Abstract
The therapeutic value of many growth factors is often hindered by the narrow therapeutic index and sustained concentrations required for efficacy. Controlled release approaches provide a valuable tool to achieve these goals; however, growth factor stability must be maintained. Repifermin® is a truncated form of fibroblast growth factor-10, also known as keratinocyte growth factor-2, that exhibits promise in wound healing applications; however, controlled release formulation presents a challenge for this labile protein. Taking advantage of the heparin-binding motif of this class of biopharmaceuticals, Repifermin® was effectively stabilized and packaged in polyelectrolyte complexes. In the presence of dextran sulfate, the unfolding temperature of this growth factor was increased by ~10°C as confirmed by a variety of spectroscopic techniques. Dextran sulfate with bound Repifermin® was then complexed with several polycations (chitosan, poly-L-lysine, and polyethylenimine) resulting in the formation of ~250 nm polyelectrolyte complexes that entrapped the protein with ~70–80% efficiency. Release was controlled for more than 10 days and the mitogenic activity of Repifermin® on human umbilical cord vascular endothelial cells was significantly enhanced, whereas no effect was noted for free Repifermin®.
Keywords: polyelectrolyte complex, fibroblast growth factor, nanoparticle, controlled release, stability
Introduction
Fibroblast growth factor-10 (FGF-10), also known as keratinocyte growth factor-2 (KGF-2), is a 208 amino acid polypeptide (Mw 19,000) belonging to the FGF family. These structurally related proteins bind to polyanions including heparin and are involved in a variety of physiological and pathological processes [1]. FGF-10 activities include epithelial cell proliferation and migration as well as less pronounced endothelial stimulating effects. Repifermin® (Human Genome Sciences, Inc.) is a truncated form of recombinant FGF-10 (140 amino acids, Mw 16,000) that retains the physiological and biological activity of full-length FGF-10 [2]. Unfortunately, FGF-10 is unstable, similar to FGF-1 [3], and is relatively rapidly eliminated if administered by bolus intravenous injections [4]. In addition, the mitogenic activities of this protein in vitro have been somewhat inconclusive due to its poor stability (Tm ~37°C) [3].
Administration of Repifermin® has been identified as a potential therapeutic approach in promoting angiogenesis [5, 6] and accelerating healing of wounds [7, 8]. The action of FGF-10 in vivo and in vitro is presumed to be a result of its mitogenic and chemotactic specificity for epithelial cells [9]; however, reports of angiogenesis in vivo suggested the possibility that stabilizing Repifermin® and sustaining its release may reveal in vitro mitogenic activity on endothelial cells. Growth factors such as FGF-10 commonly suffer from in vivo short half-lives, which encumbers the ability to sustain local concentrations to maintain pharmacological effects [10, 11]. These properties further hinder formulation stability since sustained treatment may be desirable in comparison to frequent bolus dosing. Recently, Middaugh and others have reported dramatically enhanced thermal stability of Repifermin® by formulating with various polyanions [3], thus, providing a premise for translating these solution based stabilization strategies into a particulate dosage form.
Recent research has been developing several strategies for controlled release vehicles that effectively deliver growth factors to overcome the current shortcomings of fibroblast growth factor therapy. In many cases, the unique ability of polyanions to bind to fibroblast growth factors has been used to develop drug delivery systems for the controlled release of heparin-binding growth factors. For instance, heparin has been well established to improve the stability of many growth factors including acidic-FGF under heat and acid treatment [12, 13]. It was also reported that the binding of basic-FGF to heparin–Sepharose beads could achieve prolonged storage in a microsphere controlled release matrix [14]. Some researchers have taken advantage of the heparin binding domain to create gel networks that bind and stabilize various FGF proteins for controlled release [15].
In this work, we employed an aqueous process to form polyelectrolyte complexes by using the biocompatible polymers dextran sulfate and chitosan. Dextran sulfate (DS) is a biodegradable negatively charged polymer, which was previously shown to bind and stabilize FGF-10 [16–19]. Three polycations including chitosan (CS), polyethylenimine (PEI), and poly-L-lysine (PLL) were employed to condense DS leading to the formation of polyelectrolyte complexes. This work is an extension of a previous publication utilizing polyelectrolyte complexes for vascular endothelial growth factor (VEGF) delivery [20], which was originally derived from technology developed by Middaugh and others [18, 21, 22]. Among the polyelectrolyte complexes studied, dextran sulfate and chitosan were especially desirable due to their biodegradability and low toxicity. In particular, dextran sulfate/chitosan nanoparticles possessed the highest FGF-10 encapsulation efficiency (~78%) and released FGF-10 for over 10 days. Based on an in vitro endothelial cell proliferation assay using FGF-10 PEC nanoparticles, it was confirmed that the labile FGF-10 proteins maintain biological activity as released from the PEC nanoparticles. Nanoscale materials capable of stabilizing FGFs provide an attractive formulation option for inclusion in topical or injectable vehicles.
Materials and Methods
Materials
Fibroblast growth factor-10 (FGF-10, KGF-2) was graciously provided by Human Genome Sciences (HGS) in a truncated form (Repifermin®), which retains the pharmacological and biological activity of full-length FGF-10. Chitosan (Mw = 15 kDa, 84% deacetylated, Polysciences, Inc.), dextran sulfate (Mw = 500 kDa, Fisher Scientific), polyethylenimine (Mw = 10 kDa, Aldrich), and poly-L-lysine (Mw = 10 kDa, Sigma) were used as obtained without further purification. Zinc sulfate heptahydrate (Sigma) was used as a nanoparticle crosslinker of sulfate side groups in some experiments. 8-Anilino-1-naphthalenesulfonic acid (ANS) was purchased from Sigma. Microsep™ centrifugal devices (Pall Life Sciences), dialysis membranes (Spectrum), side-A-lyzer dialysis cassettes (Pierce), and mannitol (Sigma) were used during particle purification.
Formulation of polyelectrolyte complexes
FGF-10 was received in HGS formulation buffer and stored at −80 °C. A sample of FGF-10 was dialyzed into PBS buffer using a dialysis cassette with a 10,000 MWCO (Pierce) overnight. The concentration of the recovered sample (~1 mg/mL) was determined by UV spectroscopy. To prepare FGF-10-loaded polyelectrolyte complex (PEC) nanoparticles, 80 μL of the recovered sample was added to 800 μL of 1% (w/v) dextran sulfate solution (in water) and stirred at 600 rpm for 30 min. Then, 1.6 mL of the appropriate polycationic solution (0.1% w/v) was added dropwise and stirred for another 5 minutes. Chitosan was dissolved in 0.1 M HAc (pH adjusted to 6 using 1 M NaOH) and added dropwise to the dextran sulfate solution. Finally, 80 μL of 1 M zinc sulfate solution was added and stirred for 30 min. The prepared particles were dialyzed into water with 5% mannitol, for 24 hours in the dark. The dialyzed sample was filtered through a 0.8 μm filter and lyophilized to obtain a dry powder.
Physical characterization of polyelectrolyte complexes
The mean particle size was determined by dynamic light scattering experiments employing a ZetaPALS Zeta Potential Analyzer (Brookhaven Instruments Corp.). An aliquot of lyophilized particles was suspended in water and three consecutive 1 minute measurements were obtained by detecting light scattering at a 90° angle. The mean effective diameter and polydispersity were determined by the method of cumulants. The zeta potential of the particles was investigated by phase analysis light scattering using the same instrument. Samples were prepared by dispersing 5 mg of the lyophilized nanoparticles in 1 mL of deionized water (Barnstead Nanopure). Three measurements were taken for each sample.
Biophysical study of FGF-10 and FGF-10 bound to DS
Circular Dichroism
Far-UV spectra were recorded using a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller (Jasco, Easton, MD). Far-UV spectra were collected at a scan rate of 10 nm/min and a resolution of 0.5 nm using a response time of 2 sec and a bandwidth of 1 nm. To monitor changes in the secondary structure of FGF-10 bound with DS, CD spectra were obtained for FGF-10 in the presence of 50-fold-weight excess of dextran sulfate. Spectra for FGF-10 bound with DS were background corrected with respect to DS in buffer. The thermal unfolding of the proteins was evaluated by monitoring the CD signal at 206 nm and 230 nm over a temperature range of 10–95°C with a resolution of 0.5 at a thermal ramp rate of 15°C/h. The protein concentration was 0.1 mg/ml for all measurements. CD signal was converted to molar ellipticity, and the thermal transitions were analyzed using the Jasco Spectra Manager and MicroCal Origin v.7.0 software.
Fourier Transform Infrared Spectroscopy
The secondary structures of FGF-10 and DS-bound FGF-10 were evaluated using Fourier transform infrared (FTIR) spectroscopy. Measurements were done using attenuated total reflectance (ATR) mode employing a 45° ZnSe trough plate with volatile liquid cover. Data were collected using a Bomem PROTA FTIR protein analyzer (Norwalk, CT) to acquire spectra using 256 scans from 700–4000 cm−1 with a resolution of 4 cm−1. A spectrum of water was subtracted from each sample spectrum by eliminating the interference of water at 2200 cm−1. The resulting spectra were smoothed with a Savitsky-Golay function. Secondary derivative analysis was employed to identify peaks in the amide-I and amide-II region. A Gaussian curve fit was performed between 1700–1500 cm−1 using the software GRAMS/AI version 7.00 (Thermo Galactic).
High-resolution Derivative UV Absorbance Spectroscopy
UV absorbance spectra of both FGF-10 and DS-bound FGF-10 were conducted using an Agilent 8453 diode array UV-visible spectrophotometer (Palo Alto, CA). Spectra were collected at an experimental resolution of 1 nm in the range of 200 to 400 nm under thermal denaturation using an integration time of 25 s over a range of 10 °C to 90 °C at 2.5 °C intervals. Temperature-induced aggregation of FGF-10 was studied simultaneously with the secondary-derivative UV data by monitoring the turbidity at 350 nm (OD350). A 5-min equilibration period was included before collection of each spectrum to allow for sufficient time to achieve thermal homogeneity. Spectral analysis was conducted using Chemstation™ software (Agilent). Second derivative spectra were calculated using a nine-point data filter and fifth degree Savitzky-Golay polynomial, and subsequently fitted to a cubic function, with 99 interpolated points per raw data point, permitting 0.01-nm resolution. Peak positions were determined from the interpolated curves using Microcal Origin™7.0. A 1 cm path length capped cuvette was used with a total sample of 200 μl of protein sample at 0.2 mg/ml. The spectrum of FGF-10-DS was also conducted in the same manner except that the DS solution was used as a blank to remove interference from DS.
Dynamic Light Scattering
The effective diameter of FGF-10 before and after binding with DS was determined by dynamic light scattering using a Brookhaven BI-9000AT digital autocorrelator and a BI-200SM goniometer (Brookhaven Instruments Corporation) equipped with a 50 mW He-Ne laser operating at 532 nm. All the solutions were filtered through 0.22 um filters to remove any extraneous large particles. Data were collected at a scattering angle of 90 °C over a temperature range of 10–80 °C at a sample concentration of 4 mg/ml. For each sample the data were collected continuously for five 1-min intervals. The mean diameter of the sample was obtained from the Stokes-Einstein equation using the method of cumulants.
Intrinsic Fluorescence Spectroscopy
A FGF-10 protein concentration of 0.2 mg/ml was used for the intrinsic fluorescence studies. For FGF-10 bound with DS, an excitation wavelength of 295 nm was employed, and emission spectra were recorded from 290 to 440 nm using a QuataMaster spectrofluorometer equipped with a Peltier thermostated cuvette holder (Photon Technologies International). Data were collected every 2.5 °C from 10 °C to 90 °C with 300 s of thermal equilibrium at each temperature. The data were processed using Origin version 7.0 software. Initially, an 11-point Savitsky-Golay smoothing function was applied to all spectra. The peak positions were determined using first derivative analysis.
ANS Fluorescence Spectroscopy
ANS binding to FGF-10 as a function of temperature was examined at an optimized ANS:protein molar ratio of 10:1. FGF-10 samples in the presence of ANS were excited at 375 nm with a 3 nm slit width and emission monitored from 400 to 600 nm with 3 nm slit widths. The weak emission spectrum of ANS in buffer was used as a blank.
FGF-10 loading, entrapment efficiency, and release from PEC nanoparticles
FGF-10 loading into polyelectrolyte nanoparticles was calculated by an indirect method where FGF-10 concentration was determined in purified supernatant and subtracted from the initial amount entered into the experiment. Nanoparticles were centrifuged at 15,000 g for 45 min and the supernatant separated for analysis. The amount of FGF-10 in the original FGF-10 sample used to make particles and the supernatant were determined by UV spectroscopy (Agilent 8453) based on the standard curve. To determine the FGF-10 release kinetics, ~30 mg of lyophilized sample was resuspended in 1 ml of PBS buffer (pH 7.0) and shaken at 200 rpm at 37 °C. At specified times, the vial was centrifuged at 13,000 rpm for 10 min and the supernatant was collected and analyzed by BCA assay (Pierce).
Stability of FGF-10 loaded PEC nanoparticles
To study the colloidal stability of FGF-10 loaded PEC nanoparticles, 50 mg of lyophilized nanoparticle powders were redispersed in 1 ml of DMEM medium supplemented with 10% serum and was incubated at room temperature. Particle size and polydispersity was determined every 30 min over a range of 12 hours (Brookhaven Instruments Corp.).
Cytotoxicity of DS, CS, PEI and PLL
The determination of cell viability is a common assay to evaluate the in vitro cytotoxicity of biomaterials. In the present study, cell viability was assessed using a CellTiter 96® AQueous Cell Proliferation assay kit (Promega) based on the standard [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) assay where the MTS tetrazolinium compound is bioreduced into a colored formazan product by metabolically active cells. Briefly, human umbilical cord vascular endothelial cells (HUVEC) were seeded in 96-well tissue culture plates at a density of 8,000 cells/well and cultured in DMEM medium supplemented with 10% heat inactivated bovine serum. Cells were then incubated with 100 μL of sample containing cell culture medium for another 24 hours. At the completion of the experiment, 20 μL of MTS and phenazine methosulfate (PMS) were added and incubated for four hours before reading the absorbance at 490 nm with a 96-well plate reader (Molecular Devices Corporation). Cell viability was expressed as a percentage of absorbance relative to control, the control comprising cells not exposed to the particular material. Experiments were performed in eight replicate wells for each sample.
Mitogenic bioassay of FGF-10 and FGF-10 loaded PEC nanoparticles
The bioactivity of FGF-10 either in solution or released from PEC nanoparticles was determined using the cell proliferation assay as described above. Briefly, HUVEC cells were seeded in 96-well tissue culture plates in DMEM medium supplemented with 2% heat inactivated bovine serum at a density of 2,000 cells/well. Cells were incubated with 20 ng/mL of FGF-10 either in solution or in nanoparticles. The Cell Titer 96® AQueous Cell Proliferation assay kit (Promega) was used to quantify the cell proliferation after incubation with FGF-10 or FGF-10 loaded nanoparticles. The cell proliferation assay was determined at different time points over a period of 5 days post-treatment. At ~5 days, cells began to reach confluency and contact inhibition of cell proliferation confounded further analysis. Statistical significance was calculated using one-way analysis of variance (ANOVA) with Tukey comparisons post-test.
Results
Stabilization of FGF-10 by binding dextran sulfate
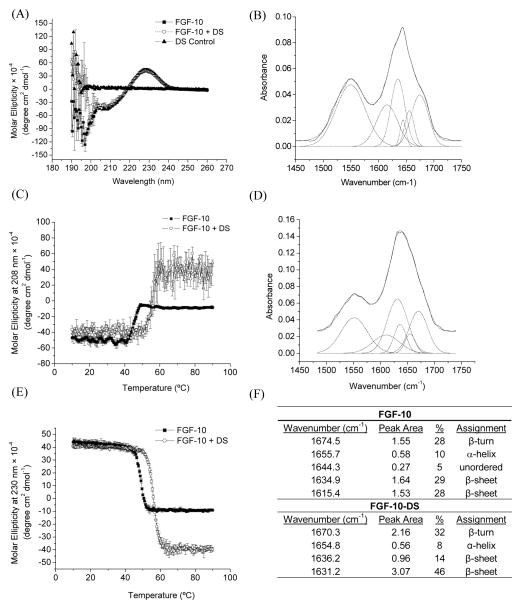
Far-UV CD was employed to explore the secondary structure of FGF-10 in the presence and absence of dextran sulfate and within nanoparticles. The unbound form of FGF-10 possessed a negative peak around 206 and a positive peak at 230 nm (Figure 1A). The negative peak at 206 nm and positive peak at 230 nm reflect the presence of β–trefoil structure and the optically active Trp residues [3]. The secondary structure was quantified by deconvolution of the CD spectrum using the program CTSSTR provided by Dichroweb software. The result revealed a high percentage of β-sheet structure (42%) and β-turn structure (18%). FGF-10 bound to dextran sulfate presented similar CD spectra (β-sheet: 45%, β-turn: 19%), which overlapped quite well with the unbound form of FGF-10, indicating insignificant changes of the secondary structure of FGF-10 after binding to dextran sulfate (Figure 1A).
Figure 1.
(A) CD spectra of FGF-10, FGF-10 bound with DS, and DS. (C) The effect of temperature on the molar ellipticity of FGF-10 as monitored at 208 nm by CD (n=3). (E) The effect of temperature on the molar ellipticity of FGF-10 as monitored at 230 nm by CD (n=3). FTIR spectrum of (B) FGF-10 and (D) FGF-10 bound with DS. (F) Structural assignment for FGF-10 and FGF-10 bound with DS based on FTIR deconvolution.
Temperature-induced changes in the secondary structure of FGF-10 were also evaluated by monitoring changes in the CD signal at 206 nm and 230 nm (Figure 1C and E). For FGF-10, a transition temperature at around 40 °C was observed for the CD signal at both of the wavelengths. In contrast, after binding FGF-10 with DS, the transition temperature was improved to ~50 °C, suggesting that DS effectively enhances the thermal stability of the secondary structure of FGF-10. The high molecular weight of DS employed did not lead to any large aggregates (indicative of protein-polymer crosslinking) within the solution and stabilized FGF-10 to a similar degree as other reported polyanions of lower molecular weight [3].
FTIR was employed in this study to further explore the structures of FGF-10 in the presence and absence of dextran sulfate. The FGF-10 spectrum presents the typical signature amide I (1700-1600 cm−1) and amide II bands (1600-1500 cm−1) of the protein (Figure 1B). These bands were deconvoluted to determine the secondary structure composition of FGF-10 in the presence and absence of DS. This analysis demonstrated that FGF-10’s secondary structure consists primarily of β-sheet (57%) and β-turn (28%) (Figure 1F), which was relatively consistent with the CD analysis. The spectrum of FGF-10 bound with DS (Figure 1D) were deconvoluted and also demonstrated mainly β-sheet (50%) and β-turn (32%) structures, thus, confirming that there was negligible alteration of secondary structure upon FGF-10 binding.
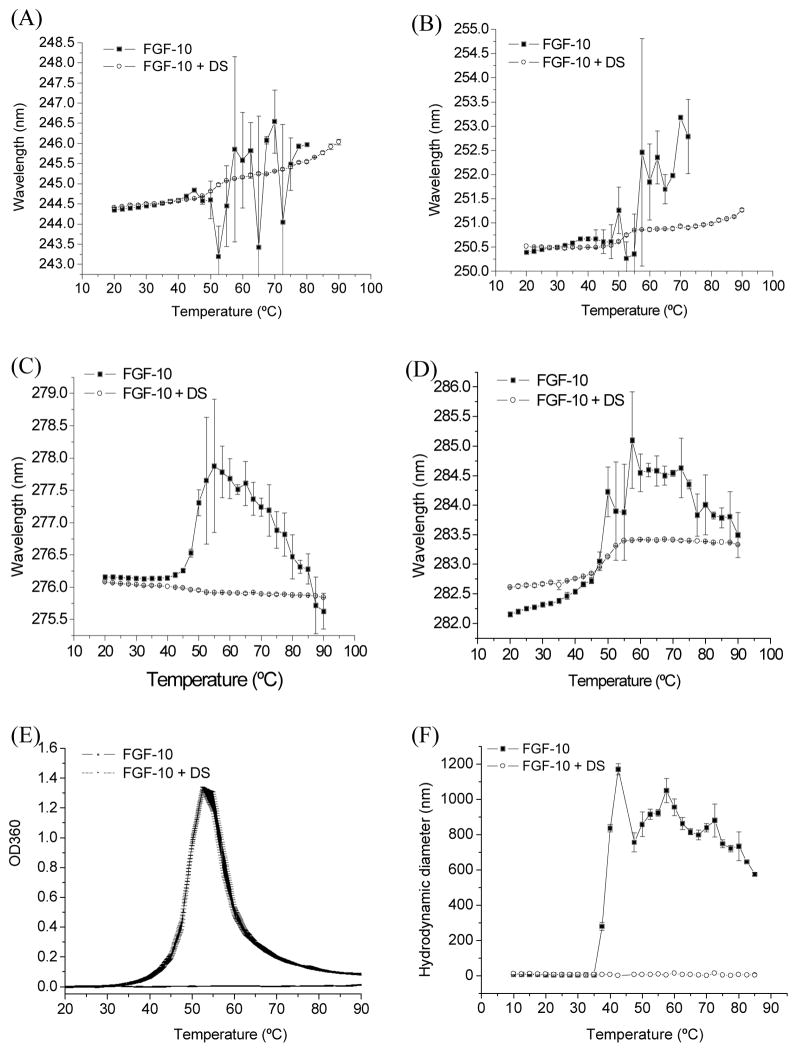
The effect of temperature on a protein’s secondary-derivative UV spectra reflects the changes in the microenvironment of aromatic protein residues providing a mechanism to evaluate the tertiary structure of a protein. The four distinct negative peaks of the second-derivative UV absorbance spectrum of FGF-10 were monitored as a function of temperature. At 10 °C, the negative peaks occurred at around 244 nm (Phe), 250 nm (Phe), and 276 (Tyr), and 282 (Tyr/Trp). With the increase of temperature, the peak position for these amino acids all shifted to longer wavelengths (Figure 2A–D) followed by very significant fluctuations with a transition temperature around 40 °C. The prominent variation in the spectrum was attributed to the aggregation of the protein at higher temperatures. For all three amino acid peaks, FGF-10 bound with DS generally showed a decreased red shift and less variation over the temperature range studied. These results suggested that the tertiary structure of FGF-10 was thermally stabilized through binding to dextran sulfate. UV data further confirmed that that a major source of FGF-10 instability was protein aggregation. Both the turbidity assay at 360 nm and the dynamic light scattering study (Figure 2E and F) indicated that dextran sulfate could effectively inhibit the aggregation of FGF-10. During the temperature range studied, no significant aggregation of FGF-10 was observed when DS was present.
Figure 2.
UV absorbance revealed the effect of temperature on the second derivative peak (A) at 245 nm, (B) 250 nm (Phe), (C) at 276 nm (Tyr) and (D)282 nm (Tyr/Trp) (for FGF-10 and FGF-10 bound with DS (n=3). (E) Temperature induced aggregation as reflected by OD at 360 nm (n=3). (F) Mean hydrodynamic diameter of FGF-2 and FGF-10 bound with DS as a function of temperature determined by dynamic light scattering measurements (n=3).
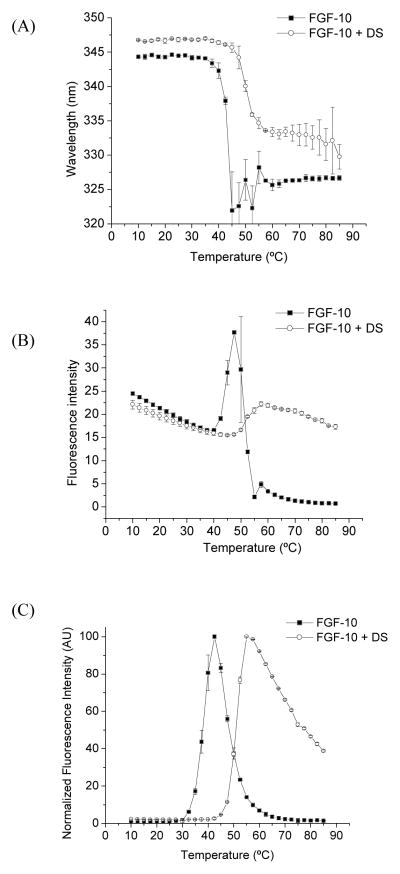
FGF-10 protein has two Trp residues. The Trp intrinsic fluorescence spectroscopy was employed to evaluate the stability of tertiary structure of FGF-10 at increasing temperatures with and without DS (Figure 3A and B). Starting at ~36 °C, the peak position starts to blue shift indicating that the Trp residue was exposed to a more hydrophobic environment, which could also be attributed to the aggregation of the protein. Binding FGF-10 with DS enhanced the transition temperature to about 45 °C. 1,8-ANS is essentially nonfluorescent in water, but becomes appreciably fluorescent when bound to protein non-polar regions; therefore, ANS was used as a sensitive extrinsic probe of protein unfolding. When FGF-10 protein was thermally stressed, the ANS fluorescence increased substantially at 32 °C (Figure 3C), due to the initial unfolding of the protein at this temperature. On the other hand, for FGF-10 bound with DS, the transition temperature for ANS fluorescence was enhanced to 42 °C. According to other reports, FGF-10 may exist as molten globule state at the transition temperature as indicated by the enhanced exposure of hydrophobic core regions that are inaccessible to the ANS dye in the native structure [3]. This assay provides strong evidence to validate that the thermal stability of the tertiary structure of FGF-10 protein was improved though binding to dextran sulfate.
Figure 3.
Tryptophan fluorescence peak position (A) and intensity (B) as a function of temperature for FGF-10 and FGF-10 bound with DS (n=3). (C) ANS dye-binding fluorescence intensity as a function of temperature for FGF-10 and FGF-10 bound with DS (n=3).
Physicochemical characterization of polyelectrolyte complexes and FGF-10
In the present study, dextran sulfate was complexed with three polycations (chitosan, polyethylenimine, and poly-L-lysine) to evaluate the effect of these biomaterials on particle size, polydispersity, zeta potential, and FGF-10 encapsulation efficiency. Our previous work optimized PEC nanoparticle fabrication conditions by varying polyelectrolyte molecular weights and concentrations to form ~200–300 nm particles [20]. The results for chitosan, polyethylenimine or poly-L-lysine complexed with dextran sulfate to encapsulate FGF-10 coincided with our previous report on vascular endothelial growth factor. A Brookhaven ZetaPALS Zeta Potential Analyzer was used to analyze the particle size and size distribution of ~5 mg/mL solutions of the complexes in deionized water (Table 1). Poly-L-lysine nanoparticles demonstrated the smallest particle size under these particular fabrication conditions. PEC formulations exhibited moderate polydispersity values (< 0.24) and possessed negative surface charges. The encapsulation of FGF-10 slightly increased the particle size and decreased the zeta potentials of PECs.
Table 1.
Properties of polyelectrolyte nanoparticles.
| Nanoparticle formulation | Diameter (nm) | Polydispersity | Zeta potential (mV) | Entrapment efficiency (%) |
|---|---|---|---|---|
| CS-DS | 239 ± 4 | 0.18 ± 0.02 | −18.4 ± 0.3 | |
| FGF-10 CS-DS | 306 ± 10 | 0.23 ± 0.02 | −15.5 ± 0.5 | 78.3 ± 6.4 |
| PEI-DS | 209 ± 3 | 0.10 ± 0.01 | −16.0 ± 1.9 | |
| FGF-10 PEI-DS | 257 ± 1 | 0.04 ± 0.01 | −15.2 ± 0.2 | 68.4 ± 3.0 |
| PLL-DS | 173 ± 3 | 0.14 ± 0.02 | −18.7 ± 2.3 | |
| FGF-10 PLL-DS | 210 ± 2 | 0.17 ± 0.03 | −10.3 ± 2.3 | 75.2 ± 3.7 |
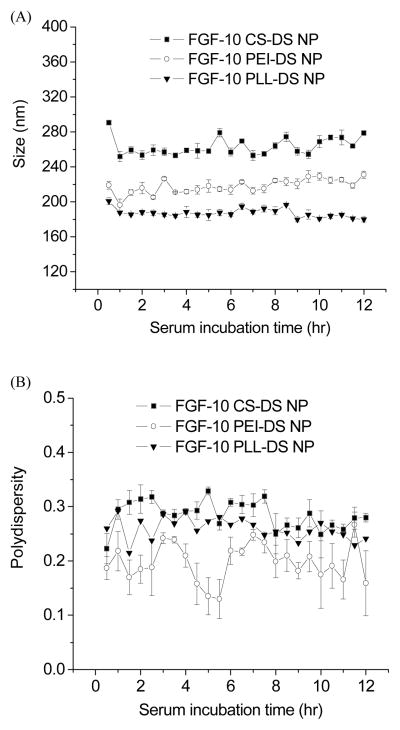
All three PEC nanoparticles made from polycations (chitosan, polyethylenimine, and poly-L-lysine) were stable in the presence of serum (Figure 4). When these nanoparticles were incubated with DMEM cell culture medium in the presence of 10% serum, the particle size remained unchanged, indicating that there was no significant agglomeration of PEC nanoparticles when incubated with serum.
Figure 4.
(A) Particle size and (B) polydispersity for FGF-10 CS-DS (■), FGF-10 PEI-DS (○) and FGF-10 PLL-DS (▼) nanoparticles as a function of time incubated in the presence of 10% serum (n=3).
FGF-10 entrapment efficiency, release kinetics and bioactivity
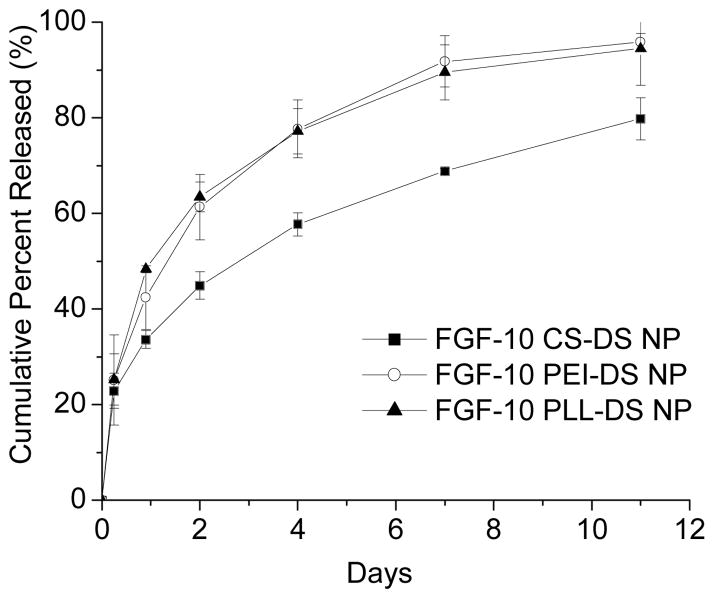
PEC nanoparticles prepared using chitosan showed the highest entrapment efficiency of 78.3% (Table 1). In all cases, the release profiles of FGF-10 from nanoparticles demonstrated an initial burst release of ~12 % of FGF-10 in the first 2 h followed by a slower sustained release (Figure 5). PEI-DS and PLL-DS entrapped FGF-10 protein at efficiency of 68.4 and 75.2 %, respectively (Table 1). Nearly all (≥80%) of the encapsulated protein was released over a period of 11 days for all formulations. The profiles also suggested the likelihood of concentration-driven release.
Figure 5.
FGF-10 release profiles for CS-DS (■), PEI-DS (○), and PLL-DS (▼) showed similar trends proportional to the encapsulation efficiency of each formulation (n=3).
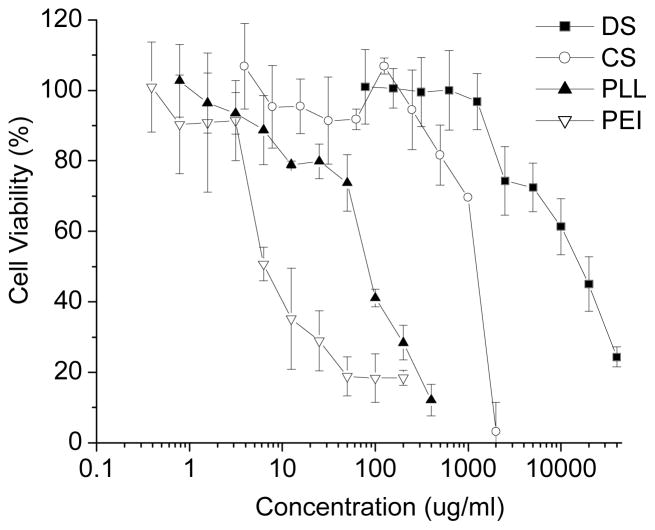
The cytotoxicity of individual components of the PEC nanoparticles were tested against HUVEC cells. Among all the materials used, DS and CS were the least toxic to these cells, with IC50 values around 10 mg/ml and 1 mg/ml, respectively (Figure 6). These two materials may be regarded as relatively biocompatible and prefered for intravenous injections as compared to polyethlenimine and poly-L-lysine.
Figure 6.
Cytotoxicity of CS, PEI, PLL and DS towards HUVEC as determined by the MTS assay. (n=8).
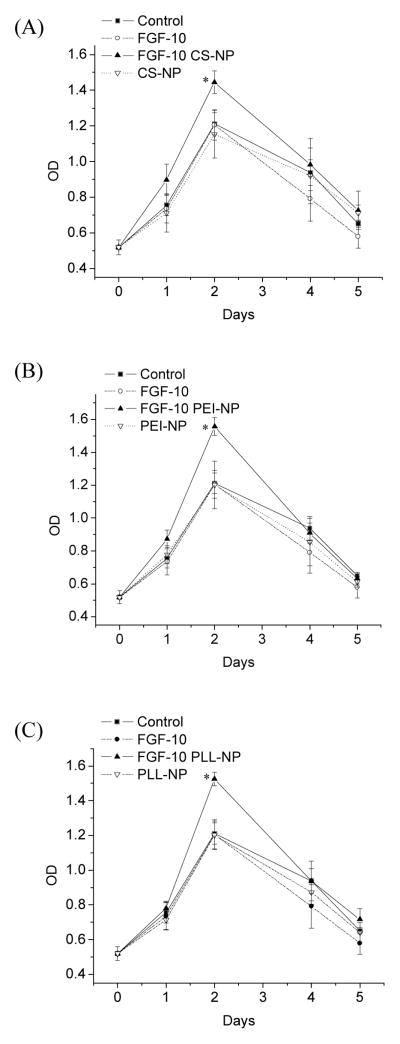
FGF-10 bioactivity was determined in solution or as released from PEC nanoparticles using an in vitro HUVEC proliferation bioassay. A cell viability curve was employed to present the proliferation assay (Figure 7). HUVEC treated with FGF-10 solution (20 ng/mL) showed a negligible increase in cell proliferation that was not statistically significant from controls in the absence of FGF-10 during the 5-day incubation period. On the other hand, cells treated with FGF-10-loaded nanoparticles generally demonstrated a sustained significant increase in the cell proliferation activity at day 2 (p < 0.05 for CS, PEI, PLL nanoparticles at day 2). Control nanoparticles (blanks) as well as controls employing no nanoparticles or FGF-10 demonstrated significantly lower proliferation activity as compared to FGF-10 nanoparticles at day 2. At day 4 and 5, the cells begin to die as a result of contact inhibition combined with the exhausting of nutrients, since the cell culture medium was not changed during the 5-day incubation.
Figure 7.
Sustained HUVEC proliferation effect of FGF-10 in nanoparticles. HUVEC were incubated with a solution of FGF-10 or FGF-10 entrapped in PEC nanoparticles for 5 days. The optical density at 490 nm was recorded using an MTS assay. (A) CS-DS nanoparticles (B) PEI-DS nanoparticles (C) PLL-DS nanoparticles (n=5).
Discussion
FGF-10 has been shown to stimulate repair of injured epithelial tissues. A phase II clinical trial on Repifermin® indicated that intraveneous injection of this form of FGF-10 is effective in reducing mucositis [23]. Another phase II clinical trial of Repifermin® applied topically to skin with venous ulcers demonstrated that a 26 week treatment with Repifermin® promoted improved wound healing as compared to placebo [8]; however, a clinical trial performed after this did not demonstrate similar efficacy. For the treatment of wounds and chronic ulceration (e.g. cancer therapy-induced mucositis), it may be desired to achieve controlled-release of this class of therapeutics to reduce injection or application frequency and increase patient comfort and compliance. Controlled release may also improve safety and efficacy of a drug by avoiding peak concentrations and achieving prolonged concentrations at therapeutically effective levels. Nevertheless, any effective controlled release technology must safeguard a protein’s integrity while achieving the desired pharmacokinetic profile.
The formulation of unstable proteins such as FGF-10 into controlled release polymer matrices or particles can be problematic. Schwendeman and others have utilized the heparin-binding strategy as an excipient approach to formulate growth factors into poly(DL-lactic-co-glycolic acid) (PLGA) mini-cylinders and microspheres [24]. Many of the traditional controlled release polymers such as PLGA, however, are hydrophobic and can induce protein denaturation at the polymer interface [25]. Blockade of this interface is possible by utilizing proteins such as bovine serum albumin, which are known to adsorb to hydrophobic surfaces. In this aspect, hydrophilic materials may be preferred to mitigate protein denaturation. Unfortunately, hydrophilic nanoparticles will often release active ingredients rapidly as a result of short diffusional path lengths required for escape. Larger gel matrices can increase the duration of controlled release, especially if a binding event is utilized to hold proteins such as FGF inside the material [15, 26, 27]. Therefore, heparin-FGF binding may be leveraged for stability and controlled release, especially as the geometry of the delivery vehicle decreases to the nanoscale [28, 29].
In this study, an aqueous process was employed to form ionic complexes by using anionic polymers dextran sulfate and cationic polymers such as chitosan, PEI and PLL. DS binds and stabilizes heparin-binding growth factors such as VEGF165 and FGF-2 [26] that exhibit a cationic pocket that is conserved in many medicinal proteins [30–33]. In addition, DS has been shown to form stable polyelectrolyte complexes with multiple polycations. Here, these particulate systems were optimized to stabilize the growth factor FGF-10 and control the release of active protein. Based on this report, our previous work, and articles from Middaugh and others, research to date strongly indicates that the PEC nanoparticles will enhance the stability of labile proteins, especially proteins containing a heparin-binding domain. Although FGF-10 was a very unstable protein as indicated from its low transition temperature in CD, UV, and fluorescence data, dextran sulfate effectively stabilized the secondary and tertiary structure of FGF-10 and inhibited aggregation. This binding event also facilitated FGF-10 entrapment during the formation of polyelectrolyte complexes. Dextran sulfate and chitosan PECs were especially attractive based on their low cytotoxicity.
The orchestration of FGF signaling is complex with evidence of multiple receptor interactions and multiple functions from single growth factors. In many cases, the persistence of a narrow range of protein concentration is required for activity, deviation from a specified range leading to inactivity, other activities, or the opposite activity of the protein [34]. In addition, many FGFs can rapidly lose activity due to poor stability and/or proteolytic modification [35]. Accurately leveraging the therapeutic value of these proteins requires careful consideration of stabilization strategies, protection from proteolytic modification or degradation, and methods to accurately control release. In some cases, efforts to deduce the precise activity of some growth factors have been confounded by the inability to systematically study them in vitro. The mitogenic activity of Repifermin® towards endothelial cells reported here may not have been uncovered if not for a controlled release delivery system.
Conclusions
Self-assembled polyelectrolyte complexed nanoparticles were successfully formulated to stabilize and entrap FGF-10 using a method of coacervation of oppositely charged polyelectrolytes. Dextran sulfate effectively stabilized FGF-10 as indicated from various biophysical techniques. The resulting nanoparticles efficiently entrapped and sustained the release of FGF-10. Additionally, FGF-10-loaded PEC nanoparticles significantly stimulated endothelial cell proliferation in comparison to FGF-10 in solution delivered at the same dose, which showed no effect. HUVEC cell proliferation could be attributed to maintainence of a higher concentration of the bio-active FGF-10 in the culture medium coupled with improved stability. This study provides insight into a nanoparticle approach for stabilizing and delivering FGF-10 in the endeavor of developing intravenous or localized injection formulations of growth factors for tissue healing.
Acknowledgments
This work was partially supported through a Scientist Development Grant from the American Heart Association and through an Innovation Award from the Juvenile Diabetes Research Foundation. We also acknowledge support from NIH grant P20 RR016443 and R03 AR054035, the Department of Defense (W81XWH-07-1-0021), and a grant from the Cystic Fibrosis Foundation. We would like to thank Human Genome Sciences, Inc. for graciously supplying FGF-10. We also extend our thanks to Russ Middaugh for helpful discussions and for the use of equipment.
References
- 1.Emoto H, Tagashira S, Mattei MG, Yamasaki M, Hashimoto G, Katsumata T, Negoro T, Nakatsuka M, Birnbaum D, Coulier F, Itoh N. Structure and expression of human fibroblast growth factor-10. J Biol Chem. 1997;272(37):23191–23194. doi: 10.1074/jbc.272.37.23191. [DOI] [PubMed] [Google Scholar]
- 2.Ruben SM, Jimenez P, Duan DR, Rampy MA, Mendrick D, Zhang J, Ni J, Moore PA, Coleman TA, Gruber JR, Dillon PJ, Gentz RL. Construction of deletion and substitution mutants of human keratinocyte growth factor-2 and their use to promote wound healing. 98-23082 6077692, 19980213. Application: US US Patent. 2000
- 3.Derrick T, Grillo AO, Vitharana SN, Jones L, Rexroad J, Shah A, Perkins M, Spitznagel TM, Middaugh CR. Effect of polyanions on the structure and stability of repifermin (keratinocyte growth factor-2) J Pharm Sci. 2007;96(4):761–776. doi: 10.1002/jps.20797. [DOI] [PubMed] [Google Scholar]
- 4.Sung C, Parry TJ, Riccobene TA, Mahoney A, Roschke V, Murray J, Gu ML, Glenn JK, Caputo F, Farman C, Odenheimer DJ. Pharmacologic and pharmacokinetic profile of repifermin (KGF-2) in monkeys and comparative pharmacokinetics in humans. AAPS PharmSci. 2002;4(2):E8. doi: 10.1208/ps040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiwata T, Naito Z, Lu YP, Kawahara K, Fujii T, Kawamoto Y, Teduka K, Sugisaki Y. Differential distribution of fibroblast growth factor (FGF)-7 and FGF-10 in L-arginine-induced acute pancreatitis. Exp Mol Pathol. 2002;73(3):181–190. doi: 10.1006/exmp.2002.2472. [DOI] [PubMed] [Google Scholar]
- 6.Gillis P, Savla U, Volpert OV, Jimenez B, Waters CM, Panos RJ, Bouck NP. Keratinocyte growth factor induces angiogenesis and protects endothelial barrier function. J Cell Sci. 1999;112(Pt 12):2049–2057. doi: 10.1242/jcs.112.12.2049. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez PA, Rampy MA. Keratinocyte growth factor-2 accelerates wound healing in incisional wounds. J Surg Res. 1999;81(2):238–242. doi: 10.1006/jsre.1998.5501. [DOI] [PubMed] [Google Scholar]
- 8.Robson MC, Phillips TJ, Falanga V, Odenheimer DJ, Parish LC, Jensen JL, Steed DL. Randomized trial of topically applied repifermin (recombinant human keratinocyte growth factor-2) to accelerate wound healing in venous ulcers. Wound Repair Regen. 2001;9(5):347–352. doi: 10.1046/j.1524-475x.2001.00347.x. [DOI] [PubMed] [Google Scholar]
- 9.Aaronson SA, Bottaro DP, Miki T, Ron D, Finch PW, Fleming TP, Ahn J, Taylor WG, Rubin JS. Keratinocyte growth factor. A fibroblast growth factor family member with unusual target cell specificity. Ann N Y Acad Sci. 1991;638:62–77. doi: 10.1111/j.1749-6632.1991.tb49018.x. [DOI] [PubMed] [Google Scholar]
- 10.Cleland JL, Duenas ET, Park A, Daugherty A, Kahn J, Kowalski J, Cuthbertson A. Development of poly-(D,L-lactide--coglycolide) microsphere formulations containing recombinant human vascular endothelial growth factor to promote local angiogenesis. J Control Release. 2001;72(1–3):13–24. doi: 10.1016/s0168-3659(01)00258-9. [DOI] [PubMed] [Google Scholar]
- 11.Street J, Bao M, deGuzman L, Bunting S, Peale FV, Jr, Ferrara N, Steinmetz H, Hoeffel J, Cleland JL, Daugherty A, van Bruggen N, Redmond HP, Carano RA, Filvaroff EH. Vascular endothelial growth factor stimulates bone repair by promoting angiogenesis and bone turnover. Proc Natl Acad Sci U S A. 2002;99(15):9656–9661. doi: 10.1073/pnas.152324099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan H, Vitharana SN, Chen T, O’Keefe D, Middaugh CR. Effects of pH and polyanions on the thermal stability of fibroblast growth factor 20. Mol Pharm. 2007;4(2):232–240. doi: 10.1021/mp060097h. [DOI] [PubMed] [Google Scholar]
- 13.Gospodarowicz D, Cheng J. Heparin protects basic and acidic FGF from inactivation. J Cell Physiol. 1986;128(3):475–484. doi: 10.1002/jcp.1041280317. [DOI] [PubMed] [Google Scholar]
- 14.Edelman ER, Mathiowitz E, Langer R, Klagsbrun M. Controlled and modulated release of basic fibroblast growth factor. Biomaterials. 1991;12(7):619–626. doi: 10.1016/0142-9612(91)90107-l. [DOI] [PubMed] [Google Scholar]
- 15.Zisch AH, Lutolf MP, Hubbell JA. Biopolymeric delivery matrices for angiogenic growth factors. Cardiovasc Pathol. 2003;12(6):295–310. doi: 10.1016/s1054-8807(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Miyazaki Y, Yakou S, Takayama K. Sustained release of highly water-soluble drugs with micelle forming ability from polyionic matrix tablets. Pharmazie. 2007;62(1):41–45. [PubMed] [Google Scholar]
- 17.Sarmento B, Ribeiro A, Veiga F, Ferreira D. Development and characterization of new insulin containing polysaccharide nanoparticles. Colloids Surf B Biointerfaces. 2006;53(2):193–202. doi: 10.1016/j.colsurfb.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Tiyaboonchai W, Woiszwillo J, Middaugh CR. Formulation and characterization of DNA-polyethylenimine-dextran sulfate nanoparticles. Eur J Pharm Sci. 2003;19(4):191–202. doi: 10.1016/s0928-0987(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 19.Chen BL, Arakawa T, Hsu E, Narhi LO, Tressel TJ, Chien SL. Strategies to suppress aggregation of recombinant keratinocyte growth factor during liquid formulation development. J Pharm Sci. 1994;83(12):1657–1661. doi: 10.1002/jps.2600831204. [DOI] [PubMed] [Google Scholar]
- 20.Huang M, Vitharana SN, Peek LJ, Coop T, Berkland C. Polyelectrolyte complexes stabilize and controllably release vascular endothelial growth factor. Biomacromolecules. 2007;8(5):1607–1614. doi: 10.1021/bm061211k. [DOI] [PubMed] [Google Scholar]
- 21.Tiyaboonchai W, Woiszwillo J, Middaugh CR. Formulation and characterization of amphotericin B-polyethylenimine-dextran sulfate nanoparticles. J Pharm Sci. 2001;90(7):902–914. doi: 10.1002/jps.1042. [DOI] [PubMed] [Google Scholar]
- 22.Tiyaboonchai W, Woiszwillo J, Sims RC, Middaugh CR. Insulin containing polyethylenimine-dextran sulfate nanoparticles. Int J Pharm. 2003;255(1–2):139–151. doi: 10.1016/s0378-5173(03)00055-3. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler G. Repifermin. Human Genome Sciences/GlaxoSmithKline. IDrugs. 2001;4(7):813–819. [PubMed] [Google Scholar]
- 24.Zhu G, Mallery SR, Schwendeman SP. Stabilization of proteins encapsulated in injectable poly (lactide-co-glycolide) Nat Biotechnol. 2000;18(1):52–57. doi: 10.1038/71916. [DOI] [PubMed] [Google Scholar]
- 25.Butler SM, Tracy MA, Tilton RD. Adsorption of serum albumin to thin films of poly(lactide-co-glycolide) J Control Release. 1999;58(3):335–347. doi: 10.1016/s0168-3659(98)00173-4. [DOI] [PubMed] [Google Scholar]
- 26.Liu LS, Ng CK, Thompson AY, Poser JW, Spiro RC. Hyaluronate-heparin conjugate gels for the delivery of basic fibroblast growth factor (FGF-2) J Biomed Mater Res. 2002;62(1):128–135. doi: 10.1002/jbm.10238. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Cai S, Shu XZ, Shelby J, Prestwich GD. Release of basic fibroblast growth factor from a crosslinked glycosaminoglycan hydrogel promotes wound healing. Wound Repair Regen. 2007;15(2):245–251. doi: 10.1111/j.1524-475X.2007.00211.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, Go DH, Bae JW, Lee SJ, Park KD. Heparin conjugated polymeric micelle for long-term delivery of basic fibroblast growth factor. J Control Release. 2007;117(2):204–209. doi: 10.1016/j.jconrel.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Chung YI, Tae G, Hong Yuk S. A facile method to prepare heparin-functionalized nanoparticles for controlled release of growth factors. Biomaterials. 2006;27(12):2621–2626. doi: 10.1016/j.biomaterials.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 30.Jones LS, Yazzie B, Middaugh CR. Polyanions and the proteome. Mol Cell Proteomics. 2004;3(8):746–769. doi: 10.1074/mcp.R400008-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Joshi SB, Kamerzell TJ, McNown C, Middaugh CR. The interaction of heparin/polyanions with bovine, porcine, and human growth hormone. J Pharm Sci. 2007 doi: 10.1002/jps.21056. [DOI] [PubMed] [Google Scholar]
- 32.Kamerzell TJ, Joshi SB, McClean D, Peplinskie L, Toney K, Papac D, Li M, Middaugh CR. Parathyroid hormone is a heparin/polyanion binding protein: binding energetics and structure modification. Protein Sci. 2007;16(6):1193–1203. doi: 10.1110/ps.062613807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salamat-Miller N, Fang J, Seidel CW, Assenov Y, Albrecht M, Middaugh CR. A network-based analysis of polyanion-binding proteins utilizing human protein arrays. J Biol Chem. 2007;282(14):10153–10163. doi: 10.1074/jbc.M610957200. [DOI] [PubMed] [Google Scholar]
- 34.Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7(4):311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 35.Burgess WH, Maciag T. The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]