Abstract
Altered choline phospholipid metabolism is a hallmark of cancer, leading to malignant choline metabolite profiles consisting of low glycerophosphocholine (GPC) and high phosphocholine (PC) in human breast cancers. Glycerophosphocholine phosphodiesterase (GPC-PDE) catalyzes the degradation of GPC to free choline and glycerol-3-phosphate. The gene(s) encoding for the GPC-PDE(s) responsible for GPC degradation in breast cancers have not yet been identified. Here we have demonstrated for the first time that the GPC-PDE encoded by glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) is associated with breast cancer malignancy. Two human breast cancer cell lines (n=8 and 10) and primary human breast tumor samples (n=19) were studied with combined magnetic resonance spectroscopy (MRS) and qRT-PCR to investigate several isoforms of GDPD expression with respect to choline phospholipid metabolite levels. Out of five GDPDs tested, GDPD5 was found to be significantly overexpressed in highly malignant estrogen receptor negative (ER−) compared to weakly malignant estrogen receptor positive (ER+) human breast cancer cells (P=0.027) and breast tumors from patients (P=0.015). GDPD5 showed significantly positive correlations with PC (P<0.001), total choline (tCho) (P=0.007) and PC/GPC (P<0.001) levels in human breast tumors. GDPD5 showed a trend towards negative correlation with GPC levels (P=0.130). Human breast cancers with malignant choline metabolite profiles consisting of low GPC and high PC levels highly co-expressed GDPD5, choline kinase alpha (CHKA), and phosphatidylcholine-specific phospholipase D1 (PLD1), while cancers containing high GPC and relatively low PC levels displayed low co-expression of GDPD5, CHKA, and PLD1. GDPD5, CHKA and PLD1 were significantly overexpressed in highly malignant ER− tumors in our patient cohort. Our study identified GDPD5 as a GPC-PDE that likely participates in regulating choline phospholipid metabolism in breast cancer, which possibly occurs in cooperation with CHKA and PLD1.
Keywords: choline phospholipid metabolism, glycerophosphodiester phosphodiesterase domain containing 5, choline kinase, magnetic resonance spectroscopy, breast cancer
Introduction
The choline containing metabolites free choline (Cho), glycerophosphocholine (GPC), and phosphocholine (PC) are important water-soluble intermediates in phosphatidylcholine (PtdCho) metabolism. An aberrant choline metabolite profile with high PC and elevated total choline-containing metabolites (tCho) is typically observed in cancer [1, 2] and is increasingly being used as an adjunct for diagnosis and treatment evaluation of breast cancer [3–7]. Malignant transformation of breast epithelial cells leads to an increase in PC and relatively lower GPC levels resulting in an increased PC/GPC ratio and elevated tCho levels [8]. Choline phospholipid metabolism consists of a complex network of biosynthetic and catabolic pathways controlled by several regulatory enzymes that may be potential targets for anticancer therapy [5]. Overexpression and increased enzyme activity in cancer versus corresponding normal cells or tissue has been demonstrated for the choline phospholipid enzymes choline kinase alpha (CHKA) [9–12], PtdCho-specific phospholipase D (PLD) [13–15], PtdCho-specific phospholipase C [14, 15], and the choline transporters organic cation transporter-2 and high affinity choline transporter-1 [16–18]. CHKA phosphorylates Cho to PC at the first step of the Kennedy pathway, which is the major biosynthetic pathway for de novo PtdCho synthesis in mammalian cells [5]. CHKA plays an important role in oncogenic transformation and carcinogenesis in cancers of various origins [19–21]. In breast cancers, CHKA is frequently overexpressed in human breast cancer tissue compared to normal breast tissue from the same patient [22]. CHKA inhibition [9, 10, 23] and CHKA silencing by RNA interference (RNAi) [11, 12] decreased cellular PC levels and reduced proliferation in human breast cancer models. CHKA inhibitors are currently in clinical phase I trials for toxicity testing and pharmacokinetic profile assessment in cancer patients. PLD, which degrades PtdCho to Cho, plays a significant role in oncogene transformation [13].
While recent research has focused on the molecular causes leading to elevated PC levels in cancer, relatively little effort has been made toward elucidating the molecular causes resulting in the cellular GPC levels detected in several cancers. In brain tumors, GPC was shown to be the dominant choline component in low compared to high grade gliomas [24]. Elevated GPC was detected in lung [25] and prostate [26] cancer tissues compared to noninvolved tissues. Relatively low GPC compared to PC is typically observed in breast [8] and ovarian cancers [14]. GPC is generated by sequential hydrolysis of PtdCho by phosphoplipase A2 (PLA2) and lysophopsholipase A1 (Lyso-PLA1). Besides being a membrane breakdown product of PtdCho, GPC is also an abundant osmoprotective osmolyte [27, 28]. GPC is degraded into Cho and glycerol-3-phosphate by glycerophosphocholine phosphodiesterase (GPC-PDE, E.C. 3.1.4.2). To the best of our knowledge, the gene(s) for enzymes with GPC-PDE activity have not yet been identified in human breast cancer cells and tumors. It is important to understand the molecular causes of the GPC levels detected in breast cancer because it is a part of the total choline signal that is increasingly being used in diagnosis and treatment monitoring of breast cancer patients [3–7]. In addition, the enzymes responsible for GPC-PDE activity in cancers may be good therapeutic targets for anticancer treatment.
The human glycerophosphodiester phosphodiesterase domain (GDPD) containing family consists of 5 isoforms, with GDPD1 expressed as 3 transcript variants, GDPD2 as 4 transcript variants, and GDPD3, GDPD4, and GDPD5 as 1 transcript variant each. The functional role of GDPD5 (also referred to as GDE2) was shown to be necessary for motor neuron differentiation in chick spinal cord [29, 30]. GDPD5 is a ubiquitously expressed transmembrane protein, found in mouse lung, heart, brain, kidney, and testis [31]. Multiple sequence alignments demonstrated 90.4% amino acid sequence homology of human and mouse GDPD5 [32]. GDPD5 is ubiquitously expressed in human tissues, but relatively low in kidney and prostate [32]. Down-regulation of GDPD5 by RNAi efficiently increased GPC levels in mouse renal cells, while overexpression of GDPD5 contributed to GPC reduction by increasing GPC-PDE activity, suggesting that GDPD5 confers GPC-PDE activity [33]. GDPD1 (GDE4) has been detected in human ovary and small intestine [34]. Molecular cloning and sequence analysis of GDPD1 showed more than 80% amino acid homology between humans and other mammals [35]. Unlike GDPD5, overexpression of GDPD1 did not promote neurite formation [35]. GDPD2 (GDE3) is involved in differentiation, actin cytoskeleton modulation, and morphological changes of mouse osteoblasts [36, 37]. The role of GDPD3 is still largely unknown, while GDPD4 (GDE6) is predominantly overexpressed in spermatocytes of mouse testis, which suggests that it plays a role in male germ cell differentiation [31].
To date, the levels of GDPD1-5 gene expression in human breast cancer cells and tumors have not yet been determined. As candidate gene(s) for GPC-PDE, it is of interest to investigate the role of GDPDs in choline phospholipid metabolism in breast cancer. In the present study, we have investigated for the first time the expression levels of GDPD1-5 in human breast cancer cell lines and tumor samples from breast cancer patients. The levels of GDPD1-5 expression were related to choline phospholipid metabolite levels detected with magnetic resonance spectroscopy (MRS) and the levels of CHKA and PLD1 expression in two breast cancer groups with distinct estrogen receptor status to investigate their role in regulating choline phospholipid metabolism and their association with a malignant phenotype.
Materials and Methods
Human breast epithelial and breast cancer cell lines
We used nonmalignant MCF-12A human mammary epithelial cells, estrogen-sensitive weakly metastatic human MCF-7 breast cancer cells, and estrogen-independent highly metastatic human MDA-MB-231 breast cancer cells in our cell studies. All cell lines were obtained from American Type Culture Collection (Rockville, MD) and used within 6 months of obtaining them from ATCC. The cell lines were tested and authenticated by ATCC by two independent methods: the ATCC cytochrome C oxidase I PCR assay and short tandem repeat profiling using multiplex PCR. Cell culture of these cell lines was performed as previously described [38].
Dual-phase extraction of cells, MRS acquisition, and quantification
Approximately 107 cells per extract were harvested by trypsinization with 0.25 % Trypsin-EDTA Solution (Sigma-Aldrich). Cells were counted in a dilution of trypan blue as a vital stain for quantification. Both lipid and water-soluble cell extract fractions were obtained using a dual-phase extraction method based on methanol/chloroform/water (1:1:1; v/v/v) as previously described [1]. The methanol/water phase contains the water-soluble metabolites, such as Cho, PC, and GPC, while the chloroform phase contains the cellular lipids, such as PtdCho. Fully relaxed high-resolution 1H MR spectra were obtained on a Bruker Avance 500 (11.7 T) spectrometer (Bruker BioSpin Corp.) using a 5-mm HX inverse probe. The water-soluble and lipid fractions were dissolved in deuterated solvents containing 0.24×10−6 mol 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP; Sigma-Aldrich) and 2.17×10−7 mol tetramethylsilane (TMS; Cambridge Isotope Laboratories, Inc.), respectively, as internal concentration and chemical shift standards. The fully relaxed high-resolution 1H MR spectra were acquired using the following acquisition parameters: 45° flip angle, 6,000 Hz sweep width, 12.7 s repetition time, 32 K time domain size, and 128 scans. The spectra were processed using the MestReC 4.9.9.6 software (Mestrelab Research). Signal integrals of the −N(CH3)3 signals of free Cho (3.209 ppm), PC (3.227 ppm) and GPC (3.236 ppm) in the water-soluble fractions and of PtdCho (3.220 ppm) in the lipid fractions were determined and normalized to cell number and cell volume as previously described [1]. The formula used for this normalization was [metabolite] = { I(metabolite) × standard } / { I(standard) × cell number × cell volume }, in which [metabolite] represents the intracellular concentration of the metabolite of interest expressed in mM [1]. I(metabolite) represents the signal integral of the metabolite of interest divided by the number of protons, and (standard) represents the amount of TSP (water-soluble metabolites) or TMS (lipids) used in mol divided by the number of protons. The number of cells in each sample (cell number) was counted before extraction, and the average cell volumes used in this formula were 9.8×103 μm for MCF-12A, 6.7×103 μm for MCF-7, and 8.4×103 μm for MDA-MB-231 as previously determined [1].
Total RNA isolation and qRT-PCR of cells
MCF-12A, MCF-7, and MDA-MB-231 cells were cultured in 10-cm diameter dishes to approximately 60% confluence, at which time total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The concentration and purity of RNA yield was determined with a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific) by measuring the absorption ratio A260/280. Total RNA (1 μg) was used for preparation of cDNA by SuperScript III First-Strand Synthesis SuperMix (Invitrogen). GDPD1-5 and CHKA expression levels in MCF-12A, MCF-7, and MDA-MB-231 were measured by qRT-PCR using the iCycler real-time PCR detection system (Bio-Rad) and iQ SYBR Green Supermix (Quanta BioSciences). For each sample, 2 μL of 1:10 diluted cDNA, 2 μM of the paired sense and antisense primers (see Table 1) and 10 μL of SYBR Green were used. The final reaction volume was filled up to 20 μL with sterile water. Relative quantification of gene expression was achieved in triplicates. Normalization was performed to housekeeping genes, human ribosomal protein 36B4 and hypoxanthine phosphoribosyltransferase 1 (HPRT1). The relative fold change in gene expression of GDPD1-5 was calculated based on the threshold cycle (ct) using the ΔΔct method [39] as R = 2 Δ(Δct), where Δct = ct(GDPD1-5/CHKA) ct(housekeeping gene), and Δ(Δct) = Δct(GDPD1-5/CHKA_MCF-7 or _MDA-MB-231) Δct(GDPD1-5/CHKA_MCF-12A). The Δct levels are inversely proportional to the amount of measured mRNA. Gene specific primers for GDPD1-5 and CKHA (Invitrogen) were designed using the Primer-BLAST tool (http://www.ncbi.nlm.nih.gov) and are listed in Table 1.
Table 1.
Sense (S) and antisense (AS) primer sequences used for qRT-PCR of GDPD1-5
| Gene | Cell lines | Tumor samples from patients |
|---|---|---|
| GDPD1-S | 5’-GAGCTTCTGTGGGAGTACGG-3’ | 5’-AAGCAGCGATTCCTCAGTA-3’ |
| GDPD1-AS | 5’-CGCAGGAAGAAGATGGAGAG-3’ | 5’-AGTCCAATTCTAGCATATCAGTTC-3’ |
| GDPD2-S | 5’-TGCTGCTGACAAGGATCAAC-3’ | 5’-ACTGTGTTTGAGACTGAT-3’ |
| GDPD2-AS | 5’-CCCCTGAAGCATTCCACTTA-3’ | 5’-GGAATACAGAGGCTACAT-3’ |
| GDPD3-S | 5’-CCACACACCATGTCCAGAAG-3’ | 5’-TTCGTGAGACGCTATGAC-3’ |
| GDPD3-AS | 5’-GAATGCCTCGAGCAGTTAGG-3’ | 5’-ATCCTCGGCTTATTGTGAA-3’ |
| GDPD4-S | 5’-CCCAAGAGTGGAAAGGAACA-3’ | 5’-TCAGCATAACTGTGATGGT-3’ |
| GDPD4-AS | 5’-GCTTCAAAGGTGTGACAGCA-3’ | 5’-CACTTGTAGACCTCTTAACCT-3’ |
| GDPD5-S | 5’-AGCAGTCACCATGCTCTCCT-3’ | 5’-CTACAACCCTGAGCAGAT-3’ |
| GDPD5-AS | 5’-AAACACCACGGTGAAGAAGG-3’ | 5’-AACATACGGAGAGCACAT-3’ |
| CHKA-S | 5’-GATCCGAACAAGCTCAGAAAGAAAATG-3’ | 5’-GATCCGAACAAGCTCAGAAAGAAAATG-3’ |
| CHKA-AS | 5’-CGGCTCGGGATGAACTGCTC-3’ | 5’-CGGCTCGGGATGAACTGCTC-3’ |
| PLD1-S | - | 5’-ATGGAATCCGAATTGATAATCTT-3’ |
| PLD1-AS | 5’-AGCATTCTCTTGGATAGCA-3’ | |
|
| ||
| Housekeeping genes
| ||
| 36B4-S | 5’-GATTGGCTACCCAACTGTTGCA-3’ | |
| 36B4-AS | 5’-CAGGGGCAGCAGCCACAAAGGC-3’ | |
|
| ||
| HPRT1-S | 5’-CCTGGCGTCGTGATTAGTGATG-3’ | |
| HPRT1-AS | 5’-CAGAGGGCTACAATGTGATGGC-3’ | |
All GDPD primers were designed to be gene specific and not transcript specific.
Protein lysates of cells, gel electrophoresis, and immunoblotting
Approximately 3 × 106 cells at 70% confluence were homogenized with lysis buffer containing protease inhibitor cocktail (P8340, Sigma-Aldrich) as previously described [11]. Thirty micrograms of total protein, as determined by a modified Lowry assay (Bio-Rad), was loaded in each lane, and two lanes were loaded with molecular weight standard (BenchMark, Life Technologies). Ten percent SDS-PAGE and immunoblotting were performed as previously described [11]. Membranes were incubated for 1 hour with a 1:500 dilution of primary affinity-purified rabbit polyclonal anti-GDPD5 antibody (Cat. No. AP10992c, Abgent Inc.) followed by incubation with horseradish peroxidase–conjugated second-step antibody (Amersham Life Science), and visualization using the Supersignal West Pico chemiluminescent substrate kit (Pierce Biotechnology) recorded on Blue Bio film (Denville Scientific). The films were scanned and densitometry was performed using the Gel-analysis-tool in ImageJ (Wayne Rasband, NIH, Bethesda, MD). We did not have sufficient tumor material to run the immunoblots on the patient samples.
Human breast tumor samples
Pre-treatment biopsies from 19 female patients with locally advanced breast cancer enrolled in a prospective study [40] were included. The study protocol was approved by the Regional Ethical Committee (Norwegian Health Region III). Each patient gave written informed consent. Tumor biopsies were obtained from open biopsy at diagnosis. Part of each tumor specimen was formaldehyde-fixed and paraffin-embedded for histopathological classification and receptor status assignment, and part of each tumor specimen was immediately snap-frozen and stored in liquid nitrogen until use. Patients were diagnosed with invasive ductal carcinoma or lobular carcinoma. Estrogen receptor (ER) status was determined by immunohistochemistry according to standard clinical pathology protocols (positive ≥ 10% of cells +). To be able to study the causes of different choline metabolite profiles, tumors were selected and grouped according to their ER status and choline metabolite profiles, resulting in group (a) ER positive (ER+)/low PC/GPC ratio and group (b) ER negative (ER−)/high PC/GPC ratio. The cut-off value between high and low PC/GPC was 1.0, and all selected samples fit into one of these two defined groups. These two tumor groups matched the characteristics of the two breast cancer cell lines in our study. Detailed patient and tumor characteristics are listed in Table 2.
Table 2.
Patient and tumor characteristics
| ER+/ low PC/GPC (n=11) | ER−/ high PC/GPC (n=8) | |
|---|---|---|
| Age | 47.8±9.7 | 44.7±10.9 |
|
| ||
| Tumor size (mm) | 64.6×64.7 ± 17.4×19.6 | 79.3×75.8 ± 24.3×25.3 |
|
| ||
| UICC classifier | IIB/IIIA/IIIB (3/6/2) | IIB/IIIA/IIIB (1/4/3) |
|
| ||
| Sample weight (mg) | 14.3±2.8 | 15.1±1.8 |
| Tumor cells | >1000 | >1000 |
| RIN-value | 7.5±1.1 | 7.7±1.4 |
Values are mean±standard deviation (SD); RIN, RNA integrity number; UICC, Union for International Cancer Control.
Ex vivo HR MAS MRS of breast tumor specimens and metabolite quantification
Each tumor specimen was analyzed by imprint cytology and May-Grünwald-Giemsa staining to assess tumor cell content prior to MRS analysis [41]. All samples had a high total number of tumor cells (n ≥ 1000). Each sample (14.6 ± 2.4 mg) was immediately transferred to a 30 μL disposable insert (Bruker Biospin Corp.) filled with 3 μL PSB in D2O containing 98.8 mM TSP, which was inserted into a 50 μL HR-MAS rotor (Bruker BioSpin Corp.). High resolution magic angle spinning (HR MAS) spectra were acquired on a Bruker Avance 600 (14.1 T) (Bruker BioSpin Corp.) spectrometer, at 5 kHz spinning rate and 4 °C to minimize tissue degradation as previously described [42]. For all samples, we performed pulse-acquire experiments, which included the electronic reference to access in vivo concentration (ERETIC) sequence for quantification [43]. Spectra were acquired using the following acquisition parameters: water saturation using a 60 dB continuous wave for 15 seconds, followed by a 60° pulse for excitation, 16.7 ppm sweep width, 3.28 s acquisition time, 64 K time domain size, 18.28 s repetition time, and 64 scans. Chemical shifts were calibrated relative to the TSP signal at 0 ppm. Spectra were processed using the XWIN-NMR 3.5 software (Bruker NMR). Peak areas of Cho (3.209 ppm), PC (3.227 ppm) and GPC (3.236 ppm) were fitted as Voigt curves using the PeakFit program (v 4.12, SeaSolve Software Inc.). Absolute concentrations of individual metabolites were determined and normalized to tumor sample weight as previously described [43].
Total RNA isolation and qRT-PCR of human tumor specimens
Subsequent to the ex vivo HR MAS analysis, total RNA was isolated from the same tumor specimen for each sample using rotor-stator homogenizer and the RNeasy Mini Kit (Qiagen). Quantity and quality measurements of total RNA and qRT-PCR were performed as described above for cells. All samples had sufficient total RNA concentration (> 6.9 μg). The RNA integrity number (RIN), which was measured by electropherogram and gel-like image using Bioanalyzer 2100 (Agilent), was 7.6 ± 1.2 for all samples on a scale of 1 to 10 with 10 being the best. Hypoxanthine phosphoribosyltransferase 1 was used as housekeeping gene. The relative fold change in gene expression of GDPD1-5, CHKA and PLD1 was calculated relative to the sample with lowest GDPD5 expression, which was done because no normal breast tissue was available from the breast cancer cases in this study. Gene specific primers for GDPD1-5, CHKA, and PLD1 (Invitrogen) were designed using the Primer-BLAST tool (http://www.ncbi.nlm.nih.gov) and are listed in Table 1.
Statistical analysis
The normality of data was evaluated by the Kolmogorov-Smirnov test (SPSS 16.0 Inc.). Grubbs test was performed to detect significant statistical outliers for each gene set independently (significant level alpha = 0.05) (GraphPad Software). Maximally one outlier was discarded per set. An unpaired two-tailed t-test was performed to detect significant differences between the tested groups. Pearson correlation analysis (SPSS 16.0 Inc.) was also performed between gene and metabolite data, as well as Western Blot and metabolite data. The protein content used to normalize Western Blot data was constant for each cell line. The cell volume used to normalize MR spectra of cell extracts was also constant for each cell line. Both Western Blot data and MR data were normalized to the cell count of the respective sample, and therefore, both types of data can be compared. P values of < 0.05 were considered to be significant. P values of 0.15 > P > 0.05 were considered to be a trend towards significance [44].
Results
Co-expression of GDPD5 and CHKA in cell extracts and correlation to choline containing metabolite levels
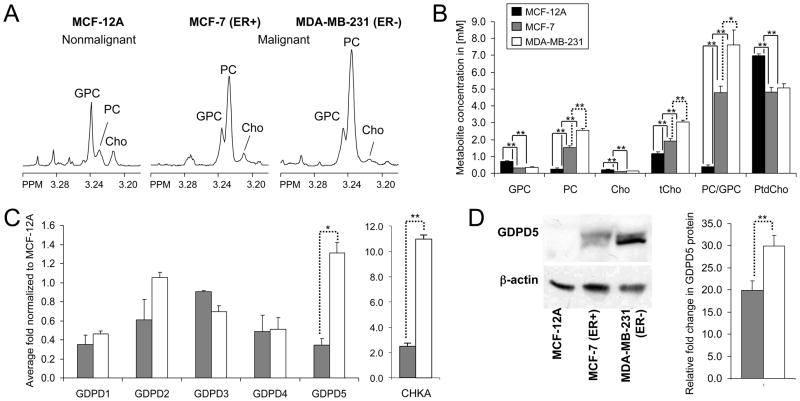
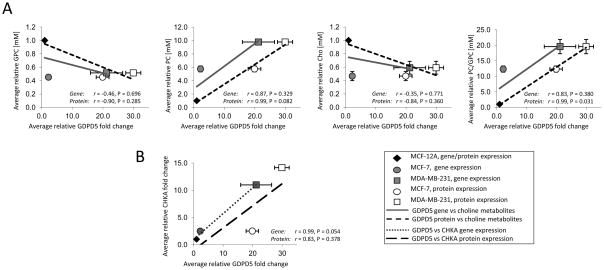
The representative choline metabolite profiles obtained from 1H MR spectra in Fig. 1A show the characteristic switch from high GPC and low PC to low GPC and high PC in malignant compared to nonmalignant mammary epithelial cells. Metabolite quantification of nonmalignant MCF-12A (n = 7) and the breast cancer cell lines MCF-7 (n = 10) and MDA-MB-231 (n = 8) cells is summarized in Fig. 1B. The PC, tCho, and PC/GPC levels increased progressively with increasing malignancy. MDA-MB-231 cells exhibited significantly higher levels of PC (P < 0.001), tCho (P < 0.001), and PC/GPC ratio (P = 0.017) compared to MCF-7 cells. The relative fold changes (R = 2 Δ(Δct)) in gene expression of GDPD1-5 obtained from MCF-12A, MCF-7, and MDA-MB-231 are presented in Fig. 1C (n = 2 × 3 technical replicates). CHKA gene expression obtained from MCF-12A (n = 3), MCF-7 (n = 4), and MDA-MB-231 (n = 3) are also presented in Fig. 1C (n × 3 technical replicates). GDPD1-5 expression levels were present in nonmalignant and malignant mammary epithelial cell lines. GDPD5 had the highest amount of mRNA in all tested cell lines compared to the other GDPDs (data not shown). GDPD5 and CKHA mRNA levels were significantly overexpressed (P = 0.027 and P < 0.001, respectively) in the malignant estrogen-independent highly metastatic cell line MDA-MB-231 compared to the estrogen-sensitive weakly metastatic breast cancer cell line MCF-7. Fig. 1D shows the immunoblots of GDPD5 protein, in which the relative fold change in GDPD5 protein expression was significantly higher (P = 0.001) in MDA-MB-231 compared to MCF-7 cells (n = 3, each). Pearson correlation analysis was performed with the average number of samples for each of the three cell lines. Positive correlations were found between GDPD5 mRNA and protein levels versus PC levels (r = 0.87, P = 0.329 and r = 0.99, P = 0.082, respectively) and GDPD5 mRNA and protein levels versus PC/GPC ratios (r = 0.83, P = 0.380 and r = 0.99, P = 0.031) in nonmalignant and malignant mammary epithelial cells (Fig. 2A). GDPD5 mRNA and protein levels showed negative correlations with GPC (r = −0.46, P = 0.696 and r = −0.90, P = 0.285) and Cho (r = −0.35, P = 0.771 and r = −0.84, P =0.360). As a consequence of using the average values, the p-values are not statistically significant (P > 0.05) for several of the correlations, even for those with high Pearson’s correlation coefficient (r). PtdCho showed no significant correlations with GDPD5 mRNA and GDPD5 protein levels. The CHKA protein levels in the three breast cell lines studied have previously been reported by Glunde et at [11]. GDPD5 versus CHKA mRNA and protein levels (n = 3) showed positive correlations (r = 0.99, P = 0.054 and r = 0.83, P = 0.378, Fig. 2B), and both GDPD5 and CHKA increased with increasing malignancy in the three tested breast cell lines.
Figure 1.
A, representative 1H MR spectra of the choline region from hydrophilic extracts of MCF-12A, MCF-7, and MDA-MB-231 cells. B, metabolite concentrations in [mM] quantified from 1H MR spectra of MCF-12A (black bars), MCF-7 (gray bars), and MDA-MB-231 (white bars) cells. C, relative fold change in gene expression of GDPD1-5 and CHKA obtained from MCF-7 and MDA-MB-231 cells normalized to MCF-12A cells. D, representative GDPD5 immunoblots and relative fold change in GDPD5 protein levels normalized to MCF-12A cells. β-actin was used as loading control. Values are mean±standard error (SE). *P < 0.05, **P < 0.01.
Figure 2.
A, Correlation of GDPD5 mRNA expression as a function of R = 2 Δ(Δct) and protein levels expressed as relative fold change normalized to MCF-12A versus relative choline phospholipid metabolite concentrations in human breast cancer cells normalized to MCF-12A. B, Correlation of GDPD5 versus CHKA mRNA and protein levels expressed as relative fold change normalized to MCF-12A. Values are mean±standard error (SE). The gene, protein, and metabolite levels were normalized to MCF12-A, and therefore the symbols for gene and protein expression data of MCF-12A superimpose.
GDPD5 expression in tumors and correlation to choline containing metabolite levels
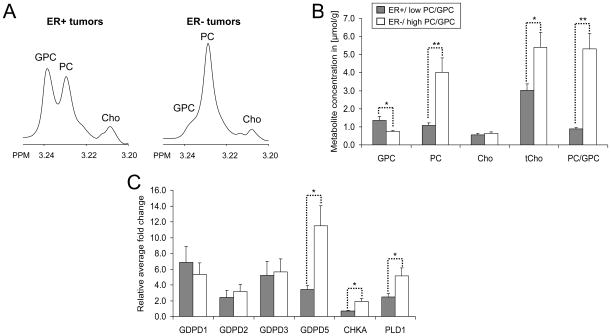
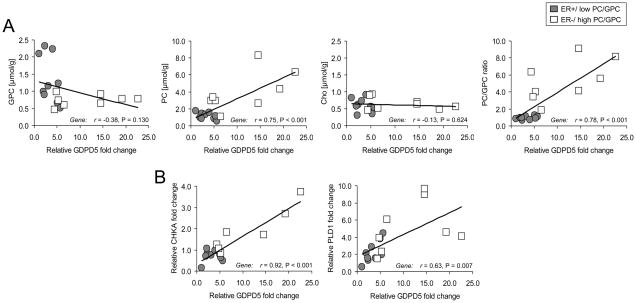
Representative choline metabolite profiles obtained from ex vivo HR MAS spectra and the corresponding metabolite quantifications are shown in Fig. 3, A and B, respectively. The resolution of HR MAS spectra acquired from tumor specimens is comparable to the spectral resolution of cell extracts, which makes it feasible to combine the investigation of individual choline metabolite levels with gene expression analysis within the same sample. The correlation factor (r2) which describes the quality of the quantification was > 0.99 for all spectra. As expected, ER−/high PC/GPC (n = 8) tumors exhibited significantly higher levels of PC (P = 0.008), tCho (P = 0.028), and PC/GPC ratio (P = 0.001) and lower levels of GPC (P = 0.016) compared with ER+/low PC/GPC tumors (n = 10). No significant difference in Cho levels (P = 0.378) was observed between these two tumor groups. The relative fold change in gene expression of GDPD 1, 2, 3, and 5 obtained from breast tumors with ER+/low PC/GPC and ER−/high PC/GPC are presented in Fig. 3C (n=17 × 3 technical replicates). GDPD4 had a very high ct value (> 31 cycles), which indicated a high chance of non-specific binding and was therefore excluded. GDPD5 was significantly overexpressed in the ER−/high PC/GPC tumors compared to the ER+/low PC/GPC tumors (P = 0.015). Significant positive correlations were found between GDPD5 expression versus PC levels (r = 0.75, P < 0.001) and GDPD5 expression versus PC/GPC ratios (r = 0.78, P < 0.001) in human breast tumors (Fig. 4A). A significant positive correlation was also found between GDPD5 expression versus tCho levels (r = 0.63, P = 0.007, data not shown). The GDPD5 expression showed a trend towards negative correlation with GPC levels (r = −0.38, P = 0.130). No correlation was found between GDPD5 expression and Cho levels (r = −0.13, P = 0.624). Although GDPD2 expression was not significantly different between the ER+ and ER− tumor groups, GDPD2 expression positively correlated with GDPD5 expression (r = 0.61, P = 0.015) and negatively correlated with Cho levels (r = −0.55, P = 0.028), data not shown.
Figure 3.
A, representative ex vivo HR MAS spectra of the choline region from ER+/low PC/GPC and ER−/high PC/GPC tumors. B, metabolite concentrations [μmol/g] quantified from HR MAS spectra of ER+/low PC/GPC (gray bars) and ER−/high PC/GPC tumors (white bars). C, relative fold change in gene expression of GDPD1, 2, 3, and 5, CHKA and PLD1 obtained from ER−/high PC/GPC tumors and ER+/low PC/GPC tumors normalized to the sample with the lowest GDPD5 expression. Values are mean±standard error (SE). *P < 0.05, **P < 0.01.
Figure 4.
A, correlation of GDPD5 expression versus choline phospholipid metabolite concentrations in human breast tumors. B, correlation of GDPD5 versus CHKA and GDPD5 versus PLD1 expression in human breast tumors.
Co-expression of GDPD5, CHKA, and PLD1 and their correlation with choline phospholipid metabolism
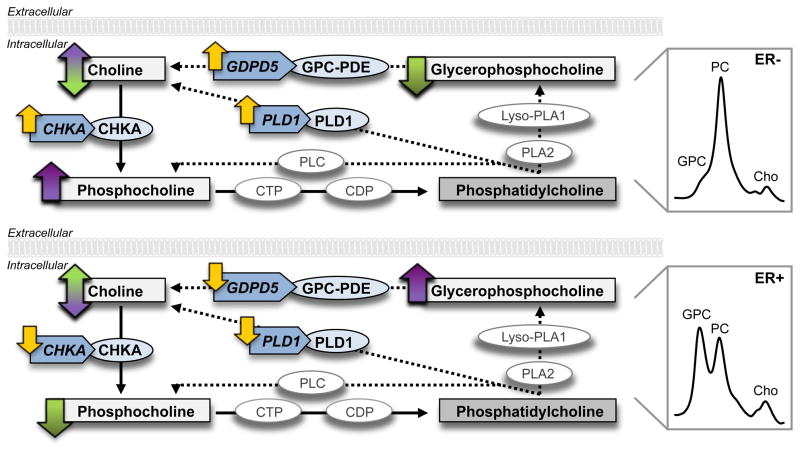
Similar to the GDPD5 expression, CHKA and PLD1 were significantly overexpressed in ER−/high PC/GPC tumors compared to ER+/low PC/GPC tumors (P = 0.021 and P = 0.042, respectively, Fig. 3C). Notably, significant correlations were found between GDPD5 versus CHKA (r = 0.92, P < 0.001) and GDPD5 versus PLD1 (r = 0.63, P = 0.007) in human breast tumors (Fig. 4B). High expression of GDPD5, CHKA, and PLD1 was detected in human ER− tumors, which displayed choline metabolite profiles consisting of low GPC and high PC levels as determined by HR MAS MRS. Low expression of GDPD5, CHKA, and PLD1 was observed in human ER+ tumors with choline metabolite profiles consisting of high GPC and relatively low PC levels (Fig. 5).
Figure 5.
A, high co-expression of GDPD5, CHKA, and PLD1 was associated with choline metabolite profiles consisting of low GPC and high PC levels in human ER−/high PC/GPC breast tumors, as determined by HR MAS MRS. B, low co-expression of GDPD5, CHKA, and PLD1 was accompanied by high GPC and relatively low PC levels in human ER+/low PC/GPC breast tumors. The Cho level remained the same in both tumor groups. GPC-PDE, glycerophosphocholine phosphodiesterase; PLC, phosphatidylcholine-specific phospholipase C; CTP, phosphocholine cytidylyltransferase; CDP, diacylglycerol cholinephosphotransferase; PLA2, phosphoplipase A2; Lyso-PLA1, lysophospholipase A1.
Discussion
Here we have shown for the first time that GDPD5 is overexpressed in highly metastatic human ER−/high PC/GPC breast cancer cells as well as in ER−/high PC/GPC breast tumor samples from patients as compared to ER+/low PC/GPC cells and tumors. We demonstrated that GDPD5 expression positively correlates with PC levels and PC/GPC ratios, and shows a trend towards negative correlation with GPC levels, which was consistent in cell lines and tumors. GDPD5 expression also positively correlates with CHKA in human breast epithelial and cancer cell lines, and CHKA and PLD1 expression in human breast tumor samples. Our studies were performed with combined MRS, qRT-PCR, and immunoblotting to investigate the potential role of GDPD1-5 as GPC-PDE encoding genes and to determine their relationship with CHKA and PLD1 in choline phospholipid metabolism.
The expression of GDPD5 was significantly increased in highly metastatic ER− MDA-MB-231 breast cancer cells compared to weakly metastatic ER+ MCF-7 breast cancer cells, while GDPD1-4 were not significantly differentially expressed. The PC and tCho levels were shown to increase progressively with increasing malignancy. These findings are in agreement with a previous study, where malignant transformation of breast epithelial cells led to an increased PC/GPC ratio and tCho levels [8]. The increased membrane PtdCho level in nonmalignant MCF-12A cells compared to MCF-7 and MDA-MB-231 breast cancer cells may be caused by the larger cell volume of this cell line, possibly requiring more plasma membrane and hence PtdCho. The membrane PtdCho pool of cells is rather large compared to the pools of the water-soluble metabolites Cho, PC, and GPC. The PtdCho concentration has not been implicated in malignant transformation. The protein levels of GDPD5 correlate well with GDPD5 mRNA expression levels. The expression of GDPD5 increased progressively with increasing PC and PC/GPC ratio levels, suggesting an association between GDPD5 expression and breast cancer malignancy.
High levels of PC detected in breast cancer cells can be caused by enhanced generation of PC by CHKA [1, 18, 21] or increased degradation of PtdCho by phospholipase C and D [1, 15]. Enhanced choline transport activities can also contribute to the increased PC levels [16–18]. Our data suggest that GDPD5, which encodes an enzyme with GPC-PDE activity, may additionally contribute to the elevated PC levels in breast cancer. Increased expression of GDPD5-mediated GPC-PDE activity in breast cancer cells likely results in a higher degradation rate of GPC to form Cho and glycerol-3-phosphate. The produced free choline is then most likely immediately used by CHKA and recycled to synthesize PC. Increasing CHKA protein levels have been shown to correlate with increasing malignancy in breast cancer cell lines [11]. The positive correlation between GDPD5 versus CHKA mRNA and protein levels along with their concerted increase with increasing malignancy demonstrated here suggests that these two enzymes may be co-regulated. The expression of GDPD5 showed a modest inverse correlation with GPC levels, which supports that GDPD5 is a GPC-PDE that is at least in part responsible for the relatively low GPC levels and high PC levels in breast cancer cells.
To further evaluate the correlation between GDPD5 and choline phospholipid metabolites observed in breast cancer cells, tumor biopsies from locally advanced breast cancer patients were studied by a combination of ex vivo HR MAS MRS and qRT-PCR analyses. HR MAS MRS analysis enabled the quantification of choline compounds from intact breast tumor tissues prior to gene expression analysis, which is an excellent method for combined metabolomics and genomics studies in the same tumor tissues. In good agreement with our results of breast cancer cells in culture, GDPD5 was significantly overexpressed in human ER−/high PC/GPC breast tumors compared to ER+/low PC/GPC breast tumors. The expression of GDPD5 showed a positive correlation with PC, tCho, and PC/GPC levels, while no significant correlation was found between GDPD1, 2 or 3 and choline phospholipid metabolites in these tumors. As a candidate gene for GPC-PDE, we expected the GDPD5 expression to inversely correlate with the GPC level. However, high expression of GDPD5 only showed a trend towards decreased GPC levels in our study. This observation suggests the possibility of additional genes responsible for the altered GPC level observed in breast cancer, such as for example nucleotide pyrophosphatases/phosphodiesterases 6 (NPP6), which is a transmembrane protein with choline-specific GPC-PDE activity. Human NPP6 has mainly been detected in kidney and brain [45]. NPP6 preferably hydrolyzed choline-containing phospholipids or phosphodiesters such as lysoPtdCho, sphingosylphosphorylcholine, lysoplatelet-activating factor, platelet-activating factor, and GPC [45]. To date, the exact role of NPP6 is still unknown. In future studies, we will evaluate if NPP6 may be responsible for GPC-PDE activity in cancer as well. A previous study showed that PLA2 and Lyso-PLA1, the enzymes responsible for the degradation of PtdCho to generate GPC, were significantly underexpressed in MDA-MB-231 breast cancer cells compared to nonmalignant MCF-12A cells resulting in decreased GPC levels in breast cancer cells detected by 1H MRS [1]. Thus, the enzyme activity of PLA2 and Lyso-PLA1 may also play an important role in regulating the GPC levels in cancer.
Our data demonstrated that GDPD5, CHKA and PLD1 were expressed in a concerted manner in human breast cancers. High co-expression of GDPD5, CHKA and PLD1 observed in ER− breast tumors was associated with choline metabolite profiles characterized by low GPC and high PC levels. Conversely, low co-expression of GDPD5, CHKA and PLD1 observed in ER+ breast tumors was accompanied by high GPC and low PC levels (Fig. 5). These data suggest that Cho is being depleted by CHKA to synthesize PC once it is generated by GDPD5 and PLD1, which might explain the unchanged Cho level observed at high and low co-expression levels of GDPD5, PLD1 and CHKA. These findings indicate that GDPD5 may contribute as a GPC-PDE to the regulation of choline phospholipid metabolism in breast cancer. Elevated PC levels and relatively low GPC levels, which are characteristic of the malignant choline metabolite profile in breast cancers, are most likely the result of collectively overexpressed/activated enzymes such as GDPD5, CHKA, and PLD1, and possibly others.
Our results also showed that GDPD5, CHKA, and PLD1 were significantly overexpressed in ER− breast tumors compared to ER+ tumors. In agreement with our findings, significantly higher CHKA expression and CHKA activity were previously shown to be associated with ER− as compared to ER+ breast carcinomas [22]. Overexpression of PLD1 was previously demonstrated in an ER-negative MCF-7 cell line variant [46]. In addition to validating these previous findings regarding CHKA and PLD1, we have for the first time shown that elevated GDPD5 is associated with negative ER status in breast cancers. ER status is an important prognostic factor in breast cancer, which is routinely used for treatment planning in the clinic. Patients with ER+ tumors typically benefit from anti-estrogen drugs such as the non-steroidal ER antagonist tamoxifen (Nolvadex, Soltamax), which was shown to result in a reduction of breast cancer recurrences [47]. The current recommendation for tamoxifen therapy after surgery for patients with ER+ tumors is 5 years, which has contributed to a decrease in the breast cancer death rate [48]. Unfortunately, ER− breast cancer patients are unresponsive to anti-estrogen drugs. Therefore, identification of new targets for treating ER− breast cancer is needed to improve treatment and eventually increase the survival rate for patients with this more aggressive type of breast cancer.
The Ras G-protein/MAP kinase signaling pathway regulates many genes involved in cellular proliferation, differentiation, and apoptosis [49]. Ras oncogenes can affect the activity of CHK and PLD, resulting in increased PC levels [50]. CHKA and PLD can play essential roles in oncogenic transformation and carcinogenesis in cancer [13, 19–21]. GDPD5 was identified as a retinoic acid responsive gene that is necessary to drive motor neuron differentiation in vivo [29] through interaction with the G-protein subunit Gαi2 [30]. Overexpression of GDPD5 induced a dose-dependent suppression of the serum responsive element controlling the transcription of MAP kinases [32]. These observations suggest that GDPD5 has diverse functions in cellular signal transduction pathways, which are frequently deregulated during oncogenic transformation and in the resulting cancers.
Targeting enzymes in choline phospholipid metabolism previously proved successful as anticancer treatment strategy in preclinical studies. MN58b was recently developed to selectively inhibit CHK [23]. MN58b treatment reduced PC and tCho levels and inhibited proliferation in cancer cells and tumor xenograft models [23]. RNAi-mediated silencing of CHKA reduced proliferation and tumor growth along with PC and tCho levels in human breast cancer cells and breast tumor xenografts [11, 12]. Novel CHKA inhibitors are currently being evaluated in cancer patients in clinical phase I trials for toxicity and pharmacokinetic profile testing. Successful inhibition of phospholipase D and prevention of tumor cell invasion was achieved with ketoepoxides in HT1080 human fibrosarcoma cells [51]. Future studies examining the effect of silencing GDPD5 alone or in combination with CHKA and/or PLD1 in breast cancer cells and xenograft models will be pursued, as targeting GDPD5 alone or in combination with CHKA and/or PLD1 might provide potential anticancer therapies. Experiments with NPP6, another candidate gene for GPC-PDE, will also be explored.
Conclusions
To conclude, our study demonstrated for the first time an increased expression of GDPD5 in highly malignant ER− human breast cancers cell lines and patient tumors that display malignant choline metabolite profiles consisting of low GPC and high PC levels. Our results identified GDPD5 as a GPC-PDE that is, together with CHKA and PLD1, overexpressed in highly malignant ER− breast cancer. Our data suggest that GDPD5 is most likely involved in membrane phosphatidylcholine metabolism and is at least partially responsible for the malignant choline metabolite profile detected by MRS in breast cancer.
Acknowledgments
We thank Tiffany R. Greenwood and Dr. Beathe Sitter for technical laboratory support. We thank Dr. Dieter Leibfritz and Dr. Venu Raman for helpful discussions in this project.
This work was supported by the National Institutes of Health (NIH) grant R01 CA134695.
Abbreviations (used in the paper three or more times)
- CHKA
choline kinase alpha
- Cho
free choline
- ct
threshold cycle
- ER
estrogen receptor
- GDPD
glycerophosphodiester phosphodiesterase domain containing
- GPC
glycerophosphocholine
- GPC-PDE
glycerophosphocholine phosphodiesterase
- HR MAS
high resolution magic angle spinning
- Lyso-PLA1
lysophospholipase A1
- MRS
magnetic resonance spectroscopy
- NPP6
nucleotide pyrophosphatases/phosphodiesterases 6
- PC
phosphocholine
- PLA2
phosphoplipase A2
- PLD
phosphatidylcholine -specific phospholipase D
- PtdCho
phosphatidylcholine
- RNAi
RNA interference
- tCho
total choline-containing metabolites
- TSP
3-(trimethylsilyl)propionic-2,2,3,3-d4 acid
Footnotes
Declaration of Competing Interests
The author(s) declare that they have no competing interests.
Authors' contributions
MDC carried out all experiments with human breast tumor samples, performed all human-related data analyses, and drafted the manuscript. MD carried out all cell culture experiments and performed all cell-related data analyses. BK, FV, and MMG supervised and participated in performing all qRT-PCR experiments. PEL collected all human breast tumor samples and performed all related clinical analyses. ZMB and ISG helped in designing and coordinating the study. KG conceived, designed, and coordinated the study, and helped draft the manuscript. All authors read and approved the final manuscript.
References
- 1.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 2.Glunde K, Bhujwalla ZM. Metabolic tumor imaging using magnetic resonance spectroscopy. Semin Oncol. 2011;38:26–41. doi: 10.1053/j.seminoncol.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz-Brull R, Lavin PT, Lenkinski RE. Clinical utility of proton magnetic resonance spectroscopy in characterizing breast lesions. J Natl Cancer Inst. 2002;94:1197–1203. doi: 10.1093/jnci/94.16.1197. [DOI] [PubMed] [Google Scholar]
- 4.Meisamy S, Bolan PJ, Baker EH, Bliss RL, Gulbahce E, Everson LI, Nelson MT, Emory TH, Tuttle TM, Yee D, Garwood M. Neoadjuvant chemotherapy of locally advanced breast cancer: predicting response with in vivo (1)H MR spectroscopy--a pilot study at 4 T. Radiology. 2004;233:424–431. doi: 10.1148/radiol.2332031285. [DOI] [PubMed] [Google Scholar]
- 5.Glunde K, Jacobs MA, Bhujwalla ZM. Choline metabolism in cancer: implications for diagnosis and therapy. Expert Rev Mol Diagn. 2006;6:821–829. doi: 10.1586/14737159.6.6.821. [DOI] [PubMed] [Google Scholar]
- 6.Manton DJ, Chaturvedi A, Hubbard A, Lind MJ, Lowry M, Maraveyas A, Pickles MD, Tozer DJ, Turnbull LW. Neoadjuvant chemotherapy in breast cancer: early response prediction with quantitative MR imaging and spectroscopy. Br J Cancer. 2006;94:427–435. doi: 10.1038/sj.bjc.6602948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baek HM, Chen JH, Nalcioglu O, Su MY. Proton MR spectroscopy for monitoring early treatment response of breast cancer to neo-adjuvant chemotherapy. Ann Oncol. 2008;19:1022–1024. doi: 10.1093/annonc/mdn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 9.Hernandez-Alcoceba R, Fernandez F, Lacal JC. In vivo antitumor activity of choline kinase inhibitors: a novel target for anticancer drug discovery. Cancer Res. 1999;59:3112–3118. [PubMed] [Google Scholar]
- 10.Rodriguez-Gonzalez A, Ramirez de Molina A, Fernandez F, Ramos MA, del Carmen Nunez M, Campos J, Lacal JC. Inhibition of choline kinase as a specific cytotoxic strategy in oncogene-transformed cells. Oncogene. 2003;22:8803–8812. doi: 10.1038/sj.onc.1207062. [DOI] [PubMed] [Google Scholar]
- 11.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 12.Krishnamachary B, Glunde K, Wildes F, Mori N, Takagi T, Raman V, Bhujwalla ZM. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer Res. 2009;69:3464–3471. doi: 10.1158/0008-5472.CAN-08-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- 14.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D'Ascenzo S, Millimaggi D, Pavan A, Dolo V, Canevari S, Podo F. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–9376. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 15.Iorio E, Ricci A, Bagnoli M, Pisanu ME, Castellano G, Di Vito M, Venturini E, Glunde K, Bhujwalla ZM, Mezzanzanica D, Canevari S, Podo F. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2010;70:2126–2135. doi: 10.1158/0008-5472.CAN-09-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz-Brull R, Degani H. Kinetics of choline transport and phosphorylation in human breast cancer cells; NMR application of the zero trans method. Anticancer Res. 1996;16:1375–1380. [PubMed] [Google Scholar]
- 17.Katz-Brull R, Seger D, Rivenson-Segal D, Rushkin E, Degani H. Metabolic markers of breast cancer: enhanced choline metabolism and reduced choline-ether-phospholipid synthesis. Cancer Res. 2002;62:1966–1970. [PubMed] [Google Scholar]
- 18.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 19.Nakagami K, Uchida T, Ohwada S, Koibuchi Y, Morishita Y. Increased choline kinase activity in 1,2-dimethylhydrazine-induced rat colon cancer. Jpn J Cancer Res. 1999;90:1212–1217. doi: 10.1111/j.1349-7006.1999.tb00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez J, Bonilla F, Rosell R, Lacal J. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 21.Ramirez de Molina A, Banez-Coronel M, Gutierrez R, Rodriguez-Gonzalez A, Olmeda D, Megias D, Lacal JC. Choline kinase activation is a critical requirement for the proliferation of primary human mammary epithelial cells and breast tumor progression. Cancer Res. 2004;64:6732–6739. doi: 10.1158/0008-5472.CAN-04-0489. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J, Bonilla F, Sanchez JJ, Lacal JC. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–4322. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 23.Al-Saffar NM, Troy H, Ramirez de Molina A, Jackson LE, Madhu B, Griffiths JR, Leach MO, Workman P, Lacal JC, Judson IR, Chung YL. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of the choline kinase inhibitor MN58b in human carcinoma models. Cancer Res. 2006;66:427–434. doi: 10.1158/0008-5472.CAN-05-1338. [DOI] [PubMed] [Google Scholar]
- 24.Righi V, Roda JM, Paz J, Mucci A, Tugnoli V, Rodriguez-Tarduchy G, Barrios L, Schenetti L, Cerdan S, Garcia-Martin ML. 1H HR-MAS and genomic analysis of human tumor biopsies discriminate between high and low grade astrocytomas. NMR Biomed. 2009;22:629–637. doi: 10.1002/nbm.1377. [DOI] [PubMed] [Google Scholar]
- 25.Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Humpfer E, Spraul M, Carreira IM, Melo JB, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J Proteome Res. 2010;9:319–332. doi: 10.1021/pr9006574. [DOI] [PubMed] [Google Scholar]
- 26.Swanson MG, Keshari KR, Tabatabai ZL, Simko JP, Shinohara K, Carroll PR, Zektzer AS, Kurhanewicz J. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magn Reson Med. 2008;60:33–40. doi: 10.1002/mrm.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon ED, Zablocki K, Jung KY, Peters EM, Garcia-Perez A, Burg MB. Osmoregulation of GPC:choline phosphodiesterase in MDCK cells: different effects of urea and NaCl. Am J Physiol. 1995;269:C35–41. doi: 10.1152/ajpcell.1995.269.1.C35. [DOI] [PubMed] [Google Scholar]
- 28.Zablocki K, Miller SP, Garcia-Perez A, Burg MB. Accumulation of glycerophosphocholine (GPC) by renal cells: osmotic regulation of GPC:choline phosphodiesterase. Proc Natl Acad Sci U S A. 1991;88:7820–7824. doi: 10.1073/pnas.88.17.7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao M, Sockanathan S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science. 2005;309:2212–2215. doi: 10.1126/science.1117156. [DOI] [PubMed] [Google Scholar]
- 30.Periz G, Yan Y, Bitzer ZT, Sockanathan S. GDP-bound Galphai2 regulates spinal motor neuron differentiation through interaction with GDE2. Dev Biol. 2010;341:213–221. doi: 10.1016/j.ydbio.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogusa Y, Fujioka Y, Komatsu R, Kato N, Yanaka N. Isolation and characterization of two serpentine membrane proteins containing glycerophosphodiester phosphodiesterase, GDE2 and GDE6. Gene. 2004;337:173–179. doi: 10.1016/j.gene.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 32.Lang Q, Zhang H, Li J, Yin H, Zhang Y, Tang W, Wan B, Yu L. Cloning and characterization of a human GDPD domain-containing protein GDPD5. Mol Biol Rep. 2008;35:351–359. doi: 10.1007/s11033-007-9093-3. [DOI] [PubMed] [Google Scholar]
- 33.Gallazzini M, Ferraris JD, Burg MB. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proc Natl Acad Sci U S A. 2008;105:11026–11031. doi: 10.1073/pnas.0805496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu M, Ji C, Xu J, Zou X, Wang L, Zheng H, Jin F, Wang Y, Gu S, Ying K, Xie Y, Mao Y. A novel splice variant of human gene GDPD1 is mainly expressed in human ovary and small intestine. Int J Mol Med. 2003;12:1003–1007. [PubMed] [Google Scholar]
- 35.Chang PA, Shao HB, Long DX, Sun Q, Wu YJ. Isolation, characterization and molecular 3D model of human GDE4, a novel membrane protein containing glycerophosphodiester phosphodiesterase domain. Mol Membr Biol. 2008;25:557–566. doi: 10.1080/09687680802537605. [DOI] [PubMed] [Google Scholar]
- 36.Yanaka N, Imai Y, Kawai E, Akatsuka H, Wakimoto K, Nogusa Y, Kato N, Chiba H, Kotani E, Omori K, Sakurai N. Novel membrane protein containing glycerophosphodiester phosphodiesterase motif is transiently expressed during osteoblast differentiation. J Biol Chem. 2003;278:43595–43602. doi: 10.1074/jbc.M302867200. [DOI] [PubMed] [Google Scholar]
- 37.Corda D, Kudo T, Zizza P, Iurisci C, Kawai E, Kato N, Yanaka N, Mariggio S. The developmentally regulated osteoblast phosphodiesterase GDE3 is glycerophosphoinositol-specific and modulates cell growth. J Biol Chem. 2009;284:24848–24856. doi: 10.1074/jbc.M109.035444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–545. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Chrisanthar R, Knappskog S, Lokkevik E, Anker G, Ostenstad B, Lundgren S, Berge EO, Risberg T, Mjaaland I, Maehle L, Engebretsen LF, Lillehaug JR, Lonning PE. CHEK2 mutations affecting kinase activity together with mutations in TP53 indicate a functional pathway associated with resistance to epirubicin in primary breast cancer. PLoS One. 2008;3:e3062. doi: 10.1371/journal.pone.0003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangia A, Chiriatti A, Chiarappa P, Incalza MA, Antonaci G, Pilato B, Simone G, Tommasi S, Paradiso A. Touch imprint cytology in tumor tissue banks for the confirmation of neoplastic cellularity and for DNA extraction. Arch Pathol Lab Med. 2008;132:974–978. doi: 10.5858/2008-132-974-TICITT. [DOI] [PubMed] [Google Scholar]
- 42.Sitter B, Bathen T, Tessem M, Gribbestad I. High-resolution magic angle spinning (HR MAS) MR spectroscopy in metabolic characterization of human cancer. Prog Nucl Magn Reson Spectrosc. 2009;54:239–254. [Google Scholar]
- 43.Sitter B, Bathen TF, Singstad TE, Fjosne HE, Lundgren S, Halgunset J, Gribbestad IS. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010;23:424–431. doi: 10.1002/nbm.1478. [DOI] [PubMed] [Google Scholar]
- 44.Desbiens NA. A novel use for the word “trend” in the clinical trial literature. The American journal of the medical sciences. 2003;326:61–65. doi: 10.1097/00000441-200308000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Sakagami H, Aoki J, Natori Y, Nishikawa K, Kakehi Y, Arai H. Biochemical and molecular characterization of a novel choline-specific glycerophosphodiester phosphodiesterase belonging to the nucleotide pyrophosphatase/phosphodiesterase family. J Biol Chem. 2005;280:23084–23093. doi: 10.1074/jbc.M413438200. [DOI] [PubMed] [Google Scholar]
- 46.Gozgit JM, Pentecost BT, Marconi SA, Ricketts-Loriaux RS, Otis CN, Arcaro KF. PLD1 is overexpressed in an ER-negative MCF-7 cell line variant and a subset of phospho-Akt-negative breast carcinomas. Br J Cancer. 2007;97:809–817. doi: 10.1038/sj.bjc.6603926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 48.Hackshaw A, Roughton M, Forsyth S, Monson K, Reczko K, Sainsbury R, Baum M. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol. 2011;29:1657–1663. doi: 10.1200/JCO.2010.32.2933. [DOI] [PubMed] [Google Scholar]
- 49.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002;21:937–946. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- 51.Pai JK, Frank EA, Blood C, Chu M. Novel ketoepoxides block phospholipase D activation and tumor cell invasion. Anticancer Drug Des. 1994;9:363–372. [PubMed] [Google Scholar]