Abstract
Continuous cell lines are used frequently in reproductive biology research to study problems in early pregnancy events and parturition. It has been recognized for 50 years that many mammalian cell lines contain inter- or intraspecies contaminations with other cells. However, most investigators do not routinely test their culture systems for cross-contamination. The most frequent contributor to cross-contamination of cell lines is the HeLa cell isolated from an aggressive cervical adenocarcinoma. We report on the discovery of HeLa cell contamination of the human endometrial epithelial cell line HES isolated in our laboratory. Short tandem repeat analysis of 9 unique genetic loci demonstrated molecular identity between HES and HeLa cells. In addition, we verified that WISH cells, isolated originally from human amnion epithelium, were also contaminated with HeLa cells. Inasmuch as our laboratory did not culture HeLa cells at the time of HES cell derivations, the source of contamination was the WISH cell line. These data highlight the need for continued diligence in authenticating cell lines used in reproductive biology research.
Keywords: HES cell, HeLa, cell lines, intraspecies cross-contamination
Introduction
Immortalized mammalian cell lines are invaluable in creating facile model systems for fundamental research in biology. Continuous cell lines are frequently used to recapitulate many phenotypic features of cell types that are critical for reproductive functions, including early implantation events, placental development and function, parturition, and cytokine and eicosanoid biology.1–10 Data obtained from these mammalian in vitro model systems, particularly using human cell lines, have been instrumental in shedding light on the molecules that mediate trophoblast–endometrial communications11,12 in the peri-implantation period and many other aspects of reproduction. Many of these studies have used uterine adenocarcinoma cells (eg, Ishikawa cells) or immortalized endometrial epithelial cells to probe molecular interactions during early pregnancy events.13–15
In the field of in vitro biology, there has been a periodic appreciation of the importance of rigorous cell line characterization regarding methods and consequences of immortalization and genotypic authenticity and phenotype fidelity to the in vivo cell types of interest14 but this has been less well appreciated in reproductive biology research. Early work by Nelson-Rees et al16 highlighted the discovery that many mammalian cell lines used as model culture systems, particularly in cancer, were contaminated with HeLa cells, an aggressive cervical cancer cell line that was the first human cell to be successfully grown in vitro. Nelson-Rees and Nardone later championed the suspicion of widespread contamination of many mammalian cell lines used by laboratories throughout the world by HeLa and other cell types.17–20 When early studies began to demonstrate potential cell line cross-contamination, only a few biochemical16,21 and later chromosomal markers were available.21–24 Recent techniques using DNA fingerprinting of short tandem repeats (STRs) at selected loci within the genome offer a simple, inexpensive, and rapid method of authenticating cell lines.25–27
We have recently conducted an STR analysis of the endometrial cell line HES, developed in our laboratory in 1989.9 We discovered that the HES cell line exhibited a DNA fingerprint indistinguishable from that of the HeLa cell DNA fingerprint. Inasmuch as HeLa cells were not cultured in our laboratory during the derivation of HES cells but WISH cells, an amnion-derived cell line known to be HeLa contaminated, were simultaneously grown by our group; the source of this contamination was the WISH cell. In this article, we report on the identity between HES cells and HeLa cells. We suggest that this disclosure calls for the development of a policy for cell line authentication in the reproductive biology research community.
Materials and Methods
Cell Culture
All cell culture reagents were purchased from Invitrogen (Carlsbad, California) unless otherwise specified. The HES cell line was established in 1989 from the endometrial surface epithelium of a hysterectomy specimen performed for leiomyomata. Histopathologic analysis reported the specimen to be benign endometrium and the only documented lesion was fibroid tissue within the myometrium. A primary culture was initiated after enzymatic digestion with 0.25% collagenase and 100 U/mL of hyaluronidase. The HES cells were cultured in Dulbecco Modified Eagle Medium (DMEM) with 4.5 g/L d-glucose supplemented with 10% fetal bovine serum (FBS) and 50 μg/mL of gentamicin. The HeLa cells were obtained from the American Type Culture Collection (ATCC CCL-2) and cultured in DMEM/Ham F12 (DMEM/F12) supplemented with 10% FBS. Human amnion-derived (WISH) cells were also obtained from the ATCC (CCL-25). The WISH cells were maintained in DMEM/F12 supplemented with 2 mmol/L l-glutamine, 1 mmol/L sodium pyruvate, and 10% newborn calf serum (NCS). All cells were grown at 37°C in a humidified atmosphere of 5% CO2/95% air.
DNA Fingerprinting: STR Analysis
Cell lines used in this study were authenticated by DNA profiling using STR analysis on the StemElite ID platform (Promega, Madison, Wisconsin) at The John Hopkins University (JHU; Fragment Analysis Facility at the Genetic Resource Core Facility, Baltimore, Maryland; http://faf.grcf.jhmi.edu/str.html). Briefly, 106 cells were grown to confluence in 100 mm dishes under standard culture conditions described earlier and shipped to the Genetic Resource Core at JHU on dry ice. DNA was isolated and STR analysis was performed according to the manufacturer’s specifications using the following molecular markers: amelogenin (AMEL), the sex determination locus, CSF1PO, D13S317, D16S539, D5S818, D7S820, TH01, TPOX, and vWA, a unique pattern of nucleic acid repeats within the human genome (www.atcc.org). These loci are known to be polymorphic across nucleotide lengths of 3 to 7 base pairs and have been used extensively in cell line authentication, forensic DNA analysis, and paternity testing (http://www.promega.com/resources/pubhub/short-tandem-repeat-analysis-in-the-research-laboratory/).
Immunofluorescence
The HES, HeLa, and WISH cells were cultured on 12-mm glass coverslips (5 × 104/well) plated into 24-well plates in the media mentioned earlier. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and stored at 4°C in PBS containing 1% sodium azide until used for immunolabeling. Coverslips were preincubated with PBS + 5% NCS and then incubated with mouse monoclonal human anticytokeratin 8 (CK-8, 1:100) and phalloidin-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-fluorescein isothiocyanate (40 μg/mL) for 60 minutes at room temperature. After rinsing 3 times with PBS + 5% NCS, the cells were incubated with rabbit antimouse IgG-AlexaFluor-594 (Molecular Probes, Eugene, Oregon; 1:100 in PBS + 5% NCS) for 60 minutes at room temperature. After rinsing 3 times, the cells were incubated with Prolong reagent (Molecular Probes) including 4′,6-diamidino-2-phenylindole to label nuclei. Images were captured on a Nikon Eclipse TE200 inverted epifluorescence microscope interfaced with a SPOT RTC3 digital camera system (SPOT Imaging Solutions, Sterling Heights, Michigan).
Results
Morphology and Immunofluorescence
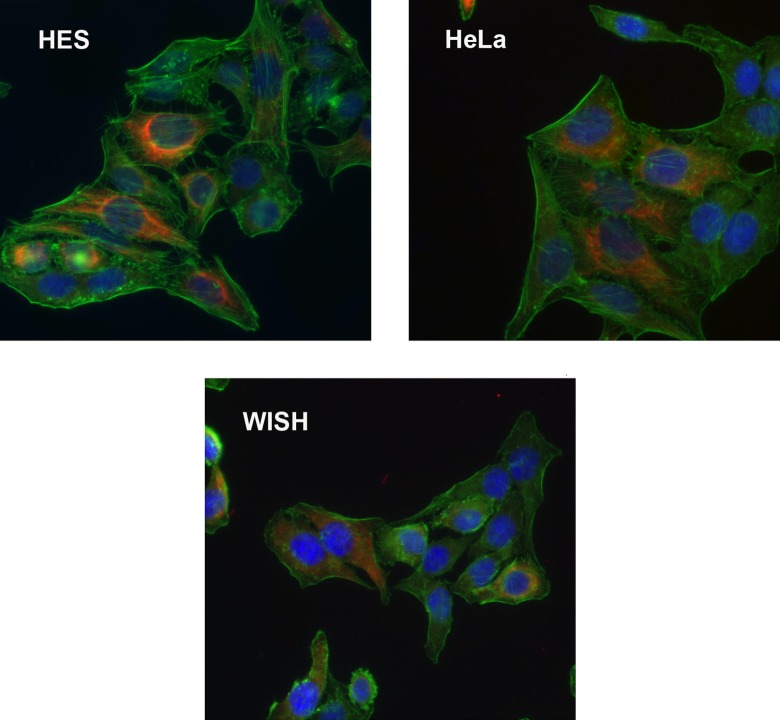
The HES, HeLa, and WISH cells grown at low density were fixed and labeled with anti-CK-8 and phalloidin to identify the keratin and actin cytoskeletons, respectively. The 3 cell types are of epithelial origin and could not be discriminated simply by staining for conventional cell markers (Figure 1). Each cell type expressed a keratin intermediate filament cytoskeleton that ramified from the perinuclear region and a filamentous actin cytoskeleton characteristic of epithelial cells grown on rigid substrates. Thus, morphological differences alone are not sufficient to discriminate among these epithelial cell types.
Figure 1.
Fluorescence staining of HES, HeLa and WISH cells for cytokeratin-8, (red) cytoskeletal actin (green), and nuclei (blue).
Short Tandem Repeat DNA Fingerprinting Analysis
To determine the endometrial epithelial cell authenticity of the HES cell line, we conducted a well-established DNA fingerprinting technique involving analysis of STR elements within a panel of 9 genetic loci. DNA from the HES cell line developed in our laboratory was submitted to JHU Genetic Resource Core for STR analysis using the StemElite ID platform. The genetic loci of all 9 markers showed identical genotypes with the HeLa cell standard used in JHU (Table 1). Furthermore, a WISH cell standard obtained from the ATCC and known to be contaminated by HeLa cells revealed an identical genotype at all 9 loci. In contrast, K562 cell DNA, part of the negative control panel used by the JHU Core, differed at all genetic loci. These allelic data support the conclusion that the HES cell line has acquired HeLa markers during its maintenance in culture.
Table 1.
DNA Fingerprinting Using Short Tandem Repeat Analysis.
| Submitted Samples | JHU Standards | ATCC Standards | |||||
|---|---|---|---|---|---|---|---|
| HeLa | HES | WISH | HeLa | K562 | HeLa | WISHa | |
| Marker | Allele 1,2 | Allele 1,2 | Allele 1,2 | Allele 1,2 | Allele 1,2 | Allele 1,2 | Allele 1,2 |
| AMEL | X,X | X,X | X,X | X,X | X,X | X,X | X,X |
| CSF1PO | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 | 9,10 |
| D13S317 | 13.3,13.3 | 13.3,13.3 | 13.3,13.3 | 12,13.3 | 8,8 | 12,13.3 | 13.3,13.3 |
| D16S539 | 9,10 | 9,10 | 9,10 | 9,10 | 11,12 | 9,10 | 9,10 |
| D5S818 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 | 11,12 |
| D7S820 | 8,12 | 8,12 | 8,12 | 8,12 | 9,11 | 8,12 | 8,12 |
| TH01 | 7,7 | 7,7 | 7,7 | 7,7 | 9.3,9.3 | 7,7 | 7,7 |
| TPOX | 8,12 | 8,12 | 8,12 | 8,12 | 8,9 | 8,12 | 8,12 |
| vWA | 16,18 | 16,18 | 16,18 | 16,18 | 16,16 | 16,18 | 16,18 |
Abbreviations: AMEL, amelogenin; ATCC, American Type Culture Collection; JHU, Johns Hopkins University.
a WISH cell line is known to be contaminated with HeLa cell markers.
Discussion
The current study reports on the discovery that the endometrial epithelial cell line HES, developed in our laboratory in 1989, became contaminated with HeLa cell markers subsequent to the derivation of the cell line. The STR analysis, also known as DNA fingerprinting, the gold standard for cell authentication, demonstrated identity with HeLa cells and WISH cells. The WISH cells, initially isolated at the Wistar Institute by Hayflick28 from primary amnion epithelium, are known to be contaminated with HeLa cells29 and many in the parturition research community have used them.6–8,30–32 Importantly, at the time when the HES cell line was isolated in our laboratory, we did not use HeLa cells. However, our group frequently used WISH cells for studies of eicosanoid and cytokine biology and this is almost certainly the source of contamination.
We reported in 2002 that the ED27 trophoblast-like cell line, derived from first-trimester human chorionic villi, contained HeLa cell markers using a similar polymerase chain reaction-based analysis.29 The ED27 cell line manifests many features of human extravillous trophoblast, including HLA-G expression, steroid biosynthesis, and human chorionic gonadotropin expression.29,33 Based on forensic analysis, we have concluded that the contamination of both HES and ED27 cell lines occurred via WISH cells during a single event when culture medium was changed using nonstandard techniques, thus likely initiating the cross-contamination event. Cell line authentication was not a routine procedure in most laboratories in the 1990s and there did not appear to be phenotypic drift that would have triggered our suspicion of a potential cross-contamination.
Based upon our immunofluorescence experiments to identify the cytokeratin and actin cytoskeletons, it was not possible to discriminate unequivocally between HES, WISH, and HeLa cell lines. As such, the use of morphological markers alone is not sufficient to discriminate cell types despite what might be reported as morphological differences that define one cell type from another. Nor, strictly speaking, should other phenotypic traits such as hormone synthesis or antigen expression be used as the principal criteria for identifying cell type authenticity.
Most investigators do not routinely perform genetic authentication of cell lines used in their laboratories. HeLa cells were initially isolated from a highly aggressive cervical cancer20,34 and have been the subject of extensive research in cell and cancer biology laboratories throughout the world. For nearly 50 years, there have been occasional reports of HeLa cell cross-contamination of many mammalian cell lines. An analysis of the cell lines evaluated by international cell banks revealed that the incidence of cell line misidentification in 1977 was 16% and was 18% in 1988, indicating that the problem had not substantially improved in a decade.35 A survey of data from a variety of sources indicates that this problem persists to this day (unpublished observations). Furthermore, the problem of cell line contamination is not restricted to HeLa cells, and according the ATCC, cross-contamination of continuous cell lines used in biomedical research is a pervasive issue (ATCC, Misidentified Cell Lines, www.atcc.org). In addition, as a further example of this persistent issue, the human epithelial cell line that has been immortalized by human telomerase reverse transcriptase overexpression is actually the human breast cancer cell line MCF-7.36
Based upon our own data and an extensive review of the literature, we make the following conclusions and recommendations. First, the biomedical literature is replete with important data on fundamental events that govern cell and molecular biology and in most instances, cross-contamination does not invalidate these findings. Indeed, caution should be used in the interpretation of any in vitro studies. Second, functional studies alone are not sufficient to prove cell type fidelity. Third, since STR analysis is convenient, inexpensive, and provides unequivocal genetic identities, cell line authentication should become routine in all reproductive biology laboratories. Our group has established a policy of conducting DNA fingerprinting of any cell accepted into our laboratory and for any cell line distributed by our laboratory, in addition to other routine testing such as mycoplasma. In 2007, the National Institutes of Health released a notice to the biomedical research community (NOT-OD-08-017: Notice Regarding Authentication of Cultured Cells) on the recommendation of cell line authentication but fell short of mandating such a policy for grant and manuscript submissions. In light of the potentially widespread issue of intraspecies cross-contamination, it may be prudent for professional societies in the reproductive sciences to develop formal standards for cell line authentication using a single, unified profiling platform such as STR analysis.36
Acknowledgments
We gratefully acknowledge the Johns Hopkins University Genetic Resource Core, Fragment Analysis Facility for the Short Tandem Repeat Analysis. William Ackerman is acknowledged for his assistance with the immunofluorescence studies.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Ohio State University Perinatal Research and Development Fund.
References
- 1. Hannan NJ, Paiva P, Dimitriadis E, Salamonsen LA. Models for study of human embryo implantation: choice of cell lines? Biol Reprod. 2010;82(2):235–245 [DOI] [PubMed] [Google Scholar]
- 2. Ackerman WE IV, Robinson JM, Kniss DA. Despite transcriptional and functional coordination, cyclooxygenase-2 and microsomal prostaglandin E synthase-1 largely reside in distinct lipid microdomains in WISH epithelial cells. J Histochem Cytochem. 2005;53(11):1391–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ackerman WE IV, Rovin BH, Kniss DA. Epidermal growth factor and interleukin−1β utilize divergent signaling pathways to synergistically upregulate cyclooxygenase-2 gene expression in human amnion-derived WISH cells. Biol Reprod. 2004;71(6):2079–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kniss DA, Mershon J, Su H-C, et al. Evidence of a role for protein kinase C in epidermal growth factor-induced prostaglandin E2 synthesis in amnion cells. Amer J Obstet Gynecol. 1990;163(6 pt 1):1883–1890 [DOI] [PubMed] [Google Scholar]
- 5. Harris AN, Perlman M, Schiller SL, Romero R, Mitchell MD. Characterization of prostaglandin production in amnion-derived WISH cells. Amer J Obstet Gynecol. 1988;159(6):1385–1389 [DOI] [PubMed] [Google Scholar]
- 6. Marvin KW, Hansen WR, Miller HC, Eykholt RL, Mitchell MD. Amnion-derived cells express intercellular adhesion molecule-1: regulation by cytokines. J Mol Endocrinol. 1999;22(2):193–205 [DOI] [PubMed] [Google Scholar]
- 7. Eykholt RL, Hansen WR, Potter S, Marvin KW, Mitchell MD. Cytokines regulate prostaglandin H synthase-1 transcription in human amnion-derived cells. Prostaglandins Leukot Essent Fatty Acids. 1999;61(5):323–329 [DOI] [PubMed] [Google Scholar]
- 8. Sato TA, Keelan JA, Blumenstein M, Mitchell MD. Efficacy and specificity of non-steroidal anti-inflammatory drugs for the inhibition of cytokine-stimulated prostaglandin E2 secretion by amnion-derived WISH cells. Prostaglandins Leukot Essent Fatty Acids. 2002;66(5-6):525–527 [DOI] [PubMed] [Google Scholar]
- 9. Desai NN, Kennard EA, Kniss DA, Friedman CI. Novel human endometrial cell line promotes blastocyst development. Fertil Steril. 1994;61(4):760–766 [PubMed] [Google Scholar]
- 10. Xue S, Slater DM, Bennett PR, Myatt L. Induction of both cytosolic phospholipase A2 and prostaglandin H synthase-2 by interleukin-1 beta in WISH cells in inhibited by dexamethasone. Prostaglandins. 1996;51(2):107–124 [DOI] [PubMed] [Google Scholar]
- 11. Dharmaraj N, Wang P, Carson DD. Cytokine and progesterone receptor interplay in the regulation of MUC1 gene expression. Mol Endocrinol. 2010;24(12):2253–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382(pt 1):363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hohn HP, Linke M, Denker HW. Adhesion of trophoblast to uterine epithelium as related to the state of trophoblast differentiation: in vitro studies using cell lines. Mol Reprod Dev. 2000;57(2):135–145 [DOI] [PubMed] [Google Scholar]
- 14. Ho H, Singh H, Aljofan M, Nie G. A high-throughput in vitro model of human embryo attachment. Fertil Steril. 2012;97(4):974–978 [DOI] [PubMed] [Google Scholar]
- 15. Fleming H. Structure and function of cultured endometrial epithelial cells. Sem Reprod Endocrinol. 1999;17(1):93–106 [DOI] [PubMed] [Google Scholar]
- 16. Nelson-Rees WA, Flandermeyer RR, Hawthorne PK. Banded marker chromosomes as indicators of intraspecies cellular contamination. Science. 1974;184(4141):1093–1096 [DOI] [PubMed] [Google Scholar]
- 17. Nelson-Rees WA, Hunter L, Darlington GJ, O'Brien SJ. Characteristics of HeLa strains: permanent vs. variable features. Cytogenet Cell Genet. 1980;27(4):216–230 [DOI] [PubMed] [Google Scholar]
- 18. Nelson-Rees WA, Daniels DW, Flandermeyer RR. Cross-contamination of cells in culture. Science. 1981;212(4493):446–452 [DOI] [PubMed] [Google Scholar]
- 19. Nardone RM. Curbing rampant cross-contamination and misidentification of cell lines. Biotechniques. 2008;45(3):221–227 [DOI] [PubMed] [Google Scholar]
- 20. Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med. 1999;133(9):1463–1467 [DOI] [PubMed] [Google Scholar]
- 21. Auersperg N, Gartler SM. Isoenzyme stability in human heteroploid cell lines. Exp Cell Res. 1970;61(2):465–467 [DOI] [PubMed] [Google Scholar]
- 22. Beckman G, Beckman L, Ponten J, Westermark B. G-6-PD and PGM phenotypes of 16 continuous human tumor cell lines. Evidence against cross-contamination and contamination by HeLa cells. Hum Hered. 1971;21(3):238–241 [DOI] [PubMed] [Google Scholar]
- 23. Gartler SM. Apparent Hela cell contamination of human heteroploid cell lines. Nature. 1968;217(5130):750–751 [DOI] [PubMed] [Google Scholar]
- 24. Herz F, Miller OJ, Miller DA, Auersperg N, Koss LG. Chromosome analysis and alkaline phosphatase of C41, a cell line of human cervical origin distinct from HeLa. Cancer Res. 1977;37(9):3209–3213 [PubMed] [Google Scholar]
- 25. Lorenzi PL, Reinhold WL, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8(4):713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thacker J, Webb MB, Debenham PG. Fingerprinting cell lines: use of human hypervariable DNA probes to characterize mammalian cell cultures. Somat Cell Mol Genet. 1988;14(6):519–525 [DOI] [PubMed] [Google Scholar]
- 27. Dirks WG, Drexler HG. STR DNA typing of human cell lines: detection of intra- and interspecies cross-contamination. Meth Mol Biol. 2013;946:27–38 [DOI] [PubMed] [Google Scholar]
- 28. Hayflick L. The establishment of a line (WISH) of human amnion cells in continuous cultivation. Exp Cell Res. 1961;23:14–20 [DOI] [PubMed] [Google Scholar]
- 29. Kniss DA, Xie Y, Li Y, et al. ED27 trophoblast-like cells isolated from first-trimester chorionic villi are genetically identical to HeLa cells yet exhibit a distinct phenotype. Placenta. 2002;23(1):32–43 [DOI] [PubMed] [Google Scholar]
- 30. Gilmour JS, Hansen WR, Miller HC, Keelan JA, Sato TA, Mitchell MD. Effects of interleukin-4 on the expression and activity of prostaglandin endoperoxide H synthase-2 in amnion-derived WISH cells. J Mol Endocrinol. 1998;21(3):317–325 [DOI] [PubMed] [Google Scholar]
- 31. Keelan JA, Sato TA, Gupta DK, Marvin KW, Mitchell MD. Prostanoid stimulation of cytokine production in an amnion-derived cell line: evidence of a feed-forward mechanism with implications for term and preterm labor. J Soc Gynecol Invest. 2000; 7: 37–44 [DOI] [PubMed] [Google Scholar]
- 32. Hansen WR, Sato T, Mitchell MD. Tumour necrosis factor-alpha stimulates increased expression of prostaglandin endoperoxide H synthase Type 2 mRNA in amnion-derived WISH cells. J Mol Endocrinol. 1998;20(2):221–231 [DOI] [PubMed] [Google Scholar]
- 33. Ma T, Yang ST, Kniss DA. Development of an in vitro human placenta model by the cultivation of human trophoblasts in a fiber-based bioreactor system. Tissue Eng. 1999;5(2):91–102 [DOI] [PubMed] [Google Scholar]
- 34. Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nature Rev Cancer. 2002;2(4):315–319 [DOI] [PubMed] [Google Scholar]
- 35. American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: the beginning of the end. Nature Rev Cancer. 2010;10(6):441–448 [DOI] [PubMed] [Google Scholar]
- 36. Korch C, Spillman MA, Jackson TA, et al. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127(1):241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sciences USA. 2001;98(14):8012–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]