Abstract
Self-fertile hermaphrodites have evolved from male/female ancestors in many nematode species, and this transition occurred on three independent occasions in the genus Caenorhabditis. Genetic analyses in Caenorhabditis show that the origin of hermaphrodites required two types of changes: alterations to the sex-determination pathway that allowed otherwise female animals to make sperm during larval development, and the production of signals from the gonad that caused these sperm to activate and fertilize oocytes. Comparisons of C. elegans and C. briggsae hermaphrodites show that the ancestral sex-determination pathway has been altered in multiple unique ways. Some of these changes must have precipitated the production of sperm in XX animals, and others were modifying mutations that increased the efficiency of hermaphroditic reproduction. Reverse genetic experiments show that XX animals acquired the ability to activate sperm by co-opting one of the two redundant pathways that normally work in males. Finally, the adoption of a hermaphroditic lifestyle had profound effects on ecological and sexual interactions and genomic organization. Thus, nematode mating systems are ideal for elucidating the origin of novel traits, and studying the influence of developmental processes on evolutionary change.
Introduction
Darwin published “On the Origin of Species” 155 years ago [1], but his theory of natural selection remained incomplete until its integration with genetics in the modern synthesis [2]. The past 30 years have seen the beginnings of a second major integration, fusing evolutionary theory with new research in development (reviewed in [3,4]). This field of evolutionary developmental biology is best known for the discovery of orthologous genes that pattern the early embryo, but it is now branching out into many other areas.
Here, we review the evolution of self-fertility in Caenorhabditis nematodes. The convergent evolution of hermaphrodites in this genus provides an ideal way to explore both evolutionary change and the use of alternative reproductive strategies (reviewed in [5,6]). Phylogenetic analysis implies that mating systems changed recently, which makes it easier to reconstruct many of the underlying genetic events (Figure 1). Furthermore, technical considerations make this genus ideal for study. C. elegans is one of the leading models for studying sex-determination, and decades of research provide the background information needed to characterize its relatives. This task is simplified by the genome sequences of C. elegans [7] and C. briggsae [8], and the partial sequences of seven related species (Figure 1). Finally, orthologous genes can be characterized by powerful reverse genetic techniques, including RNA interference and gene-editing with transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeats (CRISPRs) [9-13], which allow the control of mating systems to be dissected in all species.
Figure 1. Hermaphrodites evolved on three independent occasions in Caenorhabditis.
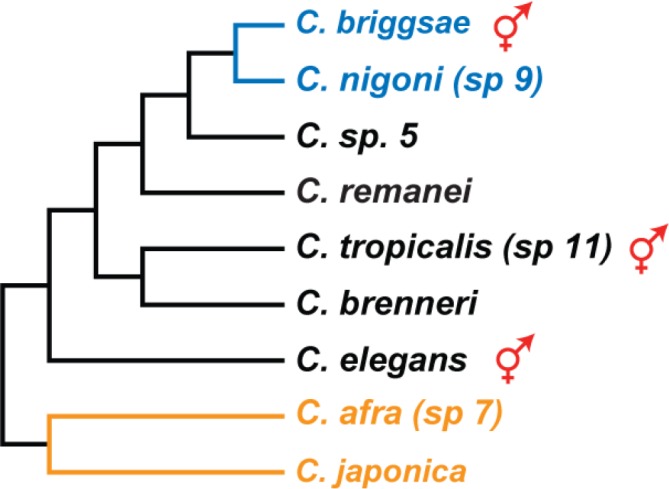
Only species with sequenced genomes are shown. Androdioecious species with males and hermaphrodites are marked with a red symbol, and the others are male/female. The species in blue are able to interbreed and produce fertile offspring, and two outgroup species are orange. Modified from Kiontke et al. [18] and Felix et al. [15].
Our discussion will focus on three major questions. First, a change in mating system requires the coordination of many genes and regulatory pathways, so we will explore how complex traits originate. Second, hermaphrodites are common in some phyla but rare in others, so we will consider whether the rules of development influence the evolution of self-fertility. Third, mating systems are central to sexual reproduction, so we will ask how self-fertility affects the evolutionary process.
How did androdioecy evolve in nematodes?
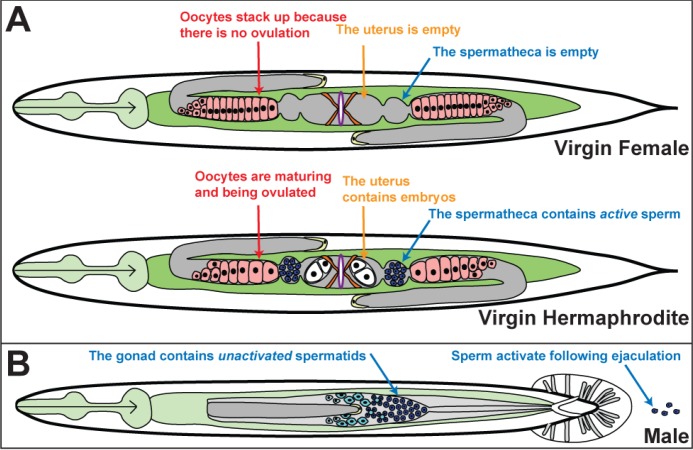
Most nematode species have males and females, just like other animals. However, some species display a rare mating system known as androdioecy, which uses males and self-fertile hermaphrodites. In androdioecious nematodes, the XO animals are normal males but XX animals are hermaphrodites (Figure 2). These hermaphrodites look like females, but the first germ cells to differentiate become sperm, which are stored in the spermathecae and used later for self-fertilization. Subsequent germ cells become oocytes. Because hermaphrodites are anatomically female, they cannot mate with each other, but do produce cross progeny if mated with males.
Figure 2. Self-fertile hermaphrodites are modified females that make and use sperm.
A. Comparison of virgin female and hermaphrodite nematodes. Ventral up, anterior to the left. Oocytes are pink and sperm are blue. In the soma, the gonad is gray, the pharynx is light green, the intestine is dark green, the sex muscles are orange, the distal tip cells are yellow and the vulva purple.
B. Male nematode. Primary spermatocytes are light blue hexagons, residual bodies are light blue circles, and spermatids are dark blue circles.
Self-fertile hermaphrodites have arisen independently many times during evolution [14]. Even within a subgroup of the genus Caenorhabditis, hermaphroditic reproduction evolved in three different species — C. elegans, C. tropicalis (formerly C. sp. 11 [15]) and C. briggsae (Figure 1) [16-18]. Comparative studies, particularly between C. elegans and C. briggsae, have elucidated the genetic control of self-fertility.
Caenorhabditis nematodes share a core set of sex-determination genes
Decades of research with C. elegans have defined a signal transduction pathway that regulates sexual development in both the somatic tissues and the germ line (Figure 3; reviewed in [19,20]). In signaling cells, the ratio of X chromosomes to autosomes controls xol-1, a gene that specifies male development. Next, XOL-1 acts through the syndecan (SDC) proteins to control the production of a hormone, HER-1 (human epidermal growth factor receptor-1), that causes cells throughout the body to adopt male fates. The ultimate target of this pathway is TRA-1, a transcription factor related to the Gli proteins [21].
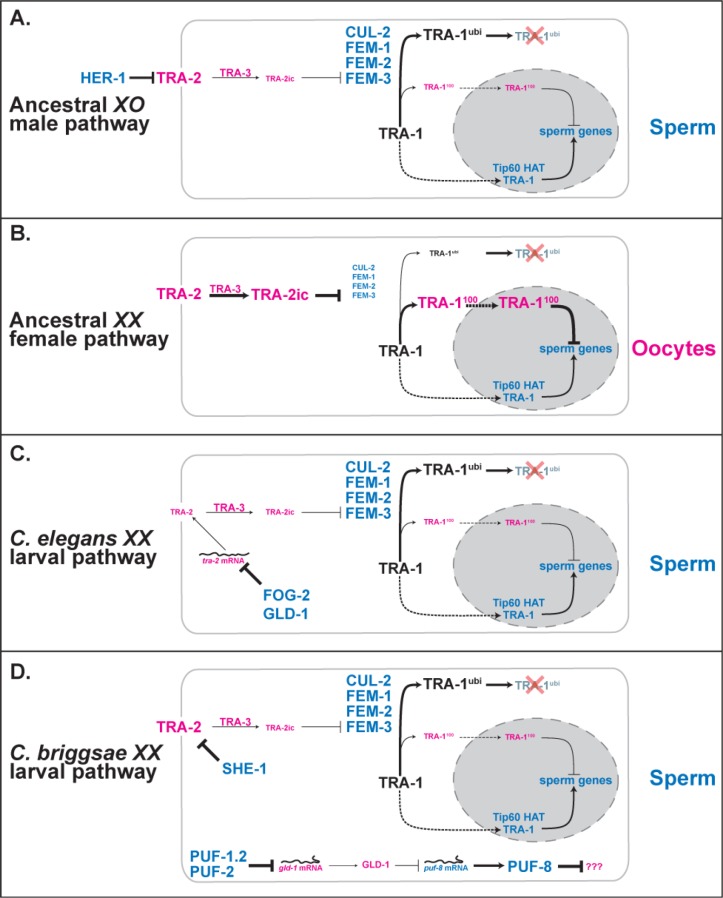
Figure 3. Modifications to the sex-determination pathway allow XX larvae to make sperm.
Proteins promoting spermatogenesis are blue, and those promoting oogenesis are pink. Positive interactions are shown as solid lines with arrowheads, negative ones as lines with bars, and nuclear import by dashed lines. Line thickness and font size represent the strength of each interaction. The TRA-2 receptor can be cleaved by the calpain protease TRA-3 to form the intracellular form TRA-2ic [111,112]. The target of the Caenorhabditis briggsae puf-1.2, puf-2, gld-1 and puf-8 pathway is not yet known, but it is likely that PUF-8 represses a gene needed for oogenesis [49,52]. Likewise, the secondary role that the three fem genes play downstream of tra-1 in C. elegans [56] and possibly in C. briggsae [113] is not shown because their targets remain unclear. Finally, additional genetic interactions are needed for adult hermaphrodites to switch back to oogenesis, which are reviewed elsewhere [20]. For other details, see the text.
In males, HER-1 binds to and inactivates its receptor, TRA-2 (Figure 3A). This interaction allows the FEM proteins and the ubiquitin ligase CUL-2 to mark TRA-1 for degradation [22] (shown as TRA-1ubi in Figure 3). However, some full-length TRA-1 remains [23] and is likely to work with the Tip60 histone acetyltransferase (HAT) complex to promote the expression of genes needed for spermatogenesis [24]. These genes include fog-1 and fog-3, which have TRA-1 binding sites in their promoters [25,26] and are required for germ cells to become sperm rather than oocytes [27,28]. FOG-1 and FOG-3 are likely to work by regulating the translation of messenger RNAs (mRNAs) [26,29,30].
In hermaphrodites, TRA-1 is cleaved to produce a repressor (TRA-1100 in Figure 3) [23], which turns off male genes like mab-3, egl-1, fog-3 and numerous other targets (Figure 3B) [25,31-33]. These repressive interactions appear to be the predominant method by which tra-1 controls somatic sex. The fact that the full-length isoform appears to activate sperm genes, whereas the cleaved form represses them, makes TRA-1 bipotential, a trait shared with many other Gli proteins [24].
Orthologs of these genes have been shown to function in the sex-determination pathway in both hermaphroditic and male/female species of Caenorhabditis. Analysis of mutants in the hermaphroditic species C. briggsae confirms that sexual development is controlled by tra-1 [34,35], tra-2 [35,36], tra-3 [35], trr-1 [24], fem-2 [37], fem-3 [37] and fog-3 [25]. Furthermore, RNA interference shows that tra-2 [38], fem-3 [39] and fog-3 [40] regulate sexual development in the male/female species C. remanei. Thus, it appears that core genes of the sex-determination pathway have been conserved throughout Caenorhabditis (reviewed in [5]). This conservation could extend farther, since an ortholog of tra-1 controls sexual development in the distant relative Pristionchus pacificus [41].
Hermaphrodites evolved through independent changes in the sex-determination pathway
Because hermaphroditism arose independently in C. elegans and C. briggsae, comparing their sex-determination pathways can reveal what types of genetic changes led to self-fertility. To date, all known mutations that allow XX larvae to produce sperm fall into one of three separate categories: the generation of novel genes by duplication, the recruitment of known germline genes to the sex-determination pathway, and the modification of core genes within the pathway (compare Figure 3C/3D with 3A/3B).
First, new genes have been created by duplication and recruited to the pathway. For example, C. elegans hermaphrodites require fog-2 to produce sperm during larval development [42]. The FOG-2 protein works with GLD-1 to block the translation of tra-2 mRNAs in the XX germline [43,44] that allows the expression of male genes needed for spermatogenesis. This system is unique to C. elegans, since fog-2 was created by a recent duplication event [44] and has no ortholog in C. briggsae [45].
A different gene, she-1, is required for C. briggsae hermaphrodites to make sperm [46]. Although SHE-1 also regulates the activity of tra-2, it does not associate with GLD-1 and its molecular function remains unknown. As with fog-2, the she-1 gene was produced by a recent duplication event. Surprisingly, both genes are distant members of the large F-box family [47]. Perhaps the adaptive radiation of this family provided opportunities for novel functions to arise.
Second, existing germline genes have been independently recruited to the sex-determination pathway. In C. elegans, the fbf genes encode conserved RNA-binding proteins that block spermatogenesis by preventing the translation of fem-3 mRNAs in the germ line [48]. By contrast, two other PUF proteins promote spermatogenesis in C. briggsae hermaphrodites by blocking the translation of gld-1 mRNAs [49]. Thus, different members of the PUF family of proteins, which normally function in the germ line, were independently recruited to the sex-determination pathway in order to control self-fertility.
The gld-1 gene also belongs to this class. GLD-1 plays numerous roles in the XX germ line [50-52]. In C. elegans, it also promotes hermaphrodite spermatogenesis by working with FOG-2 to block the translation of tra-2 mRNAs, and its physical interaction with this target is stronger than in other nematodes [43,44]. By contrast, C. briggsae GLD-1 blocks hermaphrodite spermatogenesis [45,52], in part by regulating puf-8 [52]. The ultimate target of this pathway is not yet known, but it is likely that PUF-8 represses a gene needed for oogenesis in C. briggsae [49,52]. Thus, these species recruited GLD-1 for opposing roles in sex determination.
Also, the nucleasome remodeling factor complex regulates gene expression by moving histones [53] and controls germ cell proliferation in many animals, including C. elegans [54]. In C. briggsae, it was recruited for a unique role — the control of spermatogenesis [55]. It appears to carry out this function by allowing TRA-1 access to the fog-1 and fog-3 promoters.
Third, the core pathway itself has been modified, changing the relative importance of different factors. We know that the FEM proteins and the Tip60 HAT complex are core members of the pathway, since they both influence germ cell fates in sensitive genetic backgrounds, and double mutants in either species cause synthetic feminization [24]. However, the three FEM proteins are required for spermatogenesis in C. elegans hermaphrodites [56] but dispensable in C. briggsae ones [37]. By contrast, the Tip60 HAT complex is required for spermatogenesis in C. briggsae but plays only a minor role in C. elegans [24].
One explanation for these results is that the sperm/oocyte decision might be controlled by a balance between activating and repressing activities of TRA-1 (Figure 3). In hermaphrodites, upstream regulators of the sex-determination pathway, like those described here, could alter this balance, so that larvae make sperm and adults make oocytes. Several lines of evidence support this idea. First, the analysis of mutations in the C. elegans fog-3 promoter suggests that TRA-1 both promotes and represses the expression of fog-3 [25]. Second, adult hermaphrodites normally accumulate far more of the TRA-1100 isoform than adult males [22,23]. However, C. elegans cul-2 or fem mutations create similar proportions of TRA-1100 in XO animals [22], leading to oogenesis [57]. Third, some mutations in accessory genes appear to favor activation or repression by TRA-1. For example, the Tip60 HAT complex requires TRA-1 to promote the expression of fog-3 [24]. By contrast, two WDR-5 proteins are needed for TRA-1100 to repress fog-3, and appear to work by controlling its import to the nucleus [58]. Finally, the relative importance of the FEM proteins and the Tip60 HAT complex in C. elegans and C. briggsae differ dramatically [24,37], which shows that different types of changes in the regulation of TRA-1 could lead to self-fertility.
The large number of alterations found in the sex-determination pathway (compare Figures 3C and 3D) suggests that self-fertility has been refined by selection for additional modifiers. These mutations might influence the precise amount and timing of spermatogenesis, since the analysis of weak tra-3 mutants implies that the number of sperm made by each hermaphrodite is under intense selection [59]. Thus, mutations that affect when the germ line switches to oogenesis could maximize the number of self progeny, while minimizing the delay in oogenesis.
Hermaphrodites also required changes in a sperm activation pathway
Based on these results, it seemed possible that a single mutation that altered the sexual fates of germ cells would be sufficient to make self-fertile hermaphrodites. This mutation would have to allow XX animals to make sperm as well as oocytes, perhaps by decreasing tra-2 activity. To test this hypothesis, RNA interference was used to target tra-2 in C. remanei females [60]. Some of the Cre-tra-2(RNAi) XX animals did indeed produce sperm and oocytes in a female body. Surprisingly, their spermatids failed to activate and fertilize oocytes, so they did not make progeny. Thus, alteration of the sex-determination pathway is not sufficient for self-fertility.
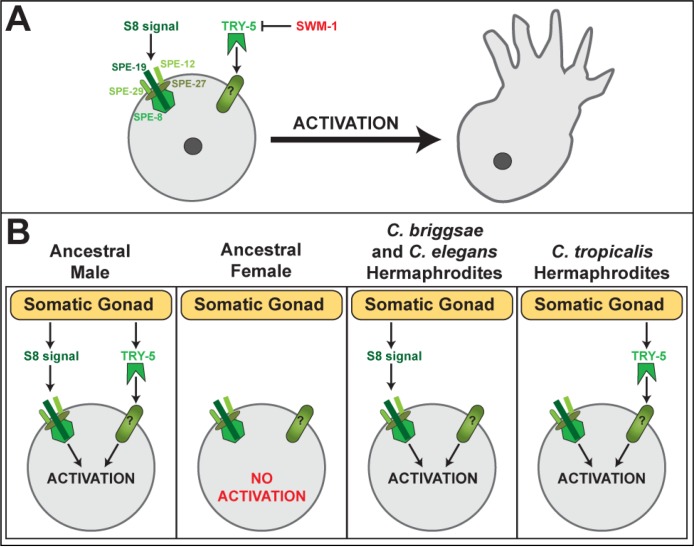
How is sperm activation controlled? C. elegans males use two redundant pathways, one dependent on SPE-8 and the other dependent on TRY-5, whereas hermaphrodites use only the SPE-8 pathway (Figure 4, reviewed in [61]). The five genes of the spe-8 group encode sperm proteins [62-66] that appear to respond to labile zinc [67]. By contrast, TRY-5 is a protease that activates sperm by cleaving unknown targets [68]. Prior to ejaculation, TRY-5 activity is kept in check by the inhibitor SWM-1 (Figure 4A) [69]. Because C. elegans hermaphrodites normally use the spe-8 pathway to activate sperm, mutants in these genes are not self-fertile. However, their spermatids can be activated by exposure to male seminal fluid in a process called trans-activation [70]. This is done experimentally by mating sterile spe-8 hermaphrodites with males, resulting in the activation of some hermaphrodite sperm and the production of self-progeny. Transactivation implies that TRY-5 targets remain functional in hermaphrodite sperm, even if they are not normally used.
Figure 4. Hermaphrodites have co-opted one of two redundant sperm activation pathways.
A. Studies of Caenorhabditis elegans and C. briggsae males show that two redundant pathways control sperm activation. One pathway uses the SPE-8 group of sperm proteins to respond to an unknown signal (denoted “S8 signal”). The other pathway uses an unknown receptor (denoted “?”) to respond to the TRY-5 protease. Males defective for both pathways are sterile.
B. C. briggsae and C. elegans hermaphrodites rely on the SPE-8 pathway to activate sperm, whereas C. tropicalis hermaphrodites rely on the TRY-5 pathway. For details, see the text (Wei et al., unpublished data).
To see if the spermatids in C. remanei tra-2(RNAi) XX animals could be activated and fertilize oocytes, they were crossed with sterile males. This resulted in the production of self-progeny, so the XX sperm functioned normally after transactivation by male seminal fluid. Likewise, simultaneous knockdown of both Cre-tra-2 and Cre-swm-1 also produced self-fertile hermaphrodites [60]. Thus, two coordinated changes are sufficient to produce self-fertility: one in the sex-determination pathway that allows XX animals to make sperm and another that causes these sperm to activate and fertilize oocytes [60].
All of the sperm activation genes from both pathways are conserved throughout Caenorhabditis (Wei et al., unpublished data). Since both pathways operate in C. briggsae males, they must have controlled sperm activation in the male/female ancestor. However, C. elegans and C. briggsae hermaphrodites use the SPE-8 pathway to control sperm activation, but C. tropicalis hermaphrodites use TRY-5 (Figure 4B) (Wei et al., unpublished data). Thus, newly evolving hermaphrodites appear to have co-opted either one system or the other, probably by expressing the appropriate signal in the somatic gonad.
What are the consequences of androdioecy in nematodes?
Once self-fertility has been acquired, it has far-reaching consequences as organisms adapt to and refine the hermaphroditic lifestyle. Work in Caenorhabditis and other androdioecious species has begun to shed light on how changes in mating systems can affect the ecology, fitness, and genomic organization of these hermaphroditic species (reviewed in [6]).
Hermaphrodites facilitate the colonization of new habitats
Although we are beginning to learn about the natural ecology of Caenorhabditis [18], we do not know what selective pressures favored the origin of self-fertility. However, the idea that hermaphrodites are better suited for colonization [71] is supported by studies of the European tadpole shrimp [72]. This species has male/female, male/hermaphrodite and hermaphrodite populations. As glaciers retreated north following the end of the last ice age, new habitats were preferentially filled by hermaphrodites. This advantage in colonization could be due to the ability of single animals to open up new territories without needing to find mates. Ecological studies with Pristionchus nematodes, which also include male/female and male/hermaphrodite species, should help test this hypothesis (reviewed in [73]).
Hermaphroditism alters the genetic structure of populations
Population structure plays a critical role in evolution, since it determines what allelic combinations will be available for selection. In large sexual populations, new mutations will most likely be present within the population in heterozygous form, so the production of favorable combinations within the same individual would be rare. However, newly evolving hermaphrodites can escape these sexual dynamics by selfing, which should make it easier to produce homozygotes with new allele combinations. This feature of self-fertile populations might accelerate the evolution of androdioecy, and also influence how alleles controlling other traits propagate within the population.
However, selfing does come at a cost. Self-fertile populations can show inbreeding depression, a decrease in fitness caused by progeny that are homozygous for harmful mutations. There is a huge reservoir of genetic diversity in male/female Caenorhabditis species [74], so when the ancestor of C. elegans began to self-fertilize, it would have faced a crisis until lethal mutations were purged from the gene pool. Many incipient hermaphrodite populations might have died out during this stage. However, once inbreeding depression was overcome, hermaphrodites should have been well adapted to their environments. Indeed, modern isolates of C. elegans actually show outbreeding depression for some traits, presumably because beneficial allele combinations have become fixed in the population [75].
Despite the impact of selfing, the existence of males in androdioecious species suggests that some out-crossing still occurs. This conclusion is bolstered by the direct observation of heterozygosity among C. elegans isolated from the wild [76,77]. Indeed, some out-crossing might be necessary for the long-term survival of the species. The existence of populations with both out-crossing and selfing animals has made Caenorhabditis ideal for exploring the role of sexual reproduction, the importance of Muller's ratchet and other mechanisms for eliminating deleterious mutations [78,79].
Androdioecious males show a decline in male fitness
In male/female species, each sex is under selective pressure to find and mate with the other. By contrast, hermaphrodites do not need males to reproduce. Indeed, wild C. elegans populations are highly skewed towards XX animals [76,77]. Consequently, many traits involved in mating have degraded from their state in the male/female ancestor. For example, C. elegans hermaphrodites no longer secrete a pheromone to attract males [80], and do not remain immobile during copulation [81]. Furthermore, males from numerous wild isolates of C. elegans have lost the ability to produce a mating plug [82]. Finally, sperm from androdioecious males are less aggressive than those from gonochoristic males, and interactions between sperm and the XX gonad have changed significantly [83]. All of these changes make C. elegans males less effective than their counterparts from gonochoristic species [80,81]. Genetic studies suggest that hermaphroditic nematodes evolved recently [84], so it is not clear if this trend will eventually result in the elimination of males altogether.
Androdioecious species show decreased sperm size
The evolution of hermaphroditism also involved two different reductions in sperm size. First, all hermaphrodites make smaller sperm than males of the same species [85,86]. This difference is probably due to a developmental bias, since sperm made by XX females following genetic manipulation are also smaller than male sperm [86]. Genetic analyses in C. elegans suggest that this bias could be due to the unsuitability of the hermaphrodite gonad and germ line for the development of large sperm.
Second, males from androdioecious species make much smaller sperm than males from gonochoristic ones [85,86]. Since larger sperm are more likely to fertilize oocytes in controlled experiments, intense sperm competition in male/female species probably favors large sperm [87,88]. Perhaps the smaller sperm in androdioecious males is another example of decreasing male effectiveness in populations that are largely composed of hermaphrodites.
Androdioecious species show a decrease in genome size
The genomes of C. elegans and C. briggsae, as well as their sets of all transcribed genes, are dramatically smaller than those of their male/female counterparts [89]. Not unexpectedly, genes with sexually dimorphic patterns of expression are most likely to have disappeared from the hermaphroditic species. However, these changes are not driven solely by selection. Crosses with C. elegans show a transmission distortion, in which shorter chromosomes are preferentially segregated to hermaphrodite progeny [90]. Thus, deletions might accumulate in selfing lineages.
A model for the origin of self-fertile hermaphrodites
Based on these studies, we propose that the evolution of self-fertile hermaphrodites in Caenorhabditis required three stages. In the first one, a small number of genetic changes combined to make self-fertile animals. The simplest possibility is that this process started with a neutral mutation causing XX animals to produce a sperm activation signal, and was completed by a mutation that caused them to make sperm as well as oocytes [60]. This sequence would avoid a stage in which XX animals wasted resources by making sperm they could not use. Even so, these incipient hermaphrodites probably had small broods and severe reproductive problems.
During the second stage, harmful recessive mutations were purged, to avoid the consequences of inbreeding. At the same time, selection would have favored modifying mutations that increased the precision and effectiveness of hermaphroditic reproduction. The impact of selfing probably explains the rapid establishment of these mutations in the population. Newly evolving androdioecious species might have had three sexes during this stage. One Rhabditis species currently makes males, females and hermaphrodites [91]. Furthermore, C. briggsae she-1 mutants are inherently temperature-sensitive, so perhaps this species once made both females and hermaphrodites too, depending on environmental conditions [46]. Eventually, females were eliminated, both because hermaphrodites are better at colonization, and because opportunities to mate with males declined as the population came to rely more on selfing.
The third stage would have been the longest, and may still be going on. In it, all aspects of the animal's behavior and genome slowly adapted to the hermaphroditic lifestyle. These changes have led to a dramatically smaller genome and lower male effectiveness in the androdioecious species of Caenorhabditis.
Given this reconstruction of events, how does the analysis of nematode mating systems fit into the broader context of evolutionary and developmental studies? Two topics are of critical importance: the role of co-option in producing new traits, and the importance of developmental biases.
Co-option plays a central role in the origin of novel traits
Some of the most detailed studies of evolutionary change involve the reduction or loss of traits. Examples include the reduction of pigmentation in beach mice [92] and pelvic reduction in stickleback fish [93]. Androdioecious mating systems are common in plants, and are thought to have arisen by a similar mechanism — the transformation of some hermaphrodites into males through the loss of female reproductive structures (reviewed in [94]).
By contrast, self-fertility in nematodes was caused by the gain of new functions in XX animals. Hermaphrodites acquired these traits by using genetic programs previously restricted to males. They co-opted the spermatogenesis program through changes in the sex-determination pathway in germ cells, and appear to have co-opted one of the sperm activation pathways by producing a male signal in the somatic gonad. Darwin first proposed that existing traits could be co-opted for new roles, based on his observation that our lungs had evolved from the swim bladder [1]. The origin of self-fertility provides molecular examples of how co-option occurs.
Furthermore, self-fertile hermaphrodites provide a model for the origin of complex traits, since their inception requires changes in two signal transduction systems (the sex-determination pathway and the sperm activation pathways), involving at least two tissues (the germ line and somatic gonad). Without co-option, this type of coordinated change would have been impossible.
Developmental biases favor androdioecy in nematodes
Darwin suspected that “laws of growth” helped shape the pattern of evolutionary change [1]. These effects are now known as developmental biases or constraints (reviewed in [95]). For example, a developmental bias caused nematode sperm to have sexually dimorphic sizes [86]. Although the general significance of this process is still being debated, it is striking that self-fertile hermaphrodites evolved independently in many species of nematodes [16-18] and branchiopod crustaceans [96-98] but never in insects or mammals (reviewed in [99]). Three factors suggest that this broad pattern of evolutionary change is caused by developmental biases.
First, it should be difficult for XX hermaphrodites to evolve in species that use a Y chromosome to specify male development because the Y usually contains sperm genes. Nematodes use an XX/XO system, so this is not a problem. Studies from the branchiopod crustacean Eulimnadia texana support this model, since it uses a ZW/ZZ system to specify sex, which also ensures that newly evolving hermaphrodites would not lack sperm genes [100]. By contrast, fruit flies and mammals have Y chromosomes that contain sperm genes (e.g. [101,102]), so it should not be surprising that neither group has produced self-fertile hermaphrodites. Instead, we note that the only androdioecious vertebrates are fish of the genus Kryptolebias [103]. In this group, males can be induced by environmental perturbations, so they also lack an XX/XY system [104]. Hence, we propose that a genetic constraint prevents the evolution of self-fertile hermaphrodites in many taxa but does not affect nematodes.
Second, the structure of the sex-determination pathway might facilitate the origin of self-fertility. All hermaphrodites in Caenorhabditis make sperm as larvae and oocytes as adults; no other arrangement has been observed. Furthermore, mutations that eliminate tra-1 cause younger animals to make sperm and older ones to make oocytes [35,105,106]. These patterns suggest that each androdioecious species has found a way to exploit a predisposition towards male fates in the larval germ line. Mutations that slightly altered the delicate balance between the activating and repressing activities of TRA-1 might have led to the production of sperm and oocytes in the same animal. Hence, the structure of the sex-determination pathway could favor the evolution of hermaphroditism in nematodes.
Finally, the presence of two redundant pathways to activate male sperm might also favor the evolution of self-fertility. Experiments using C. remanei suggest that changes in both the sex-determination and sperm activation pathways are necessary for the evolution of self-fertility [60]. Furthermore, either the SPE-8 or TRY-5 pathway can be co-opted for use in newly evolving hermaphrodites (Wei et al., unpublished data). Thus, the existence of these redundant pathways might increase the number of strategies that can produce hermaphrodites without compromising male fertility.
Parallel evolution is a major topic of research [107], and parallel changes in mating systems are common. For example, asexual mating systems have arisen several times in the fungal genus Neurospora [108]. Here, we suggest that shared developmental constraints — the XX/XO sex-determination system, the structure of the sex-determination pathway, and redundancy in the sperm activation pathways — could explain why the parallel evolution of hermaphroditism is common in nematodes.
A bright future
Studies of the origin of self-fertile hermaphroditism in Caenorhabditis have contributed significantly to our understanding of the evolution of novel, complex traits. In addition, new work is beginning to reveal the effects of androdioecy on the ecology, sexual selection, and genomes of these hermaphroditic species. Recent developments suggest that the most exciting results are yet to come. First, gene-editing techniques that use TALENs [10-12] or CRISPRs [11,13] now allow rapid and unrivaled precision in evolutionary comparisons among nematode species [55] (Wei et al., unpublished data). Second, the ability to study hybrids between the male/female species C. nigoni (formerly C. sp. 9 [15]) and the male/hermaphrodite species C. briggsae should allow sophisticated tests of evolutionary models [109]. And third, experimental evolution with nematodes is now feasible [88,110], so laboratory studies can explore how selection pressures drive sexual reproduction and the choice of specific mating systems. These technical advances will allow us to compare Caenorhabditis species with more sophistication, reconstruct ancestral and intermediate stages in the path towards self-fertility, and test population genetic theories in the laboratory.
Abbreviations
- CRISPRs
clustered regularly interspaced short palindromic repeats
- HAT
histone acetyltransferase
- HER-1
human epidermal growth factor receptor-1
- mRNA
messenger RNA
- TALENs
transcription activator-like effector nucleases
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/6/62
References
- 1.Darwin C. London: John Murray; 1859. On the Origin of Species by Means of Natural Selection, of the Preservation of Favored Races in the Struggle for Life. [Google Scholar]
- 2.Mayr E. What was the evolutionary synthesis? Trends Ecol Evol (Amst) 1993;8:31–34. doi: 10.1016/0169-5347(93)90128-C. [DOI] [PubMed] [Google Scholar]
- 3.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Haag ES, Lenski RE. Development. 2011;138:2633–7. doi: 10.1242/dev.066928. [DOI] [PubMed] [Google Scholar]
- 5.Haag ES. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. WormBook. 2005::1–14. doi: 10.1895/wormbook.1.120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas CG, Woodruff GC, Haag ES. Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends Genet. 2012;28:213–20. doi: 10.1016/j.tig.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–18. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 8.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, Coulson A, D'Eustachio P, Fitch DH, Fulton LA, Fulton RE, Griffiths-Jones S, Harris TW, Hillier LW, Kamath R, Kuwabara PE, Mardis ER, Marra MA, Miner TL, Minx P, Mullikin JC, Plumb RW, Rogers J, Schein JE, Sohrmann M, Spieth J, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:166–192. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1015706
- 9.Nuez I, Félix MA. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS ONE. 2012;7:e29811. doi: 10.1371/journal.pone.0029811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490065
- 11.Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–48. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718071542
- 12.Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol Biol Evol. 2014;31:468–73. doi: 10.1093/molbev/mst213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–34. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiontke K, Fitch David HA. The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook. 2005::1–11. doi: 10.1895/wormbook.1.11.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Félix MA, Braendle C, Cutter AD. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS ONE. 2014;9:e94723. doi: 10.1371/journal.pone.0094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho S, Jin SW, Cohen A, Ellis RE. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–20. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci USA. 2004;101:9003–8. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1019625
- 18.Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/14257036
- 19.Zarkower D. Somatic sex determination. WormBook. 2006::1–12. doi: 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis RE, Schedl T. Sex-determination in the germ line. WormBook. 2006::1–13. doi: 10.1895/wormbook.1.82.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–49. doi: 10.1016/0092-8674(92)90099-X. [DOI] [PubMed] [Google Scholar]
- 22.Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007;13:127–39. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1087689
- 23.Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006;11:733–40. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1049308
- 24.Guo Y, Chen X, Ellis RE. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 2013;9:e1003850. doi: 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718133591
- 25.Chen PJ, Ellis RE. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development. 2000;127:3119–29. doi: 10.1242/dev.127.14.3119. [DOI] [PubMed] [Google Scholar]
- 26.Jin SW, Kimble J, Ellis RE. Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev Biol. 2001;229:537–53. doi: 10.1006/dbio.2000.9993. [DOI] [PubMed] [Google Scholar]
- 27.Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–77. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin SW, Arno N, Cohen A, Shah A, Xu Q, Chen N, Ellis RE. In Caenorhabditis elegans, the RNA-binding domains of the cytoplasmic polyadenylation element binding protein FOG-1 are needed to regulate germ cell fates. Genetics. 2001;159:1617–30. doi: 10.1093/genetics/159.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi W, Ross JM, Zarkower D. mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–80. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- 32.Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–27. doi: 10.1016/S0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- 33.Berkseth M, Ikegami K, Arur S, Lieb JD, Zarkower D. TRA-1 ChIP-seq reveals regulators of sexual differentiation and multilevel feedback in nematode sex determination. Proc Natl Acad Sci USA. 2013;110:16033–8. doi: 10.1073/pnas.1312087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bono M, Hodgkin J. Evolution of sex determination in Caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics. 1996;144:587–95. doi: 10.1093/genetics/144.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490071
- 35.Kelleher DF, de Carvalho CE, Doty AV, Layton M, Cheng AT, Mathies LD, Pilgrim D, Haag ES. Comparative genetics of sex determination: masculinizing mutations in Caenorhabditis briggsae. Genetics. 2008;178:1415–29. doi: 10.1534/genetics.107.073668. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718030188
- 36.Kuwabara PE. Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics. 1996;144:597–607. doi: 10.1093/genetics/144.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006;10:531–38. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1019509
- 38.Haag ES, Kimble J. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics. 2000;155:105–16. doi: 10.1093/genetics/155.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490076
- 39.Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol. 2002;12:2035–41. doi: 10.1016/S0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718030185
- 40.Chen PJ, Cho S, Jin SW, Ellis RE. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics. 2001;158:1513–25. doi: 10.1093/genetics/158.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires-daSilva A, Sommer RJ. Conservation of the global sex determination gene tra-1 in distantly related nematodes. Genes Dev. 2004;18:1198–208. doi: 10.1101/gad.293504. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490078
- 42.Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–69. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–76. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718490079
- 45.Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:e6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490081
- 46.Guo Y, Lang S, Ellis RE. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr Biol. 2009;19:1853–60. doi: 10.1016/j.cub.2009.09.042. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1387972
- 47.Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–30. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490082
- 48.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–84. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 49.Liu Q, Stumpf C, Thomas C, Wickens M, Haag ES. Context-dependent function of a conserved translational regulatory module. Development. 2012;139:1509–21. doi: 10.1242/dev.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718030182
- 50.Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–30. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beadell AV, Liu Q, Johnson DM, Haag ES. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc Natl Acad Sci USA. 2011;108:19672–77. doi: 10.1073/pnas.1108068108. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718030179
- 53.Alkhatib SG, Landry JW. The nucleosome remodeling factor. FEBS Lett. 2011;585:3197–207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen EC, Lu X, Horvitz HR. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development. 2006;133:2695–704. doi: 10.1242/dev.02444. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Shen Y, Ellis RE. Dependence of the sperm/oocyte decision on the Nucleosome Remodeling Factor Complex was acquired during recent C. briggsae evolution. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu198. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490083
- 57.Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–35. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- 58.Li T, Kelly WG. A role for WDR5 in TRA-1/Gli mediated transcriptional control of the sperm/oocyte switch in C. elegans. Nucleic Acids Res. 2014;42:5567–81. doi: 10.1093/nar/gku221. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718329397
- 59.Hodgkin J, Barnes TM. More is not better: brood size and population growth in a self-fertilizing nematode. Proc Biol Sci. 1991;246:19–24. doi: 10.1098/rspb.1991.0119. [DOI] [PubMed] [Google Scholar]
- 60.Baldi C, Cho S, Ellis RE. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science. 2009;326:1002–5. doi: 10.1126/science.1176013. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1168083
- 61.Ellis RE, Stanfield GM. The regulation of spermatogenesis and sperm function in nematodes. Semin Cell Dev Biol. 2014;29C:17–30. doi: 10.1016/j.semcdb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minniti AN, Sadler C, Ward S. Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics. 1996;143:213–23. doi: 10.1093/genetics/143.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nance J, Minniti AN, Sadler C, Ward S. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics. 1999;152:209–20. doi: 10.1093/genetics/152.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nance J, Davis EB, Ward S. spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics. 2000;156:1623–33. doi: 10.1093/genetics/156.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muhlrad PJ. PhD thesis. University of Arizona; AZ: 2001. A genetic and molecular analysis of spermiogenesis initiation in Caenorhabditis elegans. [Google Scholar]
- 66.Geldziler B, Chatterjee I, Singson A. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 2005;283:424–36. doi: 10.1016/j.ydbio.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 67.Liu Z, Chen L, Shang Y, Huang P, Miao L. The micronutrient element zinc modulates sperm activation through the SPE-8 pathway in Caenorhabditis elegans. Development. 2013;140:2103–7. doi: 10.1242/dev.091025. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/717999681
- 68.Smith JR, Stanfield GM. TRY-5 Is a sperm-activating protease in Caenorhabditis elegans seminal fluid. PLoS Genet. 2011;7:e1002375. doi: 10.1371/journal.pgen.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13747956
- 69.Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr Biol. 2006;16:252–63. doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1030836
- 70.Shakes DC, Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- 71.Baker HG. Self-compatibility and establishment after “long-distance” dispersal. Evolution. 1955;9:347–48. doi: 10.2307/2405656. [DOI] [Google Scholar]
- 72.Zierold T, Hanfling B, Gomez A. Recent evolution of alternative reproductive modes in the ‘living fossil’ Triops cancriformis. BMC Evol Biol. 2007;7:161. doi: 10.1186/1471-2148-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490085
- 73.Sommer RJ, McGaughran A. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol Ecol. 2013;22:2380–93. doi: 10.1111/mec.12286. [DOI] [PubMed] [Google Scholar]
- 74.Barriere A, Yang SP, Pekarek E, Thomas CG, Haag ES, Ruvinsky I. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res. 2009;19:470–80. doi: 10.1101/gr.081851.108. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1163311
- 75.Dolgin ES, Charlesworth B, Baird SE, Cutter AD. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution. 2007;61:1339–52. doi: 10.1111/j.1558-5646.2007.00118.x. [DOI] [PubMed] [Google Scholar]
- 76.Barrière A, Félix M. Natural variation and population genetics of Caenorhabditis elegans. WormBook. 2005::1–19. doi: 10.1895/wormbook.1.43.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sivasundar A, Hey J. Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans. Curr Biol. 2005;15:1598–602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718490086
- 78.Loewe L, Cutter AD. On the potential for extinction by Muller's ratchet in Caenorhabditis elegans. BMC Evol Biol. 2008;8:125. doi: 10.1186/1471-2148-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009;462:350–52. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1221956
- 80.Chasnov JR, So WK, Chan CM, Chow KL. The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proc Natl Acad Sci USA. 2007;104:6730–735. doi: 10.1073/pnas.0608050104. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1081833
- 81.Garcia LR, LeBoeuf B, Koo P. Diversity in mating behavior of hermaphroditic and male-female Caenorhabditis nematodes. Genetics. 2007;175:1761–771. doi: 10.1534/genetics.106.068304. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490088
- 82.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–22. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1117867
- 83.Ting JJ, Woodruff GC, Leung G, Shin N-R, Cutter AD, Haag ES. Intense sperm-mediated sexual conflict promotes reproductive isolation in Caenorhabditis Nematodes. PLoS Biol. 2014 doi: 10.1371/journal.pbio.1001915. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cutter AD, Wasmuth JD, Washington NL. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics. 2008;178:2093–104. doi: 10.1534/genetics.107.085787. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1108093
- 85.LaMunyon CW, Ward S. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proc Biol Sci. 1999;266:263–67. doi: 10.1098/rspb.1999.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baldi C, Viviano J, Ellis RE. A bias caused by ectopic development produces sexually dimorphic sperm in nematodes. Curr Biol. 2011;21:1416–20. doi: 10.1016/j.cub.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc Biol Sci. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.LaMunyon CW, Ward S. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc Biol Sci. 2002;269:1125–28. doi: 10.1098/rspb.2002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomas CG, Li R, Smith HE, Woodruff GC, Oliver B, Haag ES. Simplification and desexualization of gene expression in self-fertile nematodes. Curr Biol. 2012;22:2167–72. doi: 10.1016/j.cub.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718490089
- 90.Wang J, Chen PJ, Wang GJ, Keller L. Chromosome size differences may affect meiosis and genome size. Science. 2010;329:293. doi: 10.1126/science.1190130. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/4324979
- 91.Chaudhuri J, Kache V, Pires-daSilva A. Regulation of sexual plasticity in a nematode that produces males, females, and hermaphrodites. Curr Biol. 2011;21:1548–51. doi: 10.1016/j.cub.2011.08.009. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/13357179
- 92.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–4. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1033182
- 93.Chan YF, Marks ME, Jones FC, Villarreal GJ, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1393956
- 94.Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am Nat. 1978;112:975–97. doi: 10.1086/283342. [DOI] [Google Scholar]
- 95.Arthur W. The effect of development on the direction of evolution: toward a twenty-first century consensus. Evol Dev. 2004;6:282–88. doi: 10.1111/j.1525-142X.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 96.Sassaman C, Weeks SC. The genetic mechanism of sex determination in the conchostracan shrimp Eulimnadia texana. Am Nat. 1993;141:314–28. doi: 10.1086/285475. [DOI] [PubMed] [Google Scholar]
- 97.Weeks SC, Sanderson TF, Reed SK, Zofkova M, Knott B, Balaraman U, Pereira G, Senyo DM, Hoeh WR. Ancient androdioecy in the freshwater crustacean Eulimnadia. Proc Biol Sci. 2006;273:725–34. doi: 10.1098/rspb.2005.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zaffagnini F, Trentini M. The distribution and reproduction of Triops cancriformis (Bosc) in Europe (Crustacea Notostraca) Monitore Zool Ital (NS) 1980;14:1–8. [Google Scholar]
- 99.Weeks SC, Benvenuto C, Reed SK. When males and hermaphrodites coexist: a reiew of androdioecy in animals. Integ Comp Biol. 2006;46:449–64. doi: 10.1093/icb/icj048. [DOI] [PubMed] [Google Scholar]
- 100.Weeks SC, Benvenuto C, Sanderson TF, Duff RJ. Sex chromosome evolution in the clam shrimp, Eulimnadia texana. J Evol Biol. 2010;23:1100–106. doi: 10.1111/j.1420-9101.2010.01963.x. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/2861957
- 101.Bridges CB. Direct proof through non-disjunction that the sex-linked genes of Drosophila are borne by the X-chromosome. Science. 1914;40:107–9. doi: 10.1126/science.40.1020.107. [DOI] [PubMed] [Google Scholar]
- 102.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–24. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 103.Harrington RWJ. Oviparous hermaphroditic fish with internal self-fertilization. Science. 1961;134:1749–50. doi: 10.1126/science.134.3492.1749. [DOI] [PubMed] [Google Scholar]
- 104.Harrington RW. Environmentally controlled induction of primary male gonochorists from eggs of the self-fertilizaing hermaphrodite fish, Rivulus marmoratus. Biological Bulletin. 1967;132:174–99. doi: 10.2307/1539887. [DOI] [PubMed] [Google Scholar]
- 105.Hodgkin J. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1987;1:731–45. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- 106.Schedl T, Graham PL, Barton MK, Kimble J. Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics. 1989;123:755–69. doi: 10.1093/genetics/123.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–64. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718136971
- 108.Gioti A, Mushegian AA, Strandberg R, Stajich JE, Johannesson H. Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol Biol Evol. 2012;29:3215–26. doi: 10.1093/molbev/mss132. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718213354
- 109.Woodruff GC, Eke O, Baird SE, Félix MA, Haag ES. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics. 2010;186:997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chandler CH, Chadderdon GE, Phillips PC, Dworkin I, Janzen FJ. Experimental evolution of the Caenorhabditis elegans sex determination pathway. Evolution. 2012;66:82–93. doi: 10.1111/j.1558-5646.2011.01420.x. [DOI] [PubMed] [Google Scholar]
- 111.Barnes TM, Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996;15:4477–84. [PMC free article] [PubMed] [Google Scholar]
- 112.Sokol SB, Kuwabara PE. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 2000;14:901–6. [PMC free article] [PubMed] [Google Scholar]
- 113.Hill RC, Haag ES. A sensitized genetic background reveals evolution near the terminus of the Caenorhabditis germline sex determination pathway. Evol Dev. 2009;11:333–42. doi: 10.1111/j.1525-142X.2009.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]