Abstract
Purpose:
To evaluate the epigenetic risk linked to assisted reproductive technology (ART) at single-embryo level by analyzing the methylation status of imprinted H19, PEG1, and KvDMR1 in human preimplantation embryos.
Methods:
A total of 254 human day 3 embryos produced by ART procedures were collected. All embryos were normally fertilized but unsuitable for transfer or freezing due to poor quality. Pyrosequencing with confirmatory routine bisulfite sequencing were used to determine the DNA methylation patterns of H19 differentially methylated region (DMR) in 63 embryos, PEG1 DMR in 65 embryos, and KvDMR1 in 67 embryos.
Results:
Aberrant methylation patterns were found in 8.0% embryos at H19 DMR, 16.9% embryos at PEG1 DMR, and 10.4% embryos at KvDMR1. No methylation errors were found in corresponding sperm samples.
Conclusions:
We hypothesized that the use of poor-quality embryos may increase the risk of imprinting defects because they might have methylation errors.
Keywords: methylation, human embryo, H19, PEG1, KvDMR1
Introduction
In recent years, multiple case series and reports have been published on assisted reproductive technology (ART)-conceived children with imprinting disorders.1,2 The concerns about the possibility of epigenetic changes resulting from ART are therefore increasing. The identification of epigenetic changes at imprinted loci of ART infants has led to the suggestion that the technique itself may predispose embryos to acquire imprinting errors.3–5 The most major epigenetic mechanism of genetic imprinting is DNA methylation that regulates the expression of imprinted genes according to parental origin within discrete locations known as differentially methylated regions (DMRs).6 The H19 DMR, PEG1 DMR, and KvDMR1 are well-studied imprinted loci because their aberrant methylation may contribute to Silver-Russell syndrome (RSS)7–9 and Beckwith-Weidemann syndrome (BWS)10–12. In humans, H19 is paternally imprinted, whereas PEG1 and KvDMR1 are maternally imprinted. Previous studies, especially animal studies, revealed that some procedures used in the ART process could subject gametes and early embryos to environmental stress, resulting in the alteration of DNA methylation. For instance, in vitro maturation (IVM) and superovulation using high-dose hormone could cause methylation errors in oocytes13–15; in vitro culture may make imprinting of embryos aberrantly altered.16–18 On the other hand, sperm cells from men with severe oligozoospermia or azoospermia were found with abnormal methylation profiles, suggesting that infertility itself may be a risk factor for imprinting diseases.19–21 After fertilization, early embryos undergo a genome-wide demethylation process, but this process does not affect the differential methylation marks of imprinted genes, which were previously set in gametes. Thus, aberrant methylation in embryos could result from in vitro culture condition or as a result of epigenetic inheritance from gametes. Currently, the studies regarding human embryo methylation were fewer due to ethical consideration, and the related knowledge was mainly inferred from the hypothesis of animal studies; therefore, more human studies are needed.
In this study, we collected 254 human preimplantation embryos produced during ART procedures. We determined the methylation status of H19 DMR, PEG1 DMR, and KvDMR1 at the single-embryo level using pyrosequencing and bisulfite sequencing technique, trying to offer preliminary data to evaluate epigenetic risks linked to ART.
Materials and Methods
The study protocol was approved by the institutional ethical committee. After obtaining the informed consent from all patients, the embryos were donated for research. In our ART procedure, the gametes and embryos were manipulated using Quinn Advantage HEPES Medium, HTF Medium, and Cleavage Medium (Sage, Pasadena, California). The culture conditions were 37°C, 5% CO2, 5% O2, and 90% N2. A total of 254 day 3 embryos that were normally fertilized (2 pronuclei) were collected for the study, and all embryos were unsuitable for transfer or freezing due to poor quality. The number of embryo tested for the 3 DMRs is 80 embryos for H19 DMR, 84 embryos for PEG1 DMR, and 90 embryos for KvDMR1. The genomic DNA of single embryo was bisulfite converted using our previous protocol,22 and the last washing droplet of each embryo served as a blank.
A seminested polymerase chain reaction (PCR) was used to amplify the bisulfite-converted DNA of single embryo. To confirm that the PCR amplification had no bias, the adult leukocytes were used as controls. The bisulfite PCR, pyrosequencing, and bisulfite sequencing protocol were according to our description.3
The statistical analysis program SPSS version 13.0 was used to analyze the methylation values. The percentages of methylation were calculated as mean ± standard deviation (SD). Box plots were generated using default parameters. The bottom and the top of the box indicate the 25th and 75th percentile, respectively. The T bars extend from the boxes to at most 1.5 times the height of the box. Outliers are values that do not lie within these T bars; extreme outliers have values more extreme than 3 times the box length away from the median. The normal range of DNA methylation was determined by calculating the mean ± standard deviation (SD) of the data without the outliers or extreme outliers
Result
The bisulfite PCR amplification efficiency at single-embryo level for H19, PEG1, and KvDMR1 were 78.8% (63 of 80 embryos were successfully amplified), 77.4% (65 of 84 embryos were successfully amplified), and 74.4% (67 of 90 embryos were successfully amplified), respectively. A total of 254 negative PCR controls were tested, and none produced amplification products, indicating no PCR contamination.
For leukocytes, methylation patterns for the 3 imprinted loci demonstrated the differential methylation patterns (about half methylated and half unmethylated), indicating no bias in bisulfite PCR.
The methylation values were processed using box plots analysis (see Figure 1). For H19 DMR, 5 (8.0%) embryos showed abnormal methylation (3 embryos with demethylation profiles and 2 embryos with hypomethylation profiles); 60 embryos showed the differential methylation pattern (mean methylation: 42.7% ± 7.1%). For PEG1 DMR, 11 (16.9%) embryos exhibited aberrant methylation (7 embryos with demethylation patterns, 3 embryos with hypomethylation patterns, and 1 embryo with hypermethylation pattern). For KvDMR1, 58 embryos showed the differential methylation pattern (mean methylation: 48.6% ± 6.4%), 7 (10.4%) embryos demonstrated abnormal methylation (2 embryos with demethylation profiles, 1 embryo with hypomethylation profile, and 4 embryos with hypermethylation profiles); 62 embryos showed the differential methylation pattern (mean methylation: 38.7% ± 2.9%). These results were confirmed by the subsequent bisulfite sequencing (some results are shown in Figure 2). Table 1 summarized the results of methylation analysis of embryos with methylation errors. Comparing the rate of methylation errors of the 3 DMRs (H19: 8.0%, PEG1: 16.9%, and KvDMR1: 10.4%), no significant difference was found among the 3 DMRs groups. We also summarized and compared the day 3 embryo quality between abnormal methylation group and normal methylation group, and no significant differences were found (see Table 2).
Figure 1.
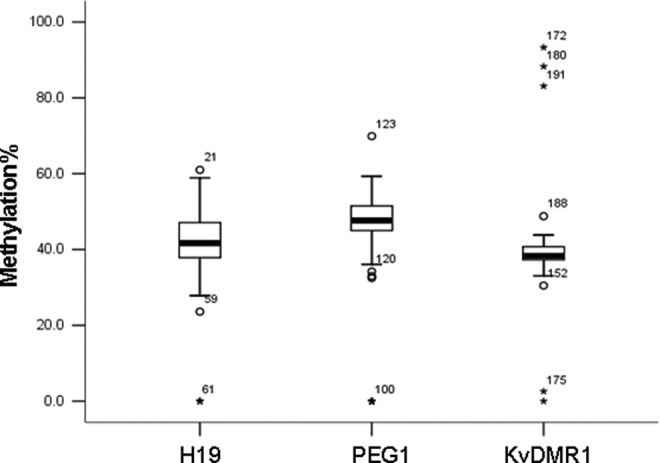
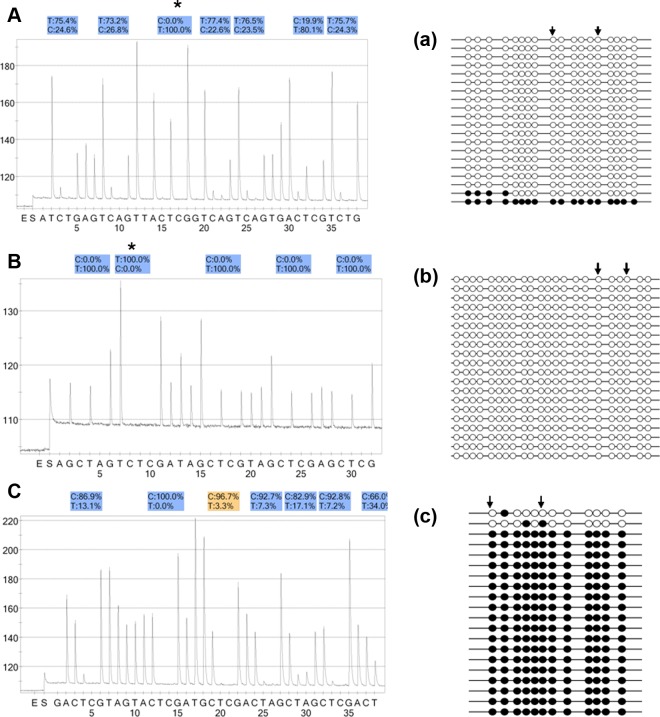
Box plot analysis showed the distribution of methylation values of 3 differentially methylated regions (DMRs) in human embryo samples. Bottom of the box indicates the 25th percentile and the top indicates the 75th percentile. Outliers, which were considered as aberrant methylation values, are shown as asterisk (for H19 DMR: embryo No. 47, 52, 59, 60, 61; for PEG1 DMR: embryo No. 64, 100, 104, 119, 120, 122, 123, 124, 126, 127, 128; for KvDMR1: embryo No. 152, 172, 175, 180, 188, 191, 194).
Figure 2.
A-C, The aberrant methylation patterns of 3 imprinted loci in human preimplantation embryos. The CpG sites were measured using pyrosequencing, and the “methylation%” was the average of “C%” of each CpG site measured. The “T%” of sites marked with an asterisk (*) represent the bisulfite conversion efficiency. a∼c, Consequent bisulfite sequencing technique confirmed the pyrosequencing results. The open and closed circles represent methylated and unmethylated CpGs, respectively. The regions between the 2 arrows indicate the CpG sites analyzed by pyrosequencing. The percentage of methylation was calculated as the percentage of methylated CpG sites. A, H19 DMR (7 CpG sites): the abnormal hypomethylation profile in No.59 embryo (23.6% methylation). a, The result of bisulfite sequencing was 6.1% methylation; (B) PEG1 differentially methylated region (DMR; 5 CpG sites): the demethylation pattern in No.100 embryo (0% methylation). b, The result of bisulfite sequencing was 0% methylation; (C) KvDMR1: (7 CpG sites): the abnormal hypermethylation pattern in No.180 embryo (88.3% methylation). c, The result of bisulfite sequencing was 91.3% methylation.
Table 1.
Results of Methylation Analysis of the 3 DMRs in Human Preimplantation Embryos Showing Aberrant Impinting.
| Case | Age | Infertile Factor | Stimulation Protocol | Insemination Mode | Embryos no. | Imprinted Loci | Day2 Quality | Day3 quality | Embryo Methylationa (%) | Sperm Methylationb (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cells | Gradeb | Cells | Gradec | |||||||||
| 12 | 33 | Tube factor | Long agonist | IVF | 47 | H19 | Uncleaved | – | 2 | 2 | 0 (0) | 94.9 |
| 18 | 31 | Tube factor | Long agonist | IVF | 52 | H19 | 2 | 1 | 3 | 1 | 0 | 95.6 |
| 19 | 29 | Tube factor | Long agonist | IVF | 59 | H19 | 4 | 3 | 6 | 4 | 23.6 (6.1) | 94.2 |
| 23 | 36 | Endometriosis | Long agonist | IVF | 60 | H19 | 3 | 2 | 5 | 4 | 27.8 | 94.5 |
| 23 | 36 | Endometriosis | Long agonist | IVF | 61 | H19 | 2 | 2 | 2 | 3 | 0 | 94.5 |
| 25 | 37 | Tube factor | Long agonist | IVF | 64 | PEG1 | 5 | 3 | 9 | 4 | 33.0 | |
| 51 | 28 | Oligospermia | Long agonist | ICSI | 100 | PEG1 | Uncleaved | — | 5 | 3 | 0 (0) | |
| 58 | 38 | Tube factor | antagonist | IVF | 104 | PEG1 | 4 | 1 | 4 | 1 | 0 | |
| 63 | 34 | Tube factor | Long agonist | IVF | 119 | PEG1 | 3 | 3 | 5 | 3 | 32.5 (21.6) | |
| 63 | 34 | Tube factor | Long agonist | IVF | 120 | PEG1 | 2 | 1 | 3 | 2 | 34.2 | |
| 79 | 25 | Azoospermia | Long agonist | ICSI | 122 | PEG1 | 4 | 2 | 4 | 3 | 0 | |
| 85 | 30 | Tube factor | Long agonist | IVF | 123 | PEG1 | 5 | 3 | 7 | 4 | 69.9 (75.0) | 0 |
| 85 | 30 | Tube factor | Long agonist | IVF | 124 | PEG1 | Uncleaved | — | 5 | 2 | 0 | |
| 90 | 32 | Tube factor | Long agonist | IVF | 126 | PEG1 | 2 | 1 | 4 | 2 | 0 | |
| 92 | 31 | Tube factor | Long agonist | IVF | 127 | PEG1 | 2 | 3 | 3 | 3 | 0 | |
| 95 | 30 | Oligospermia | Long agonist | ICSI | 128 | PEG1 | 4 | 2 | 4 | 2 | 0 | |
| 98 | 33 | Endometriosis | Antagonist | IVF | 152 | KvDMR1 | 4 | 3 | 6 | 3 | 30.5 | |
| 102 | 29 | Tube factor | Long agonist | IVF | 172 | KvDMR1 | 3 | 2 | 4 | 2 | 93.3 | 5.4 |
| 105 | 35 | Tube factor | Long agonist | IVF | 175 | KvDMR1 | Uncleaved | — | 3 | 1 | 0 | |
| 110 | 36 | Tube factor | Long agonist | IVF | 180 | KvDMR1 | 2 | 1 | 4 | 2 | 88.3 (91.3) | 4.5 |
| 112 | 30 | Tube factor | Long agonist | IVF | 188 | KvDMR1 | 5 | 2 | 6 | 4 | 48.8 | 0 |
| 112 | 34 | Tube factor | Long agonist | IVF | 191 | KvDMR1 | 4 | 1 | 4 | 1 | 83.1 | 0 |
| 116 | 36 | Tube factor | Long agonist | IVF | 194 | KvDMR1 | 4 | 2 | 5 | 2 | 2.8 (2.9) | |
Abbreviations: DMR, differentially methylated regions; ICSI, intracytoplasmic sperm injection; IVF, in vitro fertilization;
aEmbryo methylation: each value is the percentage of methylation determined by pyrosequencing. The results of routine bisulfite sequencing are given in brackets.
b Sperm methylation: DNA methylation patterns in the sperm samples used to fertilize the corresponding embryos. The values were determined by pyrosequencing.
c Embryo grading: grade 1: embryo with blastomeres of equal size, no cytoplasmic fragments; grade 2: embryo with blastomeres of equal size, fragments less than 20%; grade 3: embryo with blastomeres of distinctly unequal size, or fragments between 20%-50%; and grade 4: fragments above 50%.
Table 2.
Comparison of Embryo Quality Between Abnormal Methylation and Normal Methylation.a
| Day 3 Embryo Quality | Abnormal Methylation | Normal Methylation |
|---|---|---|
| Cell number | 4.5 ± 1.6 | 5.0 ± 2.4b |
| Percentage of grade (%) | ||
| Grade 1 | 17.4 (4/23) | 25.6 (44/172)b |
| Grade 2 | 34.8 (8/23) | 20.3 (35/172)b |
| Grade 3 | 26.1 (6/23) | 37.8 (65/172)b |
| Grade 4 | 21.7 (5/23) | 16.3 (28/172)b |
| Arrested embryos (%)c | 21.7 (5/23) | 14.5 (25/172)b |
Abbreviation: DMR, differentially methylated regions
aAbnormal methylation of the 3 DMRs were found in 23 embryos, while normal methylation in 172 embryos.
bNo significant difference (P > .05; chi-square test was used).
cThe embryos stopped developing from day 2 to day 3.
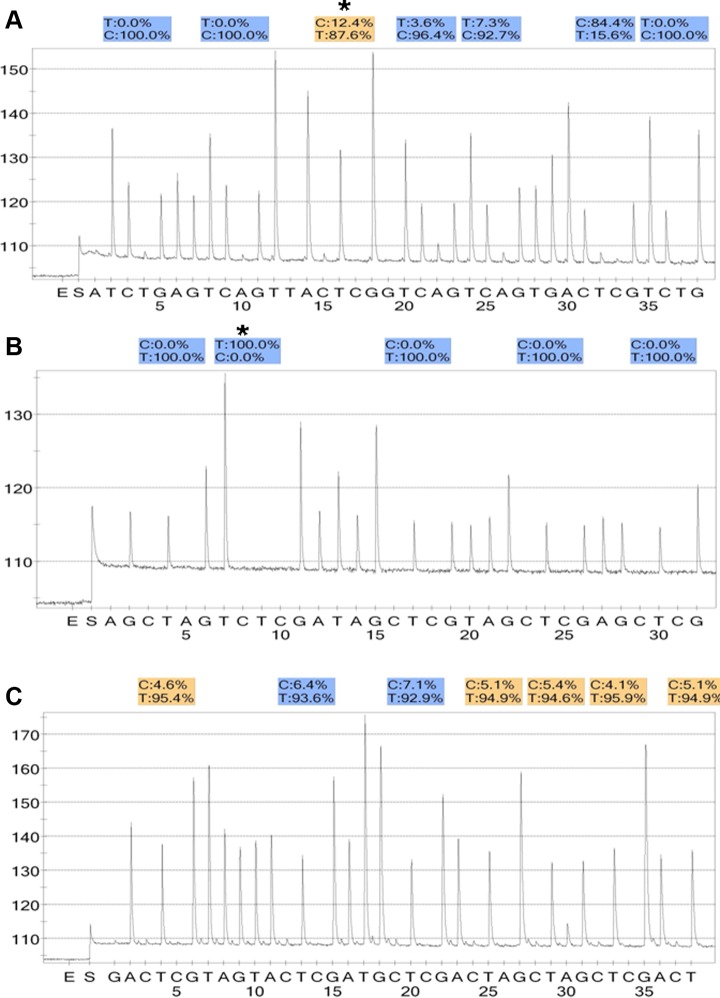
According to imprinting reprogramming mechanism, in the embryos, demethylation or hypomethylation of H19 DMR and hypermethylation of PEG1 DMR/KvDMR1 may be consequent to methylation errors in spermatozoa. We next examined DNA methylation patterns in the sperm samples used to fertilize the 10 embryos (5 embryos with demethylation or hypomethylation of H19 DMR; 1 embryo with hypermethylatikon of PEG1 DMR; 4 embryos with hypermethylation of KvDMR1). All 10 sperm samples demonstrated normal methylation patterns at the corresponding imprinted loci tested (see Figure 3).
Figure 3.
The pyrosequencing results of some sperm samples used to fertilize the corresponding embryos. A, H19 DMR (7 CpG sites): the normal hypermethylation profile (94.2%) in the sperm sample (corresponding embryo: No.60); (B) PEG1 DMR (5 CpG sites): the normal hypomethylation pattern (0%) in the sperm sample (corresponding embryo: No.123); (C) KvDMR1: (7 CpG sites): the normal hypomethylation pattern (5.4%) in the sperm sample (corresponding embryo: No.172).
Discussion
There is a growing literature evaluating the epigenetic aspects of ART, and most experiments address the effects of ovulation induction and preimplantation embryo culture. At the level of genomic CpG methylation, ART-induced epigenetic defects are convincingly observed in mouse studies. In humans, the related studies are relatively limited for ethical reasons.
After gamete fusion, embryos undergo a global paternal demethylation at zygote stage and maternal demethylation at 4-cell stage. Remethylation starts at the end of the morula stage; in blastocysts, methylation level diverges with more methylation in the inner cell mass.18,24 However, at imprinted loci, the primary imprints laid down at imprinted genes during oogenesis and spermatogenesis are resistant to this demethylation process during the cleavage divisions.
It was demonstrated that in vitro culture systems and embryo manipulations cause imprinting defects in animal models; the degree of abnormality may be related to the type of culture medium. In humans, the aberrant methylation of H19 DMR was found in surplus embryos (low quality or arrested).22,24 Dumoulin et al found that in vitro fertilization (IVF) children derived from embryos that were cultured in 2 different media showed a significant difference in birth weight, revealing an effect of culture medium on ART offspring.25 However, these studies were less and restricted to 1 gene due to the limited material. In this study, methylation errors of H19 DMR, PEG1 DMR, and KvDMR1 were, respectively, found in 8.0%, 16.9% and 10.4% day 3 embryos. We did not find which DMR had higher frequency of abnormal methylation. Our pilot study suggested no differences regarding day 3 embryo quality between abnormal methylation and normal methylation. Whether the methylation errors are related to embryo quality should be investigated in further studies and more strict study design is essential. No methylation errors were found in corresponding sperm samples, suggesting that the paternal epigenetic transmission was unlikely. Because the comparison using in vivo human day 3 embryos as controls cannot be made in the present study, we have no convincing evidence to confirm the epigenetic effect of in vitro culture on human embryos. In addition, the effects of IVF or development in culture medium alone are difficult to investigate. Therefore, we only hypothesized that the altered methylation found in this study are partly caused by ART manipulation, especially culture condition according to previous animal studies.
The embryos we studied were of poor quality, and the good-quality embryos were not detected due to the ethical reason, thus the relationship between abnormal methylation and embryo quality is not known. In ART procedure, the poor-quality embryos are usually abandoned by patients according to doctors’ advice. However, some patients may choose to transfer poor-quality embryos if good embryos are not available. Our results would suggest that the use of poor-quality embryos may increase the risk of imprinting defects because they might have methylation errors. The further studies are needed to extend analysis to more embryos and different types of culture medium.
Footnotes
Authors’ Note: Xiaoyun Shi, Shiling Chen, these authors contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Supported by National Natural Science Foundation of China (81170574); Comprehensive strategic sciences cooperation projects of Guangdong Province and Chinese Academy (04020416); Guangzhou Science and Technology Program key projects (11C22120737) and High-level and innovative science foundation of Guiyang (2012HK)209-52.
References
- 1. Le Bouc Y, Rossignol S, Azzi S, Steunou V, Netchine I, Gicquel C. Epigenetics, genomic imprinting and assisted reproductive technology. Ann Endocrinol (Paris). 2010;71(3):237–238 [DOI] [PubMed] [Google Scholar]
- 2. Bowdin S, Allen C, Kirby G, et al. A survey of assisted reproductive technology births and imprinting disorders. Hum Reprod. 2007;22(12):3237–3240 [DOI] [PubMed] [Google Scholar]
- 3. Shi X, Chen S, Zheng H, Wang L, Wu Y. Aberrant DNA methylation of imprinted loci in human in vitro matured oocytes after long agonist stimulation. Eur J Obstet Gynecol Reprod Biol. 2013;167(1):64–68 [DOI] [PubMed] [Google Scholar]
- 4. Zheng HY, Tang Y, Niu J, et al. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod. 2013;28(1):265–273 [DOI] [PubMed] [Google Scholar]
- 5. Zheng HY, Shi XY, Wu FR, Wu YQ, Wang LL, Chen SL. Assisted reproductive technologies do not increase risk of abnormal methylation of PEG1/MEST in human early pregnancy loss. Fertil Steril. 2011;96(1):84–89 [DOI] [PubMed] [Google Scholar]
- 6. van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18(2):171–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cocchi G, Marsico C, Cosentino A, et al. Silver-Russell syndrome due to paternal H19/IGF2 hypomethylation in a twin girl born after in vitro fertilization. Am J Med Genet A. 2013;161(10):2652–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eggermann T, Spengler S, Begemann M, et al. Deletion of the paternal allele of the imprinted MEST/PEG1 region in a patient with Silver-Russell syndrome features. Clin Genet. 2012;81(3):298–300 [DOI] [PubMed] [Google Scholar]
- 9. Binder G, Begemann M, Eggermann T, Kannenberg K. Silver-Russell syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25(1):153–160 [DOI] [PubMed] [Google Scholar]
- 10. Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2010;18(1):8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sutcliffe AG, Peters CJ, Bowdin S, et al. Assisted reproductive therapies and imprinting disorders--a preliminary British survey. Hum Reprod. 2006;21(4):1009–1011 [DOI] [PubMed] [Google Scholar]
- 12. DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22(1):26–35 [DOI] [PubMed] [Google Scholar]
- 14. Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19(1):36–51 [DOI] [PubMed] [Google Scholar]
- 15. Borghol N, Lornage J, Blachere T, Sophie GA, Lefevre A. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics. 2006;87(3):417–426 [DOI] [PubMed] [Google Scholar]
- 16. Market VB, Denomme MM, Mann MR. Loss of genomic imprinting in mouse embryos with fast rates of preimplantation development in culture. Biol Reprod. 2012;86(5):143, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu JQ, Liu JH, Liang XW, Xu BZ, Hou Y, Zhao XX. et al. Heat stress causes aberrant DNA methylation of H19 and Igf-2r in mouse blastocysts. Mol Cells. 2008;25(2):211–215 [PubMed] [Google Scholar]
- 18. Santos F, Hyslop L, Stojkovic P, et al. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum Reprod. 2010;25(9):2387–2395 [DOI] [PubMed] [Google Scholar]
- 19. Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94(2):585–594 [DOI] [PubMed] [Google Scholar]
- 20. Kobayashi H, Hiura H, John RM, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17(12):1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato A, Hiura H, Okae H, et al. Assessing loss of imprint methylation in sperm from subfertile men using novel methylation polymerase chain reaction Luminex analysis. Fertil Steril. 2011;95(1):129–34, 131–4 [DOI] [PubMed] [Google Scholar]
- 22. Chen SL, Shi XY, Zheng HY, Wu FR, Luo C. Aberrant DNA methylation of imprinted H19 gene in human preimplantation embryos. Fertil Steril. 2010;94(6):2356–2358, 2351–8 [DOI] [PubMed] [Google Scholar]
- 23. Fulka H, Mrazek M, Tepla O, Fulka JJ. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128(6):703–708 [DOI] [PubMed] [Google Scholar]
- 24. Ibala-Romdhane S, Al Khtib M, Khoueiry R, Blachere T, Guerin JF, Lefevre A. Analysis of H19 methylation in control and abnormal human embryos, sperm and oocytes. Eur J Hum Genet. 2011;19(11):1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dumoulin JC, Land JA, Van Montfoort AP, et al. Effect of in vitro culture of human embryos on birthweight of newborns. Hum Reprod. 2010;25(3):605–612 [DOI] [PubMed] [Google Scholar]