger age of smoking initiation compared to females.
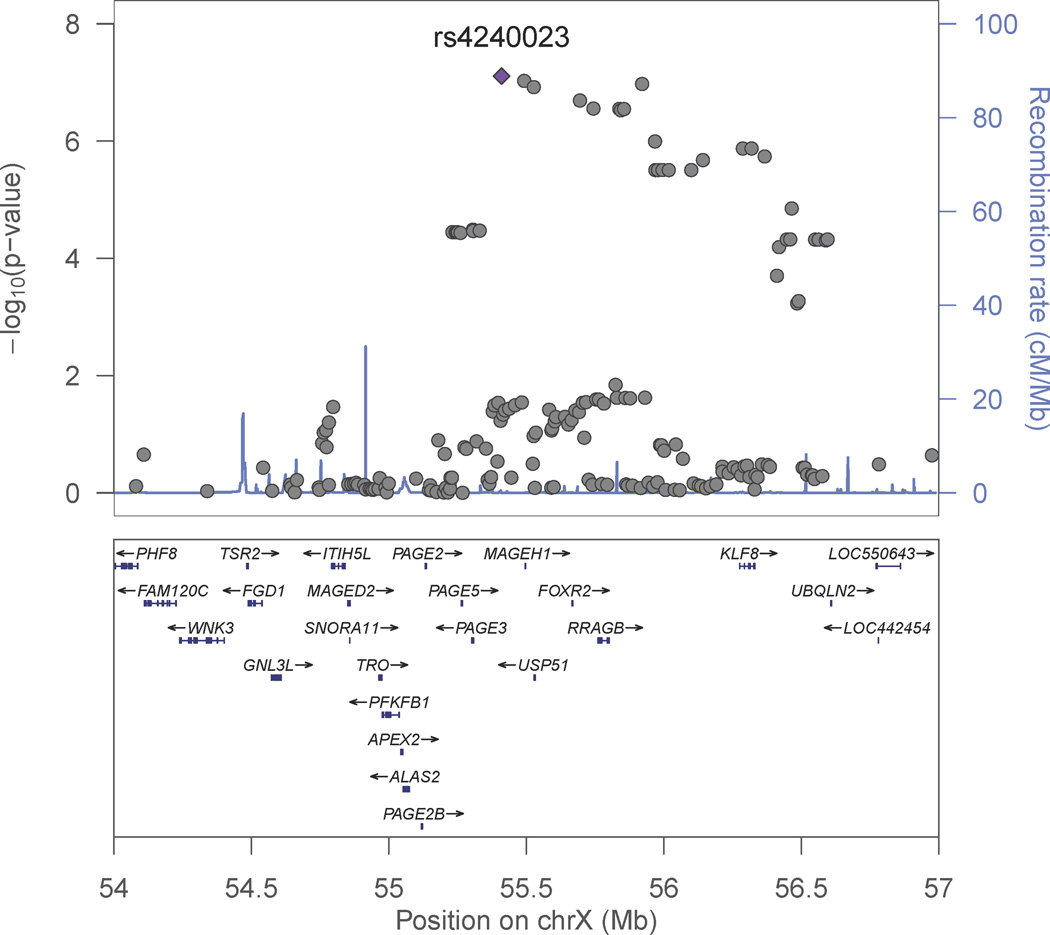
Genome-wide association analyses for smoking initiation (ever versus never) were conducted separately for men and women since the prevalence of smoking was substantially different by sex. This was primarily to address the concern that there was a lower prevalence of smoking among Bangladeshi women due to cultural norms that potentially could mask a genetic effect. The genome-wide association analysis for smoking initiation in 1,837 male ever smokers and 754 male never smokers showed associations of multiple variants with suggestive genome-wide significance (Table 2 and Figure 1A), with the strongest signal for rs2567519 (P = 1.33 × 10−7). Several SNPs on chromosome 17q21.31 were in close proximity, and Figure 2 provides a regional association plot for the top SNPs in the SLC39A11 gene. A gene-based association test for SLC39A11 based on 316 genotyped or imputed SNPs in the gene yielded a p value=3.3×10−4. The overall minor allele frequency of rs2567519 did not statistically differ between males and females (MAF=0.41 versus 0.42). The genome-wide association analysis for smoking initiation in 198 female ever smokers and 2,565 female never smokers also showed associations of multiple variants with suggestive genome-wide significance (Table 2 and Figure 1B), with the strongest signals in Xp11.21 (rs4240023, P = 7.79×10−8) and 15q26 (rs12912184, P = 9.32 × 10−8) in the region between the SLCO3A1 and ST8SIA2 genes. Several SNPs on chromosome 15q26 were in close proximity, and Figure 3A provides a regional association plot for the SNPs. A gene-based association test for SLCO3A1 based on 281 genotyped or imputed SNPs in the gene yielded a p value=0.01; whereas, the gene-based association test for ST8SIA2 based on 147 genotyped or imputed SNPs in the gene yielded a p value=0.67. The overall minor allele frequency of rs12912184 did not statistically differ between females and males (MAF=0.34 versus 0.35). The regional association plot for the signal on the X chromosome is shown in Figure 3B. See Supplementary Information Tables S1 and S2 for a summary of the top 1000 variants in relation to smoking initiation by sex. Analyses were also conducted considering betel quid chewing as part of a broader tobacco use phenotype (ever versus never); however, results were not appreciably different from those observed for tobacco smoking and are not presented here.
Table 2.
Top 10 genetic loci associated with smoking initiation (Ever versus never smoker), stratified by sex
| Nearest gene | Locus | SNP marker | MA | MAF | P value | |
|---|---|---|---|---|---|---|
| Ever smokers | Never smokers | |||||
| Males | ||||||
| SLC39A11 | 17q21.31 | rs2567519 | A | 0.39 | 0.46 | 1.33 × 10−7 |
| SLC39A11 | 17q21.31 | rs2714017 | T | 0.39 | 0.47 | 1.34 × 10−7 |
| MIR1305||TENM3 | 4q35.1 | rs11724903 | G | 0.09 | 0.13 | 2.90 × 10−7 |
| SLC39A11 | 17q21.31 | rs7406107 | G | 0.43 | 0.51 | 4.32 × 10−7 |
| SLC39A11 | 17q21.31 | rs2714016 | A | 0.39 | 0.47 | 5.06 × 10−7 |
| SLC39A11 | 17q21.31 | rs2714018 | A | 0.39 | 0.47 | 7.49 × 10−7 |
| SLC39A11 | 17q21.31 | rs4793310 | C | 0.41 | 0.48 | 7.67 × 10−7 |
| LINC00607 | 2q35 | rs4674024 | A | 0.34 | 0.41 | 1.52 × 10−6 |
| MAT2B||LOC100419716 | 5q33 | rs292404 | G | 0.13 | 0.18 | 1.72 × 10−6 |
| MAT2B||LOC100419716 | 5q33 | rs10057965 | C | 0.21 | 0.26 | 2.74 × 10−6 |
| Females | ||||||
| PAGE3||MAGEH1 | Xp11.21 | rs4240023 | A | 0.07 | 0.02 | 7.79 × 10−8 |
| SLCO3A1||ST8SIA2 | 15q26 | rs12912184 | A | 0.26 | 0.34 | 9.32 × 10−8 |
| MIR4536-1 | Xp11.21 | rs4826404 | G | 0.07 | 0.02 | 9.37 × 10−8 |
| RRAGB||LOC644924 | Xp11.21 | rs4826434 | C | 0.07 | 0.02 | 1.06 × 10−7 |
| USP51 | Xp11.21 | rs2499862 | A | 0.07 | 0.02 | 1.20 × 10−7 |
| XAGE-4 | Xp11.21 | rs4265330 | C | 0.07 | 0.02 | 2.02 × 10−7 |
| SLCO3A1||ST8SIA2 | 15q26 | rs12901615 | A | 0.26 | 0.34 | 2.21 × 10−7 |
| XAGE-4||RRAGB | Xp11.21 | rs5914419 | A | 0.07 | 0.02 | 2.78 × 10−7 |
| RRAGB||LOC644924 | Xp11.21 | rs2375467 | A | 0.07 | 0.02 | 2.81 × 10−7 |
| RRAGB||LOC644924 | Xp11.21 | rs1325570 | A | 0.07 | 0.02 | 2.84 × 10−7 |
Minor allele, MA; minor allele frequency, MAF.
Figure 1. Manhattan and QQ plots for GWAS of smoking initiation by gender.
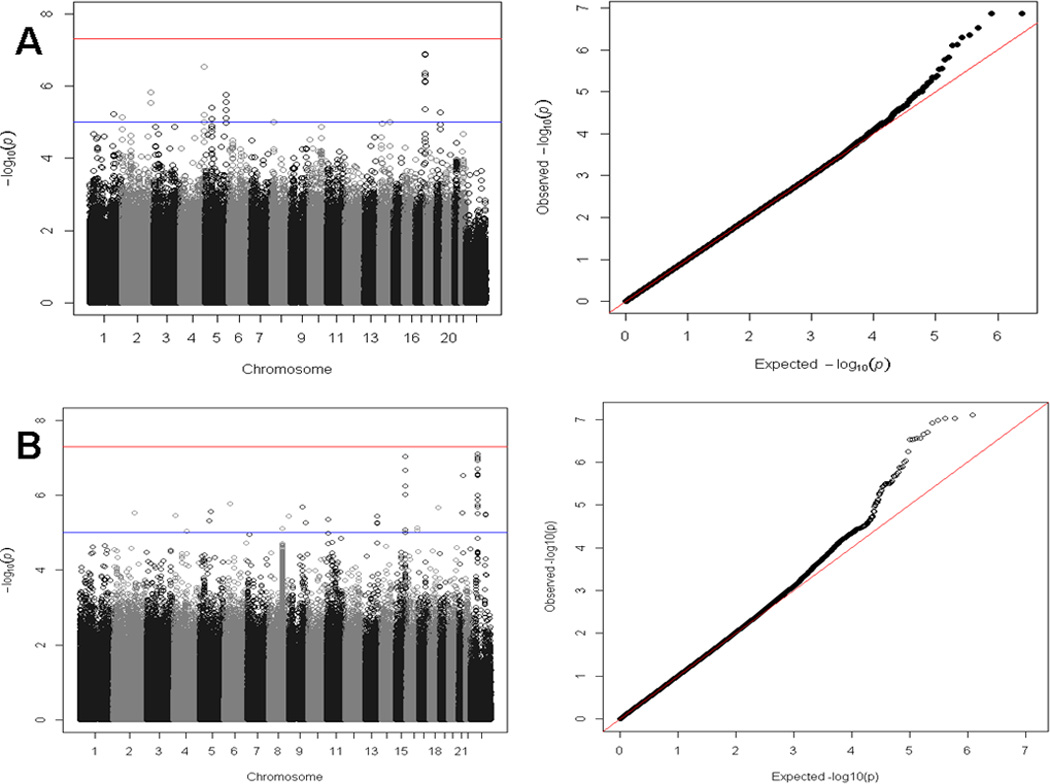
A. Ever versus never smoker among males. Depicts the Manhattan and QQ plots for the association with smoking initiation among males (adjusted for age, age×age, and genotyping batch). The spike on chromosome 17 corresponds to 7 SNPs (rs2567519, rs2714017, rs7406107, rs2714016, rs2714018, rs4793310, and rs2714011) in SLC39A11 with P<1 × 10−5.
B. Ever versus never smoker among females. Depicts the Manhattan and QQ plots for the association with smoking initiation among females (adjusted for age, age×age, and genotyping batch). The spike on chromosome 15 corresponds to 6 SNPs (rs4778038, rs11074052, rs12901615, rs12912184, rs1386775, and rs7171020) near SLCO3A1 with P<1 × 10−5. The spike on the X chromosome corresponds to 20 SNPs (rs4240023, rs4826404, rs2499862, rs4265330, rs5914419, rs2375467, rs2104429, rs1325570, rs4826434, rs4466536, rs4639650, rs17002956, rs7876496, rs5913884, rs2150887, rs12011574, rs12008339, rs5960712, rs6521385, and rs7876062) in the Xp11.21 region with P<1 × 10−5.
Figure 2. Regional association plot for 17q21 SLC39A11 region for smoking initiation among males.
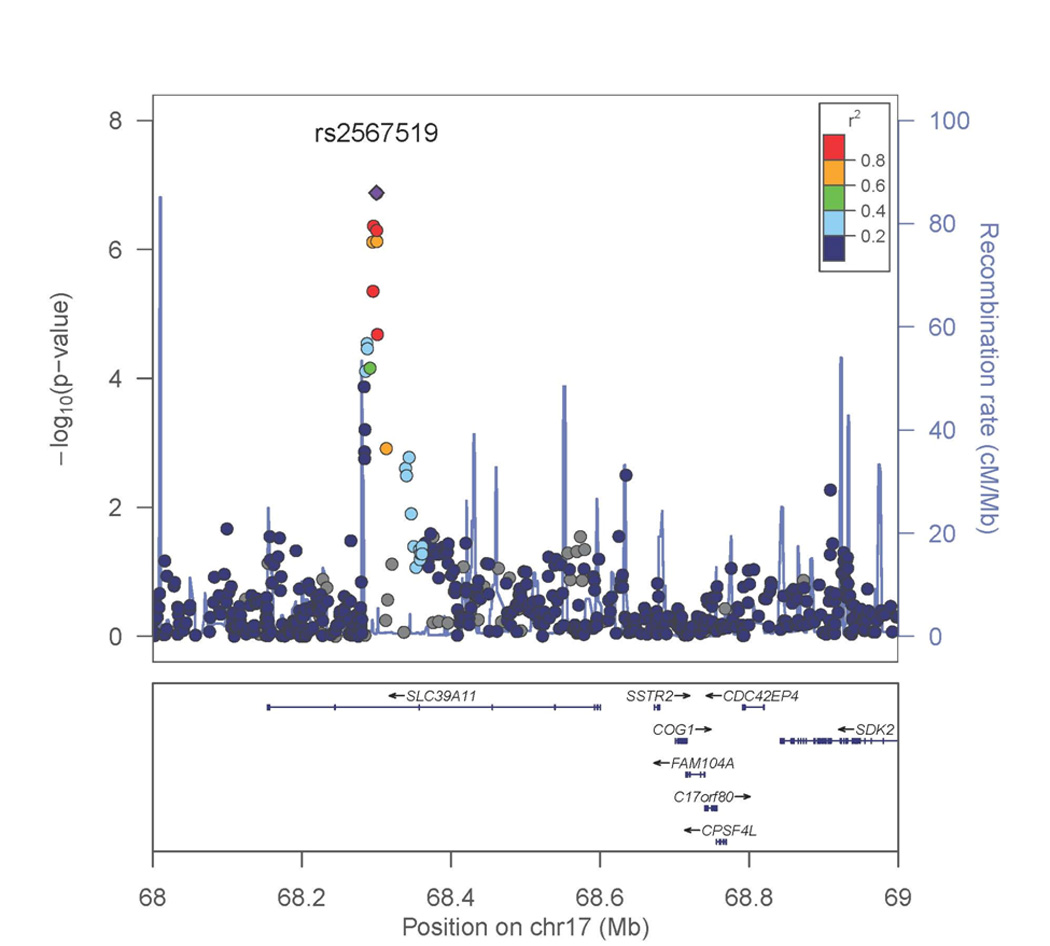
P values were generated using mixed models adjusted for age, age×age, and genotyping batch, accounting for relatedness. The strongest associated SNP is labeled.
Figure 3. Regional association plot for smoking initiation among females.
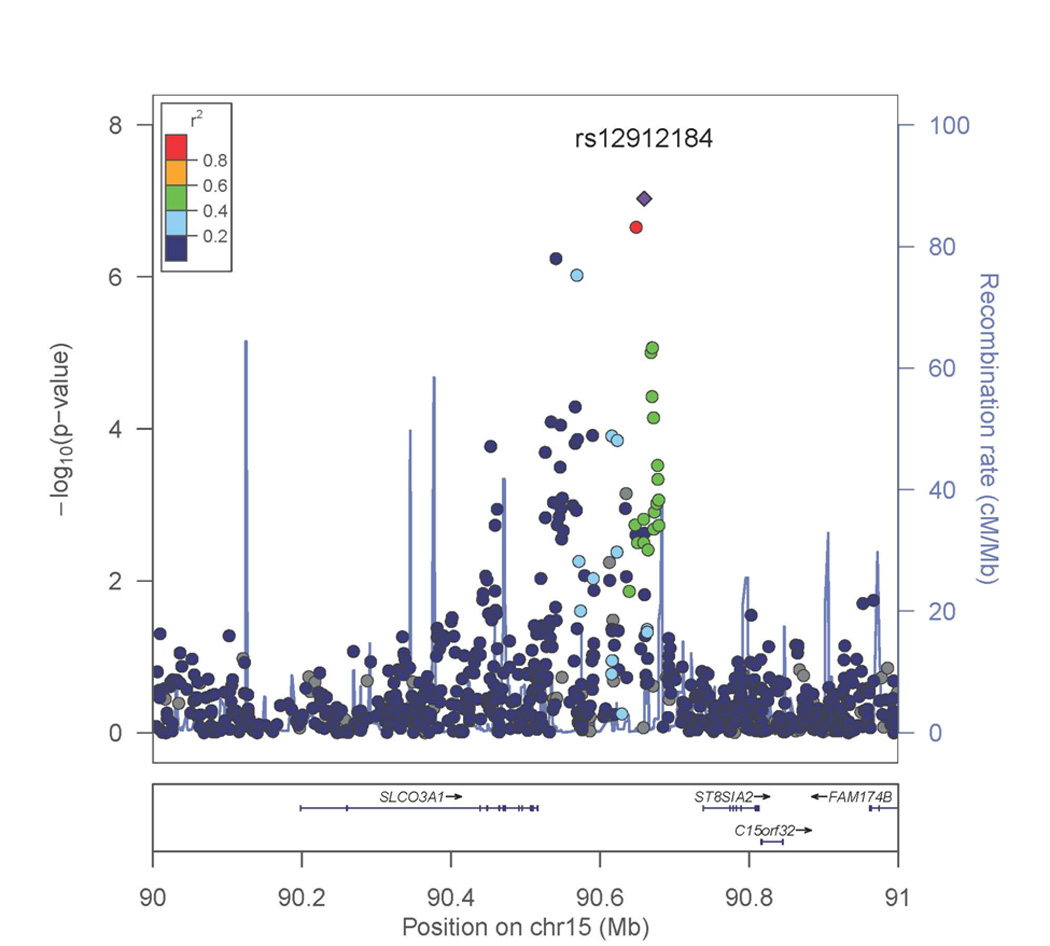
A. 15q26 region. P values were generated using mixed models adjusted for age, age×age, and genotyping batch, accounting for relatedness. The strongest associated SNP is labeled.
B. Xp11.21 region. P values were generated using mixed models adjusted for age, age×age, and genotyping batch, accounting for relatedness. The strongest associated SNP is labeled.
Analyses for smoking quantity, age of smoking initiation, and smoking cessation were conducted among ever smokers, with males and females combined (Supplementary Information, Table S3–S5). No clear genetic signals were observed for these phenotypes based on examination of the distribution of p values in the QQ plots (Supplementary Information, Figure S1A–C). In an effort to replicate findings previously reported in the 15q25.1 region associated with CHRNA5/3 for smoking quantity, we show the regional association plot of our results for this phenotype in Supplementary Information Figure S2A. The strongest signal in the region was observed for rs41280048 (p value=0.0145) upstream of CHRNA5/3 in PSMA4. However, when the study sample was restricted to male ever smokers who smoke at least 10 cigarettes a day (n=688), shown in Supplementary Information Figure S2B, the strongest signal in the region was observed for rs938682 (p value=0.0234) in CHRNA3.
The top association signals for smoking initiation were follow-up in functional analyses of gene expression using an overlapping subset of 1,799 individuals. Genome-wide eQTL results are summarized in Supplementary Information Figure S3 for rs12912184 among females and Supplementary Information Figure S4 for rs2567519 among males. Neither SNP appeared to strongly regulate mRNA levels.
DISCUSSION
In this study, we conducted genome-wide association analyses of approximately 1.2 million SNPs on four smoking behavior phenotypes in an adult Bangladeshi population. We observed evidence of gender-specific associations for transporter genes SLC39A11 (intronic variant rs2567519, P = 1.33 × 10−7) and SLCO3A1 (intergenic variant rs12912184, P = 9.32×10−8) with cigarette smoking initiation. These genes have been previously implicated with smoking initiation in a European ancestry population [8].
SLC39A11 (solute carrier family 39 (metal ion transporter), member 11), also known as ZIP11 (Zrt- and Irt-like protein 11), encodes a protein belonging to the ZIP transporter family and promotes zinc transport from the extracellular fluid or from intracellular vesicles into the cytoplasm [27]. Variants in SLC39A11 have been previously implicated with visceral and subcutaneous fat in women [28] and amyotrophic lateral sclerosis [29]. In a previous GWAS of smoking initiation in a European ancestry sample, SLC39A11 SNP rs17780310 was associated with smoking initiation (p value=2.4×10−3), and in pathway analyses was selected to be a key transporter gene associated with smoking initiation, although associated with a permutation test=0.279 after accounting for gene size [8]. Interestingly, cadmium, a constituent of tobacco smoke, has been shown to interact with zinc in relation to smoking-related outcomes [30–34].
SLCO3A1 (solute carrier organic anion transporter family, member 3A1), encodes a protein belonging to the organic anion transporting polypeptide family and is an uptake transporter that mediates sodium-independent uptake of a broad range of endogenous substances into the cell [35]. SLCO3A1 has been associated with nicotine dependence [36] and with blood pressure through gene×smoking interaction [37]. Furthermore, in a previous GWAS of smoking initiation in a European ancestry sample, SLCO3A1 SNP rs2677911 was associated with smoking initiation (p value=1.2×10−3), and in pathway analyses was selected to be a key transporter gene associated with smoking initiation, associated with a permutation test=0.066 after accounting for gene size [8].
Based on previous studies, the effect of 15q25.1 on smoking quantity is very small, accounting for 0.20–5% of the phenotypic variance of the cigarette per day phenotype [13 38–41]. Therefore, we may not have been able to detect a signal in this region due to limited power and/or due to the fact that economic factors may be more important determinants of smoking quantity for our study population. Furthermore, a weak signal in CHRNA3 was observed only when analyses were restricted to male ever smokers who smoked at least 10 cigarettes per day. Our interpretation of this is that smoking quantity in this population is quite heterogeneous, and a genetic effect may only be apparent among heavier smokers.
We acknowledge limitations of our analyses, including limited statistical power. While we identified interesting genes related to smoking initiation, no loci reached genome-wide statistical significance. We had 80% power to detect effect sizes of 1.58 for smoking initiation among males, 2.08 for smoking initiation among females, 1.73 for smoking cessation, and a β of 2.04 for smoking quantity and 1.77 for age of smoking initiation assuming a mean allele frequency of 0.2 and α of 5 × 10−8. It is possible that additional genetic effects could be revealed with a larger sample size. While self-reported smoking status has not been validated against measures such as cotinine, we have observed dose-response associations of smoking status, smoking quantity, and age of initiation in relation to mortality in this study population [42]. Therefore, we deem to have an adequate assessment of smoking phenotypes in this population based on observed associations with smoking-related endpoints. However, a strength of this study was that performing gender-specific analyses for smoking initiation enabled us to uncover new loci in genes previously implicated with smoking initiation than may potentially be related to smoking initiation in a Southeast Asian population and should be explored in future association and functional studies.
In summary, findings from this GWAS among Bangladeshi adults suggest a role for SLC39A11 and SLCO3A1 in smoking initiation in a gender-specific manner. Future studies are needed to replicate and unravel the biological mechanisms that may underlie these associations. Insights into these pathways may provide new targets for smoking cessation therapies to reduce the public health burden associated with tobacco smoking.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers P42 ES010349 and R01 CA107431).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
REFERENCES
- 1.Gandini S, Botteri E, Iodice S, Boniol M, Lowenfels AB, Maisonneuve P, Boyle P. Tobacco smoking and cancer: a meta-analysis. International journal of cancer. 2008;122(1):155–164. doi: 10.1002/ijc.23033. [DOI] [PubMed] [Google Scholar]
- 2.Cohen N. Smoking, health, and survival: prospects in Bangladesh. Lancet. 1981;1(8229):1090–1093. doi: 10.1016/s0140-6736(81)92251-0. [DOI] [PubMed] [Google Scholar]
- 3.Efroymson D, Ahmed S, Townsend J, Alam SM, Dey AR, Saha R, Dhar B, Sujon AI, Ahmed KU, Rahman O. Hungry for tobacco: an analysis of the economic impact of tobacco consumption on the poor in Bangladesh. Tobacco control. 2001;10(3):212–217. doi: 10.1136/tc.10.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li MD. The genetics of smoking related behavior: a brief review. The American journal of the medical sciences. 2003;326(4):168–173. doi: 10.1097/00000441-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Munafo MR, Johnstone EC. Genes and cigarette smoking. Addiction (Abingdon, England) 2008;103(6):893–904. doi: 10.1111/j.1360-0443.2007.02071.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu YZ, Pei YF, Guo YF, Wang L, Liu XG, Yan H, Xiong DH, Zhang YP, Levy S, Li J, Haddock CK, Papasian CJ, Xu Q, Ma JZ, Payne TJ, Recker RR, Li MD, Deng HW. Genome-wide association analyses suggested a novel mechanism for smoking behavior regulated by IL15. Molecular psychiatry. 2009;14(7):668–680. doi: 10.1038/mp.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, Ziegler RG, Hunter DJ, Chanock S, Hankinson S, Kraft P, Bergen AW. Genome-wide and candidate gene association study of cigarette smoking behaviors. PloS one. 2009;4(2):e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, Smit JH, Hoogendijk WJ, Zitman FG, Peltonen L, Kaprio J, Pedersen NL, Magnusson PK, Spector TD, Kyvik KO, Morley KI, Heath AC, Martin NG, Westendorp RG, Slagboom PE, Tiemeier H, Hofman A, Uitterlinden AG, Aulchenko YS, Amin N, van Duijn C, Penninx BW, Boomsma DI. Genome-wide association study of smoking initiation and current smoking. American journal of human genetics. 2009;84(3):367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature genetics. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schafer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Volzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Mooser V, Francks C, Marchini J. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature genetics. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon D, Kim YJ, Cui WY, Van der Vaart A, Cho YS, Lee JY, Ma JZ, Payne TJ, Li MD, Park T. Large-scale genome-wide association study of Asian population reveals genetic factors in FRMD4A and other loci influencing smoking initiation and nicotine dependence. Human genetics. 2012;131(6):1009–1021. doi: 10.1007/s00439-011-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, Brown WM, Petruzella S, Thacker EL, Kim Y, Nalls MA, Tranah GJ, Sung YJ, Ambrosone CB, Arnett D, Bandera EV, Becker DM, Becker L, Berndt SI, Bernstein L, Blot WJ, Broeckel U, Buxbaum SG, Caporaso N, Casey G, Chanock SJ, Deming SL, Diver WR, Eaton CB, Evans DS, Evans MK, Fornage M, Franceschini N, Harris TB, Henderson BE, Hernandez DG, Hitsman B, Hu JJ, Hunt SC, Ingles SA, John EM, Kittles R, Kolb S, Kolonel LN, Le Marchand L, Liu Y, Lohman KK, McKnight B, Millikan RC, Murphy A, Neslund-Dudas C, Nyante S, Press M, Psaty BM, Rao DC, Redline S, Rodriguez-Gil JL, Rybicki BA, Signorello LB, Singleton AB, Smoller J, Snively B, Spring B, Stanford JL, Strom SS, Swan GE, Taylor KD, Thun MJ, Wilson AF, Witte JS, Yamamura Y, Yanek LR, Yu K, Zheng W, Ziegler RG, Zonderman AB, Jorgenson E, Haiman CA, Furberg H. Genome-wide meta-analyses of smoking behaviors in African Americans. Translational psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumasaka N, Aoki M, Okada Y, Takahashi A, Ozaki K, Mushiroda T, Hirota T, Tamari M, Tanaka T, Nakamura Y, Kamatani N, Kubo M. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PloS one. 2012;7(9):e44507. doi: 10.1371/journal.pone.0044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahsan H, Chen Y, Parvez F, Argos M, Hussain AI, Momotaj H, Levy D, van Geen A, Howe G, Graziano J. Health Effects of Arsenic Longitudinal Study (HEALS): description of a multidisciplinary epidemiologic investigation. Journal of exposure science & environmental epidemiology. 2006;16(2):191–205. doi: 10.1038/sj.jea.7500449. [DOI] [PubMed] [Google Scholar]
- 16.Argos M, Rahman M, Parvez F, Dignam J, Islam T, Quasem I, S KH, A TH, Hossain Z, T IP, Rakibuz-Zaman M, Sarwar G, La Porte P, Harjes J, Anton K, Kibriya MG, Jasmine F, Khan R, Kamal M, Shea CR, Yunus M, Baron JA, Ahsan H. Baseline comorbidities in a skin cancer prevention trial in Bangladesh. Eur J Clin Invest. 2013;43(6):579–588. doi: 10.1111/eci.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahsan H, Chen Y, Parvez F, Zablotska L, Argos M, Hussain I, Momotaj H, Levy D, Cheng Z, Slavkovich V, van Geen A, Howe GR, Graziano JH. Arsenic exposure from drinking water and risk of premalignant skin lesions in Bangladesh: baseline results from the Health Effects of Arsenic Longitudinal Study. Am J Epidemiol. 2006;163(12):1138–1148. doi: 10.1093/aje/kwj154. [DOI] [PubMed] [Google Scholar]
- 18.Bush J, White M, Kai J, Rankin J, Bhopal R. Understanding influences on smoking in Bangladeshi and Pakistani adults: community based, qualitative study. BMJ (Clinical research ed. 2003;326(7396):962. doi: 10.1136/bmj.326.7396.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 21.Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, Paul-Brutus R, Rahaman R, Rakibuz-Zaman M, Parvez F, Ahmed A, Quasem I, Hore SK, Alam S, Islam T, Slavkovich V, Gamble MV, Yunus M, Rahman M, Baron JA, Graziano JH, Ahsan H. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS genetics. 2012;8(2):e1002522. doi: 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature genetics. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genetic epidemiology. 2008;32(2):179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 24.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics (Oxford, England) 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S. A versatile gene-based test for genome-wide association studies. American journal of human genetics. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics (Oxford, England) 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. The Journal of biological chemistry. 2006;281(34):24085–24089. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- 28.Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS genetics. 2012;8(5):e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landers JE, Melki J, Meininger V, Glass JD, van den Berg LH, van Es MA, Sapp PC, van Vught PW, McKenna-Yasek DM, Blauw HM, Cho TJ, Polak M, Shi L, Wills AM, Broom WJ, Ticozzi N, Silani V, Ozoguz A, Rodriguez-Leyva I, Veldink JH, Ivinson AJ, Saris CG, Hosler BA, Barnes-Nessa A, Couture N, Wokke JH, Kwiatkowski TJ, Jr, Ophoff RA, Cronin S, Hardiman O, Diekstra FP, Leigh PN, Shaw CE, Simpson CL, Hansen VK, Powell JF, Corcia P, Salachas F, Heath S, Galan P, Georges F, Horvitz HR, Lathrop M, Purcell S, Al-Chalabi A, Brown RH., Jr Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(22):9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazi TG, Wadhwa SK, Afridi HI, Kazi N, Kandhro GA, Baig JA, Shah AQ, Kolachi NF, Khan S. Evaluation of cadmium and zinc in biological samples of tobacco and alcohol user male mouth cancer patients. Human & experimental toxicology. 2010;29(3):221–230. doi: 10.1177/0960327109360045. [DOI] [PubMed] [Google Scholar]
- 31.Kazi TG, Wadhwa SK, Afridi HI, Kazi N, Kandhro GA, Baig JA, Shah AQ, Kolachi NF, Arain MB. Interaction of cadmium and zinc in biological samples of smokers and chewing tobacco female mouth cancer patients. Journal of hazardous materials. 2010;176(1–3):985–991. doi: 10.1016/j.jhazmat.2009.11.139. [DOI] [PubMed] [Google Scholar]
- 32.Kazi TG, Memon AR, Afridi HI, Jamali MK, Arain MB, Jalbani N, Sarfraz RA. Determination of cadmium in whole blood and scalp hair samples of Pakistani male lung cancer patients by electrothermal atomic absorption spectrometer. The Science of the total environment. 2008;389(2–3):270–276. doi: 10.1016/j.scitotenv.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 33.Napolitano JR, Liu MJ, Bao S, Crawford M, Nana-Sinkam P, Cormet-Boyaka E, Knoell DL. Cadmium-mediated toxicity of lung epithelia is enhanced through NF-kappaB-mediated transcriptional activation of the human zinc transporter ZIP8. American journal of physiology. 2012;302(9):L909–L918. doi: 10.1152/ajplung.00351.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anetor JI, Ajose F, Anetor GO, Iyanda AA, Babalola OO, Adeniyi FA. High cadmium / zinc ratio in cigarette smokers: potential implications as a biomarker of risk of prostate cancer. Niger J Physiol Sci. 2008;23(1–2):41–49. doi: 10.4314/njps.v23i1-2.54921. [DOI] [PubMed] [Google Scholar]
- 35.Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica; the fate of foreign compounds in biological systems. 2008;38(7–8):778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 36.Wang KS, Liu X, Zhang Q, Zeng M. ANAPC1 and SLCO3A1 are associated with nicotine dependence: meta-analysis of genome-wide association studies. Drug and alcohol dependence. 2012;124(3):325–332. doi: 10.1016/j.drugalcdep.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Montasser ME, Shimmin LC, Hanis CL, Boerwinkle E, Hixson JE. Gene by smoking interaction in hypertension: identification of a major quantitative trait locus on chromosome 15q for systolic blood pressure in Mexican-Americans. Journal of hypertension. 2009;27(3):491–501. doi: 10.1097/hjh.0b013e32831ef54f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, Pitkaniemi J, Peltonen L, Aromaa A, Kaprio J. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munafo MR, Timofeeva MN, Morris RW, Prieto-Merino D, Sattar N, Brennan P, Johnstone EC, Relton C, Johnson PC, Walther D, Whincup PH, Casas JP, Uhl GR, Vineis P, Padmanabhan S, Jefferis BJ, Amuzu A, Riboli E, Upton MN, Aveyard P, Ebrahim S, Hingorani AD, Watt G, Palmer TM, Timpson NJ, Group ES, Davey Smith G. Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure. Journal of the National Cancer Institute. 2012;104(10):740–748. doi: 10.1093/jnci/djs191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallfors J, Loukola A, Pitkaniemi J, Broms U, Mannisto S, Salomaa V, Heliovaara M, Lehtimaki T, Raitakari O, Madden PA, Heath AC, Montgomery GW, Martin NG, Korhonen T, Kaprio J. Scrutiny of the CHRNA5-CHRNA3-CHRNB4 smoking behavior locus reveals a novel association with alcohol use in a Finnish population based study. International journal of molecular epidemiology and genetics. 2013;4(2):109–119. [PMC free article] [PubMed] [Google Scholar]
- 41.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu F, Chen Y, Parvez F, Segers S, Argos M, Islam T, Ahmed A, Rakibuz-Zaman M, Hasan R, Sarwar G, Ahsan H. A prospective study of tobacco smoking and mortality in Bangladesh. PloS one. 2013;8(3):e58516. doi: 10.1371/journal.pone.0058516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.