Abstract
Infection-induced periodontal disease has been primarily focused on a small group of periodontal pathogens. A paradigm shift, based on data emerging from the oral microbiome project, now suggests the involvement of as-yet-unculturable and fastidious organisms. Collectively, these studies have demonstrated that there are changes in the periodontal status associated with shifts in the composition of the bacterial community in the periodontal pocket. In addition, it is likely that the emerging new pathogens may play a more significant role in the disease. One of the organisms previously unrecognized is Filifactor alocis. While this Gram-positive anaerobic rod has been identified in peri-implantitis, in endodontic infections, and in patients with localized aggressive periodontitis, its presence is now observed at significantly higher levels in patients with adult periodontitis or refractory periodontitis. Its colonization properties and its potential virulence attributes support the proposal that F. alocis should be included as a diagnostic indicator of periodontal disease. Moreover, these emerging characteristics would be consistent with the polymicrobial synergy and dysbiosis (PSD) periodontal pathogenesis model. Here, unique characteristics of F. alocis are discussed. F. alocis has specific factors that can modulate multiple changes in the microbial community and host cell proteome. It is likely that such variations at the molecular level are responsible for the functional changes required to mediate the pathogenic process.
Keywords: polymicrobial infection, periodontitis, community dynamics, oxidative stress, proteomics, host interaction
Introduction
In the United States, over 49 million people are affected by periodontitis, a chronic inflammatory condition of an infectious nature involving the tissues supporting the teeth (Oliver et al., 1991; Eke et al., 2012; Thornton-Evans et al., 2013). In addition to the heavy health care burden that this occurrence may impose, it is likely to increase given the emerging association of periodontitis with other systemic diseases, including cardiovascular diseases (Genco and Van Dyke, 2010) and rheumatoid arthritis (Bingham and Moni, 2013). Even though the human oral cavity is home to more than 700 species, only a subset of microbes, which now includes previously unrecognized and uncultivated species, is associated with disease (Dewhirst et al., 2010; Kolenbrander et al., 2010; Griffen et al., 2012). The “red complex”, consisting of Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, is well-established as being associated with adult periodontal disease (Kumar et al., 2003; Dewhirst et al., 2010). Furthermore, P. gingivalis, now designated as a “keystone” species, is able to manipulate the host immune system, thus eliciting a major effect on the composition of the oral microbial community and to the pathology of periodontitis (Darveau et al., 2012; Hajishengallis and Lamont, 2012). Because colonization of germ-free mice by P. gingivalis failed to induce the pathology associated with the disease (Hajishengallis et al., 2011), this suggests that while it may be necessary (reviewed in Darveau et al., 2012), it is insufficient to trigger the periodontitis-associated pathology. This is also observed in localized aggressive periodontitis (LAP) in young adults where Aggregatibacter actinomycetemcomitans was proposed to be a keystone pathogen that could set up the subgingival environment by producing toxins, resulting in immune paralysis and permitting the overgrowth of specific organisms at specific time-points that would otherwise be controlled and regulated by the host. Collectively, these observations have raised questions on the role other pathogens may play in the disease process. In the current emerging paradigm, periodontitis is believed to be initiated by synergistic and dysbiotic microbial communities (Hajishengallis and Lamont, 2012).
Yet-to-be-cultured Bacterial Communities
Oral microbiome studies conducted over the last several years have modified and enhanced our understanding of the oral multispecies microbial communities in health and disease. Current theories on the etiology of periodontitis favor a shift in microbial composition that is caused by a decrease in beneficial symbionts and an increase in organisms with enhanced pathogenic potential. Hence, there is an increase in microbial diversity and its composition, and thus pathogenic communities contain higher levels of fastidious and yet-to-be-cultivated taxons than previously recognized (Dewhirst et al., 2010; Griffen et al., 2012).
The previously accepted “red complex”—along with other cultivable bacteria species, such as Prevotella intermedia, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Selenomonas noxia, and Eubacterium nodatum—is associated with periodontitis (Kolenbrander et al., 2002, 2006; Saito et al., 2009). In addition, organisms such as Selenomonas, Synergistes, Desulfobulbus, TM7 (new candidate bacterial division), and Filifactor alocis have been identified as potential pathogens in a number of independent studies (Dewhirst et al., 2010; Griffen et al., 2012). Moreover, of the phylotypes identified in the oral cavity, between 20% and 60% have yet to be cultivated (Dewhirst et al., 2010; Griffen et al., 2012). This has raised questions on the relative significance of these microbes in the disease process. This review introduces the new organism F. alocis and its unique characteristics, virulence potentials, capacity to act in community dynamics, and, ultimately, its role in periodontitis.
F. alocis: General Characteristics
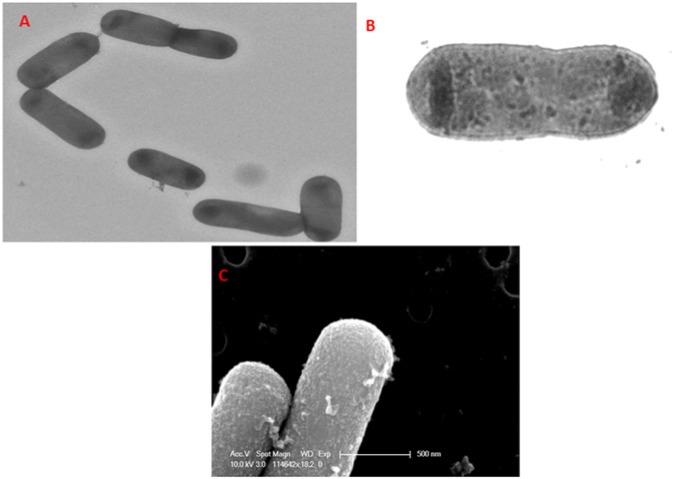
F. alocis, a Gram-positive, asaccharolytic, obligate anaerobic rod (Figs. 1A, 1B), shows surface punctations resembling minute projections (Fig. 1C). This is one of the marker organisms and is considered an important periodontal pathogen. The organism is now identified to be significant to the pathogenic structure of biofilms associated with periodontal inflammation (Kumar et al., 2003, 2006; Schlafer et al., 2010). In comparison with the other traditional periodontal pathogens, the high incidence of F. alocis in the periodontal pocket compared with its absence in healthy individuals or those who are periodontitis-resistant has highlighted its importance in the infectious disease process (Kumar et al., 2003, 2006; Wade, 2011). This organism was first isolated in 1985 from the gingival sulcus in patients with gingivitis and periodontitis and was originally classified as Fusobacterium alocis (Cato et al., 1985) then reclassified into the genus Filifactor (Jalava and Eerola, 1999).
Figure 1.
F. alocis – surface morphology. (A) Transmission electron micrograph showing F. alocis, a Gram-positive, asaccharolytic, obligate anaerobic rod. (B) Transmission electron micrograph of F. alocis showing membrane and cell wall structures. (C) Scanning electron micrograph of F. alocis showing surface punctations resembling minor projections.
F. alocis, while heterogenous, has virulence properties that may enhance its ability to survive and persist in the periodontal pocket (Aruni et al., 2011). Its relative resistance to oxidative stress and stimulated growth under those conditions are considered to be important attributes (Aruni et al., 2011). Furthermore, F. alocis has been shown to induce secretion of pro-inflammatory cytokines, triggering apoptosis of gingival epithelial cells (Moffatt et al., 2011). Additionally, colonization and survival of F. alocis in a mouse model showed pro-apoptotic local infection that is rapidly resolved by host neutrophil influx (Wang et al., 2014). Moreover, in co-culture with P. gingivalis, F. alocis showed an increased invasive capacity of HeLa cells (Aruni et al., 2011).
In silico analysis of F. alocis has shown close relatedness to Clostridium and Fusobacterium (Aruni et al., 2011). Similarities in virulence attributes among these organisms have been notably due to the presence of a battery of proteases. This characteristic, also present in F. alocis, would be consistent with the asaccharolytic nature of this organism, where specific amino acids including arginine are shown to stimulate its growth (Uematsu et al., 2003). Even though F. alocis showed low gingipain-type activity, it had increased non-gingipain protease activity (Aruni et al., 2011). The amino acids mostly utilized by F. alocis include arginine and lysine, followed by cysteine. The F. alocis arginine metabolic pathway predicts the enzymatic degradation of arginine by arginine deaminase, leading to the conversion of arginine to ornithine and ammonia (Uematsu et al., 2003). Arginine degradation could favor increase in the pH that would counteract acidic conditions generated from carbohydrate catabolism in a mixed bacterial oral flora. In the periodontal pocket, these amino acids can also be made available from the degradation of various protein substrates by other bacteria and host-derived proteases for nutritional support, survival, and virulence (Eley and Cox, 1992).
F. alocis Involvement in Other Oral Disease Conditions
The environment surrounding the sulcus of patients with peri-implantitis is well-suited for the growth of both Gram-negative and asaccharolytic anaerobic Gram-positive rods (AAGPRs). Among the AAGPRs, F. alocis is one of the most prominent bacteria (Tamura et al., 2013). F. alocis has also been discovered in the canals of root-filled teeth with periapical lesions, is associated with signs and symptoms of endodontic infections (Gomes et al., 2006), and has been identified as one of the prevalent phylotypes in cases of failed endodontic treatment (Zhang et al., 2012).
The Virulence Potential of F. alocis
Because of the presence of other microbial species, chronic inflammation, and other prevailing conditions, including fluctuations in nutrient availability, temperature, pH, and oxygen tension, F. alocis must have properties that will allow it to colonize, survive, and out-compete other traditional periodontal pathogens in the stress environment of the periodontal pocket.
Biofilm Formation
Oral biofilms are primary initiating factors of periodontal disease. Biofilm formation involving F. alocis has been demonstrated in both periodontic and endodontic cases (Schlafer et al., 2010). While some interspecies interaction can inhibit biofilm formation, P. gingivalis ATCC 33277 co-cultured with F. alocis showed significant increase in biofilm formation (Aruni et al., 2011). This enhanced biofilm-forming capacity may be due to the ability of both species to autoaggregate and express unique components. This may also indicate a symbiotic relationship between F. alocis and P. gingivalis. Thus, F. alocis and P. gingivalis, each with different growth rates, could form a mixed-species biofilm and co-exist (Aruni et al., 2011). As a result, F. alocis proteins could enable P. gingivalis to proliferate and disseminate from these biofilms, thus facilitating its virulence (Pöllänen et al., 2013).
A recent in vitro study, evaluating the community interactions of two strains of F. alocis with Streptococcus gordonii, F. nucleatum, P. gingivalis, and A. actinomycetemcomitans, which are organisms of differing pathogenic potential in the oral cavity, suggests that F. alocis is likely to interact with a variety of oral bacteria and participate in community development (Wang et al., 2013). Further, F. alocis colonization seemed to be dictated by the spatial composition of microbial microenvironments that involves quorum-sensing, and the organism may preferentially accumulate at sites rich in F. nucleatum. Streptococcus gordonii was antagonistic to the accumulation of F. alocis in a dual-species community. This was consistent with the observation that streptococci-rich dental plaques were resistant to colonization by F. alocis (Wang et al., 2013). In three-species communities of S. gordonii, F. nucleatum, and F. alocis, the antagonistic effects of S. gordonii superseded the synergistic effects of F. nucleatum toward F. alocis (Wang et al., 2013). The interaction between A. actinomycetemcomitans and F. alocis was strain-specific, and A. actinomycetemcomitans could either stimulate F. alocis accumulation or have no effect, depending on the strain. P. gingivalis and F. alocis formed heterotypic communities, with the abundance of P. gingivalis being enhanced in the presence of F. alocis (Wang et al., 2013). It is likely that F. alocis proteins induced under those conditions may facilitate adhesion and nutrient support for P. gingivalis. While the mechanism of the interaction is complex, with inhibitory and counterbalancing measures, the question of how arginine deiminase affects the community dynamics can be raised. The inhibitory effect of P. gingivalis on F. alocis was observed to be partially dependent on the minor fimbriae (Wang et al., 2013). The arginine deiminase of S. cristatus is known to suppress fimbrial production in P. gingivalis (Xie et al., 2007; Wang et al., 2009). Based on the relative abundance of F. alocis in the periodontal pocket compared with P. gingivalis, it is unclear if the F. alocis arginine deiminase is induced in that microenvironment and could have an effect on P. gingivalis fimbrial expression.
Oxidative Stress Resistance
Our studies have shown that F. alocis is relatively resistant to oxidative stress compared with P. gingivalis and that its growth is stimulated under those conditions (Aruni et al., 2011). These observations may indicate an important attribute for the survival and relative abundance of F. alocis compared with other organisms in the inflammatory microenvironment of the periodontal pocket. It is also likely that F. alocis may play a role as an “oxidative sink” to stabilize the microbial community in the microenvironment of the periodontal pocket. Preliminary observations from our laboratory suggest that the survival of P. gingivalis under hydrogen-peroxide-induced oxidative stress is enhanced in the presence of F. alocis (unpublished observations).
While the precise mechanism of oxidative stress resistance in F. alocis is yet to be elucidated, there are likely several possibilities that will need confirmation. Not only will the presence of sialidase activity in F. alocis satisfy its asaccharolytic property by breakdown of sialated glycoproteins found in saliva, but the released sialic acid can also act as a ROS scavenger to reduce the oxidative stress in the inflammatory environment of the periodontal pocket (Iijima et al., 2004). F. alocis possesses a superoxide reductase (GenBank accession no. EFE28874) that could help to facilitate its growth in the presence of hydrogen peroxide (http://www.ncbi.nlm.nih.gov/bioproject/46625).
The interaction of F. alocis with other organisms can also enhance its oxidative stress resistance and hence its virulence potential. In co-culture with P. gingivalis, an up-regulation of many proteins involved in oxidative stress resistance—such as superoxide reductase, iron-sulfur cluster protein, iron permease, ruberythrin, ferrous hydrogenase family protein, and thioredoxin family proteins—was observed in F. alocis (Aruni et al., 2012). One of the key attributes of F. alocis is the 3-methyladenine DNA glycosylase (HMPREF0389_1529), an enzyme reported to be involved in oxidative and nitrosative stress resistance in other pathogenic bacteria (Slade and Radman, 2011) (although its function under those conditions is unclear). The genome of F. alocis also includes genes that encode for a well-developed group of iron-sulfur cluster proteins and a ferrous iron transport system which are unique to this organism compared with other “red complex” bacteria. Additionally, F. alocis seems to possess a well-developed protein-sorting/-transport system which is evident by the presence of a large number of membrane proteins (Aruni et al., 2012). It is likely that surface and secretory proteins from F. alocis may play a role in this protein transport process. Together, these systems could possibly facilitate the efflux of reactive oxygen species.
Neutrophil and Macrophage Evasion
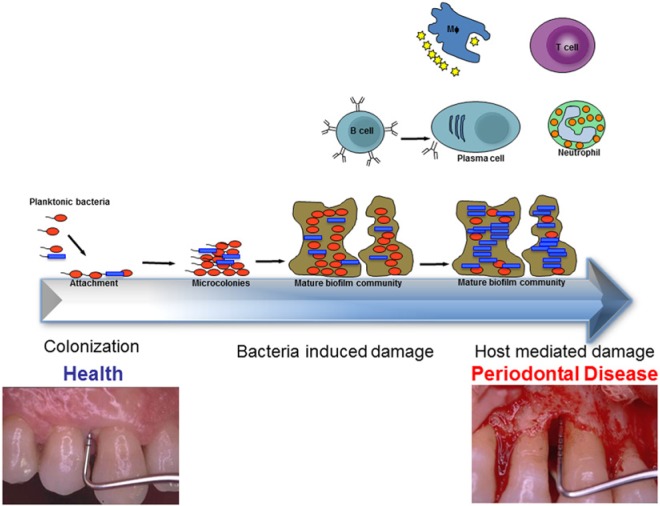
In the inflammatory response, neutrophils and macrophages are the most abundant cell types encountered at the site of infection and are responsible for the death and phagocytosis-mediated clearance of invaders (reviewed in Srinivasan, 2013); thus, immune evasion is an important mechanism in pathogen survival. In periodontitis, the attachment of planktonic bacterial species to form complex biofilm communities causes them to evade the host defense mechanisms, including neutrophils and macrophages. This affects the host, causing bacterial-induced damage that further progresses to host-mediated factors in enhancing the disease (Fig. 2). F. alocis infection in the mouse chamber model generated significant neutrophil recruitment, but selective neutrophil response was found to be dysfunctional (Wang et al., 2013). Our previous studies have shown that neutrophil-activating protein A, a possible contributor to neutrophil modulation, was abundant in the F. alocis secretome and/or associated membrane (Aruni et al., 2012). Taken together, these results suggest that F. alocis may have unique characteristics that can modulate neutrophil function leading to evasion and hence may be an important survival strategy.
Figure 2.
F. alocis as an oxidative sink. Community dynamics may play an important role in microbial survival in an oxidative stress environment of the periodontal pocket. The high abundance of F. alocis in the periodontal pocket may be consistent with the relative resistance to oxidative stress and its enhanced growth under those conditions. The beneficial relationships via the interactions with other microbes in the biofilm would stabilize the heterotypic community, including protection from the host immune response. Tissue damage observed in periodontal disease is a combination of bacterial-induced and host-mediated damage (see text for details).
Adhesion and Invasion
The genome characteristics and phenotypes attributed to F. alocis, such as adherence and invasion of host cells, are considered important to its success as a pathogen. Genes encoding for proteins such as CaaX aminopeptidases may be crucial for masking the host ubiquitin system and facilitate invasion of host cells (Price and Kwaik, 2010).
F. alocis has been shown to adhere to and invade epithelial cells (Aruni et al., 2011). These abilities, which were enhanced in the presence of P. gingivalis, may result from protein interactions modulated by bacterium-bacterium communication/signaling. A similar enhancement of invasion was observed among P. gingivalis, F. nucleatum, and P. intermedia (Saito et al., 2009), between F. nucleatum and Streptococcus cristatus (Edwards et al., 2006), and between F. nucleatum and Pseudomonas aeruginosa (Pan et al., 2009).
Filapodial projections of host microvilli were noted during F. alocis invasion (Figs. 3A, 3B) and were believed to mediate the organism’s internalization (Amano et al., 2010). In addition, vesicle-mediated internalization of P. gingivalis and F. alocis was observed during invasion of epithelial cells in co-infection studies (Aruni et al., 2011). This process may protect the pathogen after invasion and facilitate its pathogenic potential. Although membrane-ruffling mechanisms are commonly noted among Gram-positive bacteria invasion strategies, this was not the case for F. alocis. Since vesicle-mediated endocytic internalization of Gram-positive bacteria is generally mediated by types II and III exotoxins (Nitsche-Schmitz et al., 2007), it is likely that such exotoxins may contribute to the enhancement of internalization observed in the F. alocis-P. gingivalis co-culture invasion study. In fact, several exotoxins have been identified in F. alocis, but their role in invasion remains unclear.
Figure 3.

Epithelial cell interaction of F. alocis during mono and co-culture with P. gingivalis. (A) F. alocis–infected epithelial cells. The bacteria adhere to the eukaryotic cell, causing surface variations (green arrows showing adhesion of F. alocis). (B) F. alocis–infected epithelial cells showing surface variations of filamentous projections noted during co-culture with P. gingivalis strains. Orange arrows = filamentous projections; green arrows = F. alocis; blue arrows = P. gingivalis.
Proteases
To date, proteases have played a significant role in virulence modulation among the major oral pathogens (Lamont and Jenkinson, 1998; Guo et al., 2010). In Gram-positive bacteria, proteolysis plays a central role in many biological processes, such as post-translational regulation of gene expression and the processing and maturation of proteins (Laskowska et al., 1996; Gottesman et al., 1997). Expression of various surface proteins depends on proteolysis, which could strongly influence the levels of activity of proteases and their cellular localization.
The F. alocis genome possesses 15 different proteases (Aruni et al., 2012), and our study identified variations in the expression of proteases among strains of F. alocis when a low-passaged strain was compared with the type strain. Among the membrane-bound proteases of F. alocis, Caax protease (HMPREF0389_00590) could be involved in protein and/or peptide modification and secretion (Pei and Grishin, 2001). There was higher expression of Caax proteases during F. alocis co-culture (Aruni et al., 2012). Other than their metalloprotease activity, the Caax amino-terminal proteases in other oral bacteria, such as S. gordonii, have been shown to play an important role in the transport of proteins and also protect the bacteria against bacteriocins. Additionally, the Xaa–pro-dipeptidase (HMPREF0389_01538), O-sialoendopeptidase (HMPREF0389 _01445), Nlp/P60 family protein (peptidase M23/37) (HMPR EF0389_00239), and oligo endopeptidase M3 family (HMPR EF0389_00926) were shown to be present only in the membrane fraction of F. alocis. However, the protease (HMPRE F0389_00122) was identified only in the extracellular fraction. Additionally, this protease is predicted to possess a collagen peptidase function. The role of this enzyme could be important in F. alocis pathogenesis, since several oral pathogens are known to produce or induce host-derived collagenases that are implicated in tissue destruction in periodontal diseases (Kumagai et al., 2005). Some of the proteases are collagenolytic and could act as Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs), which are known to play an important role in Gram-positive bacterial virulence by mediating adherence to and colonization of host tissues as an early step toward clinical infection (Patti et al., 1994). There is also evidence to suggest that these extracellular matrix adhesion proteins can be regulated by quorum-sensing (Pinkston et al., 2011), which implies that some environmental signals can modulate their expression and hence promote adhesion and colonization.
In our preliminary studies, F. alocis co-cultured with P. gingivalis showed adhesion to epithelial cells, altering their morphology and leading to cell death. This was in contrast to mono-infections with either F. alocis or P. gingivalis that did not trigger the same morphological alteration, although these bacteria were still able to induce cell death over a longer time period (Aruni et al., 2011). Additionally, proteomic analysis of F. alocis during co-infection of epithelial cells with P. gingivalis using a tandem-mass-tagging technique revealed an increase in several membrane adhesion proteins (unpublished observations). In silico analysis of the mass spectrometry data via database search and domain prediction revealed some of these proteins to be known virulence factors; in other systems, however, the functions of several unique hypothetical proteins with transmembrane domains need to be elucidated. Taken together, this suggests that the interaction of F. alocis and P. gingivalis may result in the up-regulation of specific factor(s) that may enhance their virulence potential (Aruni et al., 2011).
Certain oral bacteria like F. nucleatum lack essential amino acid synthetic pathways and rely on the ability to import and degrade di and oligo peptides (Kapatral et al., 2002). Consistent with the asaccharolytic properties of F. alocis, several proteins that play an important role in amino acid metabolism, including many that may contribute to protein degradation, are present in its genome (http://www.ncbi.nlm.nih.gov/bioproject/46625). Even if many inherent amino acid synthesis pathways may be non-functional, the occurrence of a wide range of such dipeptidases, metalloproteases, and o-sialoglycoproteases could likely provide F. alocis with the appropriate substrates to compensate for its nutritional needs. Additionally, certain proteins—such as the oxy acyl carrier protein (HMPREF0389_ 01112), which is involved in fatty acid metabolism and not usually identified among the oral biofilm-forming pathogens, fibronectin-binding protein (HMPREF0389_00575), and dipicolinate reductase (HMPREF0389_01077), which are involved in amino acid metabolism and virulence (Berges et al., 1986)—were also identified in F. alocis (Aruni et al., 2012). Taken together, it is likely that F. alocis may be well-adapted to provide for its own nutritional needs. However, the role these systems play in bacterial community dynamics should be further elucidated.
Proteome Associated with Microbe-Host Interaction
Host responses to bacterial infections may favor survival and play a role in pathogenesis through modulation of metabolic processes. High amounts of arginine in the periodontal pocket and the abundance of F. alocis proteins involved in arginine metabolism and citrulline synthesis—such as arginine deiminase (HMPREF 0389_01584), acetyl ornithine transferase (HMPREF0389_01570) (Aruni et al., 2012), aminotransferases (HMPREF0389_01352 and HMPREF0389_01353), aminotransferase family protein (HMPREF0389_00349), arginine – tRNA ligase (HMPREF0389_ 00390), and arginine decaroboxylase (HMPREF0389_00102) (http://www.ncbi.nlm.nih.gov/bioproject/46625)—indicate that the nutritional needs of the bacterium could be adequately met during infection and are vital for its survival in the harsh microenvironment of the periodontal pocket. Furthermore, its interaction with other microbes may collectively enhance their survival. Ammonia production from arginine metabolism has been identified as an important mechanism by which oral bacteria are protected against acid killing (Burne and Marquis, 2000). Among the three key enzymes important in arginine metabolism, namely, arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase (Griswold et al., 2004), the F. alocis genome shows genes coding for arginine deiminase and carbamate kinase but not the ornithine carbamoyltransferase. It is noteworthy that P. gingivalis and F. alocis interspecies interaction resulted in the up-regulation of arginine deiminase (HMPREF_0389_01584) and carbamate kinase (HMPREF_0389_00535) in F. alocis (unpublished observations). While it may use a novel arginine catabolic pathway compared with other AAGPRs in the periodontal pocket (Uematsu et al., 2003), its relative abundance in the periodontal pocket and its enhanced ability to produce ammonia could promote species co-habitation and survival (Jakubovics et al., 2008). Because butyrate is a metabolic end-product from arginine, it could likely also have an impact on other microbial interactions, including viruses, in the oral cavity. Butyric acid produced by periodontopathic bacteria including P. gingivalis can lead to viral reactivation (Imai et al., 2012). The impact of viral infection on periodontal disease is now being recognized, since the active inflamed lesion appears to be a major site for re-activation and accumulation of Herpes virus, resulting in enhanced tissue breakdown (reviewed in Slots, 2010).
Interrogation of the F. alocis genome also revealed templates for a well-developed citrulline synthesis mechanism using arginine (Uematsu et al., 2003). Citrullination of proteins is understood to be an important post-translational modification with systemic implications (Chirivi et al., 2013). Previous studies have shown that up-regulation of peptidyl arginine deiminase (PAD) expression and the associated increase in citrullinated proteins were found in patients with rheumatoid arthritis (Foulquier et al., 2007). A mechanistic link between periodontal infection and rheumatoid arthritis has been established; collagen-induced arthritis was dependent on the expression of a unique P. gingivalis peptidylarginine deiminase (PPAD) (Maresz et al., 2013). The arginine deiminase from pathogens was shown to possess multiple regulatory roles similar to PAD function (Touz et al., 2008). Bioinformatic analyses indicate that P. gingivalis PAD has major sequence and structural homology with the F. alocis arginine deiminase enzyme (unpublished observations). It is likely that, in F. alocis, arginine deiminase could have citrullination-induced systemic implications.
It is also important to note that protein ornithine transaminase (HMPREF0389_01570), acetyl glutamate kinase (HMPREF0389_ 01569), glutamate racemase (HMPREF0389_ 00100), and aminotransferase (HMPREF0389_00478) involved in ornithine biosynthesis were identified in F. alocis. In fact, arginine deaminase (HMPREF0389_01584) involved in ornithine catabolism and urea breakdown was found in both the membrane and the extracellular fractions of F. alocis (Aruni et al., 2012), suggesting a well-developed nitrogen-assimilatory pathway that may play a role as an alternative mode of amino acid synthesis in F. alocis.
A co-infection study from our laboratory has shown that F. alocis has specific factors that could cause multiple changes in the host cell proteome (unpublished observations). It is likely that such changes may contribute to the functional modifications involved in the pathogenic process. With many unique properties, such as oxidative stress resistance (Aruni et al., 2011), and a virulence potential when associated with other red complex bacteria, F. alocis should be considered a key periodontal pathogen.
Summary
Analysis of emerging data now shows that F. alocis is a marker organism for periodontitis. It has unique characteristics that may enhance its virulence potential (Fig. 4). F. alocis could be one of the organisms that can play a pivotal role in community dynamics, establishing synergistic partnerships with other pathogenic oral bacteria during the disease state. In comparison with other Gram-positive bacteria of the oral cavity, the variations induced in the host proteome during F. alocis synergism could lead to many systemic host responses. Therefore, the significance of F. alocis putative virulence factors, which may trigger the key host response, deserves further intensive study. It is noteworthy that F. alocis is one of only a few organisms associated with both generalized and localized aggressive periodontitis (LAP) in addition to peri-implantitis and endodontic infections.
Figure 4.
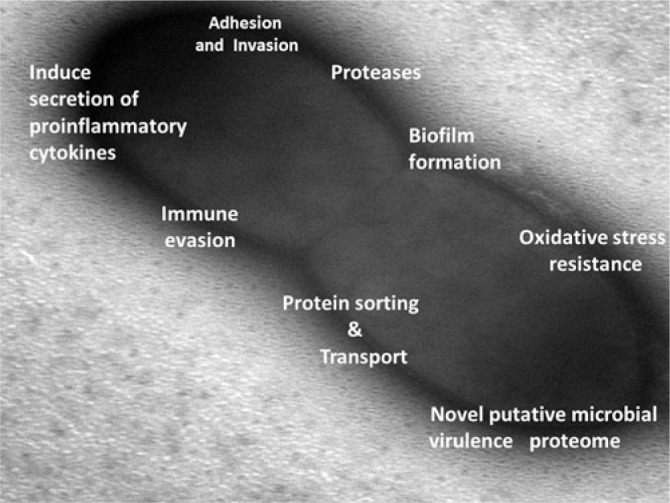
Pathogenic properties of F. alocis (see text for details).
Acknowledgments
Due to the Journal’s editorial limitations, we apologize to our colleagues/fellow scientists for not including their references in the review.
Footnotes
This work was supported by Loma Linda University and Public Health Service Grants DE13664, DE019730, DE019730 04S1, DE022508, and DE022724 from the National Institute of Dental and Craniofacial Research (NIDCR) (to H.M.F.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Amano A, Furuta N, Tsuda K. (2010). Host membrane trafficking for conveyance of intracellular oral pathogens. Periodontol 2000. 52:84-93. [DOI] [PubMed] [Google Scholar]
- Aruni AW, Roy F, Fletcher HM. (2011). Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun 79:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni AW, Roy F, Sandberg L, Fletcher HM. (2012). Proteome variation among Filifactor alocis strains. Proteomics 12:3343-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges DA, DeWolf WE, Jr, Dunn GL, Newman DJ, Schmidt SJ, Taggart JJ, et al. (1986). Studies on the active site of succinyl-CoA:tetrahydrodipicolinate N-succinyltransferase. Characterization using analogs of tetrahydrodipicolinate. J Biol Chem 261:6160-6167. [PubMed] [Google Scholar]
- Bingham CO, 3rd, Moni M. (2013). Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol 25:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Marquis RE. (2000). Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193:1-6. [DOI] [PubMed] [Google Scholar]
- Cato EP, Moore LV, Moore WE. (1985). Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol 35:475-477. [Google Scholar]
- Chirivi RG, van Rosmalen JW, Jenniskens GJ, Pruijn GJ, Raats JM. (2013) Citrullination: a target for disease intervention in multiple sclerosis and other inflammatory diseases? J Clin Cell Immunol 4:146. [Google Scholar]
- Darveau RP, Hajishengallis G, Curtis MA. (2012). Porphyromonas gingivalis as a potential community activist for disease. J Dent Res 91:816-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. (2010). The human oral microbiome. J Bacteriol 192:5002-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Grossman TJ, Rudney JD. (2006). Fusobacterium nucleatum transports non invasive Streptococcus cristatus into human epithelial cells. Infect Immun 74:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. (2012). Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 91:914-920. [DOI] [PubMed] [Google Scholar]
- Eley BM, Cox SW. (1992). Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidyl peptidase IV-like activities in gingival crevicular fluid: a comparison of levels before and after periodontal surgery in chronic periodontitis patients. J Periodontol 63:412-417. [DOI] [PubMed] [Google Scholar]
- Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al BR, Mechin MC, et al. (2007). Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum 56:3541-3553. [DOI] [PubMed] [Google Scholar]
- Genco RJ, Van Dyke TE. (2010). Prevention: reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol 7:479-480. [DOI] [PubMed] [Google Scholar]
- Gomes BP, Jacinto RC, Pinheiro ET, Sousa EL, Zaia AA, Ferraz CC, et al. (2006). Molecular analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola associated with primary endodontic infections and failed endodontic treatment. J Endod 32:937-940. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. (1997). Protein quality control: triage by chaperones and proteases. Genes Dev 11:815-823. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold A, Chen YY, Snyder JA, Burne RA. (2004). Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl Environ Microbiol 70:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Nguyen KA, Potempa J. (2010). Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 54:15-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Molec Oral Microbiol 27:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima R, Takahashi H, Namme R, Ikegami S, Yamazaki M. (2004). Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett 561:163-166. [DOI] [PubMed] [Google Scholar]
- Imai K, Ogata Y, Ochiai K. (2012). Microbial interaction of periodontopathic bacteria and Epstein–Barr virus and their implication of periodontal diseases. J Oral Biosci 54:164-168. [Google Scholar]
- Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. (2008). Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol 190:3646-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalava J, Eerola E. (1999). Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol 49(Pt 4):1375-1379. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, et al. (2002). Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr (2002). Communication among oral bacteria. Microbiol Mol Biol Rev 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. (2006). Bacterial interactions and successions during plaque development. Periodontol 2000. 42:47-79. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. (2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471-480. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Yagishita H, Yajima A, Okamoto T, Konishi K. (2005). Molecular mechanism for connective tissue destruction by dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis. Infect Immun 73:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. (2003). New bacterial species associated with chronic periodontitis. J Dent Res 82:338-344. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. (2006). Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowska E, Kuczynska-Wisnik D, Skorko-Glonek J, Taylor A. (1996). Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol 22:555-571. [DOI] [PubMed] [Google Scholar]
- Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, et al. (2013). Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD). PLoS Pathog 9:e1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. (2011). Filifactor alocis interactions with gingivalis epitheiall cells. Mol Oral Microbiol 26:365-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche-Schmitz DP, Rohde M, Chhatwal GS. (2007). Invasion mechanisms of Gram-positive pathogenic cocci. Thromb Haemost 98:488-496. [PubMed] [Google Scholar]
- Oliver RC, Brown LJ, Löe H. (1991). Variations in the prevalence and extent of periodontitis. J Am Dent Assoc 122:43-48. [DOI] [PubMed] [Google Scholar]
- Pan Y, Teng D, Burke AC, Haase EM, Scannapieco FA. (2009). Oral bacteria modulate invasion and induction of apoptosis in HEp-2 cells by Pseudomonas aeruginosa. Microb Pathog 46:73-79. [DOI] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ, Hook M. (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48:585-617. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. (2001). Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci 26:275-277. [DOI] [PubMed] [Google Scholar]
- Pinkston KL, Gao P, Diaz-Garcia D, Sillanpaa J, Nallapareddy SR, Murray BE, et al. (2011). The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J Bacteriol 193:4317-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöllänen MT, Paino A, Ihalin R. (2013). Environmental stimuli shape biofilm formation and the virulence of periodontal pathogens. Int J Mol Sci 14:17221-17237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Kwaik YA. (2010). Exploitation of poly ubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front Microbiol 1:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Inagaki S, Ishihara K. (2009). Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb Pathog 47:329-333. [DOI] [PubMed] [Google Scholar]
- Schlafer S, Riep B, Griffen AL, Petrich A, Hubner J, Berning M, et al. (2010). Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Radman M. (2011). Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. (2010). Human viruses in periodontitis. Periodontol 2000. 53:89-110. [DOI] [PubMed] [Google Scholar]
- Srinivasan PC. (2013). Neutrophils – the sentinels of periodontal innate immunity. J Clin Cell Immunol S13:1-6. [Google Scholar]
- Tamura N, Ochi M, Miyakawa H, Nakazawa F. (2013). Analysis of bacterial flora associated with peri-implantitis using obligate anaerobic culture technique and 16S rDNA gene sequence. Int J Oral Maxillofac Implants 28:1521-1529. [DOI] [PubMed] [Google Scholar]
- Thornton-Evans G, Eke P, Wei L, Palmer A, Moeti R, Hutchins S, et al. (2013). Periodontitis among adults aged >/=30 years - United States, 2009-2010. MMWR Surveill Summ 62(Suppl 3):129-135. [PubMed] [Google Scholar]
- Touz MC, Ropolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, et al. (2008). Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J Cell Sci 121(Pt 17):2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu H, Sato N, Hossain MZ, Ikeda T, Hoshino E. (2003). Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic Gram-positive rods in periodontal pockets. Arch Oral Biol 48:423-429. [DOI] [PubMed] [Google Scholar]
- Wade WG. (2011). Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol 38(Suppl 11):7-16. [DOI] [PubMed] [Google Scholar]
- Wang BY, Wu J, Lamont RJ, Lin X, Xie H. (2009). Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol 47:3902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. (2013). Oral community interactions of Filifactor alocis in vitro. PLoS One 8:e76271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jotwani R, Le J, Krauss JL, Potempa J, Coventry SC, et al. (2014). Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect Immun 82:1205-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lin X, Wang BY, Wu J, Lamont RJ. (2007). Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 153(Pt 10):3228-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hou BX, Zhao HY, Sun Z. (2012). Microbial diversity in failed endodontic root-filled teeth. Chin Med J (Engl) 125:1163-1168. [PubMed] [Google Scholar]