Abstract
The development of tools and processes used to fabricate, measure, and image nanoscale objects has lead to a wide range of work devoted to producing sensors that interact with extremely small numbers (or an extremely small concentration) of analyte molecules. These advances are particularly exciting in the context of biosensing, where the demands for low concentration detection and high specificity are great. Nanoscale biosensors, or nanobiosensors, provide researchers with an unprecedented level of sensitivity, often to the single molecule level. The use of biomolecule-functionalized surfaces can dramatically boost the specificity of the detection system, but can also yield reproducibility problems and increased complexity. Several nanobiosensor architectures based on mechanical devices, optical resonators, functionalized nanoparticles, nanowires, nanotubes, and nanofibers have been demonstrated in the lab. As nanobiosensor technology becomes more refined and reliable, it is likely it will eventually make its way from the lab to the clinic, where future lab-on-a-chip devices incorporating an array of nanobiosensors could be used for rapid screening of a wide variety of analytes at low cost using small samples of patient material.
Introduction
Advances in diagnostic technology have been essential to the progress of medicine. The ability to identify diseases and pathogens by detecting associated proteins, nucleic acid sequences, or other markers can provide biomedical researchers, disease specialists, and healthcare professionals with highly detailed knowledge of patients’ conditions, disease pathways, or the presence of contamination. However, many of the tests currently commercially available (and thus used ubiquitously) are slow, require large amounts of sample materials, and can lead to false positive or negative results. The results of these problems range from mere annoyance on behalf of the patient to misdiagnosis, inability to detect potentially lethal pathogens, and difficulty in understanding the development and propagation of diseases. Thus there is a strong push towards developing improved diagnostic technologies that would allow for rapid, trustworthy, low-cost, multiplexed screening to detect a wide range of biomaterials. The current state-of-the art diagnostic biosensors are based on several technologies, often including either the enzyme-linked immunosorbent assay (ELISA) or (for nucleic acids) amplification of a sample by polymerase chain reaction (PCR) using appropriate primers and detection methods (gel electrophoresis, radioactive or fluorescent labels, etc.). In a typical configuration, ELISA indicates the binding of analyte molecules to an immobilized probe (i.e. a “sandwich assay” configuration) by exploiting the activity of an enzyme conjugated to a detection antibody that binds to the captured analyte. With the sandwich complete, any bound enzyme is indicative of the presence of analyte, and thus enzymatic activity upon introduction of an appropriate substrate can be used to quantify the amount of analyte.1–3 On the other hand, PCR “amplifies” the signal due to the presence of analyte by selectively replicating the desired nucleotide sequences.4,5 Both of these techniques require significant sample preparation and intensive sample handling, thereby increasing the potential for error in the diagnosis, as well as the cost and time required. Moreover, in many cases, the sensitivity of the assays may not be sufficient to detect the desired levels of analyte.
To overcome the issues associated with current diagnostic techniques, a wide range of new biosensors are being developed. Several of these biosensors rely on nanotechnological platforms. While what in particular constitutes “nanotechnology” is hardly well-defined, there is some general consensus that the term refers to devices or processes that occur and can be controlled at scales below the wavelength of visible light (sometimes the threshold is instead given as 100nm). At this scale, physical processes are often manifested differently than at the macroscale due to quantum effects (e.g. quantum dots, tunnel junctions, etc.). Moreover, surface-to-volume ratios become extremely large, a fact that is beneficial for sensors that often interact with analyte only on a surface, but exploit a change in the sensor volume. It is at this scale that the analyte molecules of interest exist and interact, and thus it is appealing to attempt to probe them with technologies that are controllable at this scale. The chief motivation for driving these technologies to nanoscale architectures is increased overall sensitivity, though the mechanisms for this improvement are somewhat specific to the particular nanobiosensor type. The field of nanobiosensors is vast, and this review aims to cover the more common configurations of nanobiosensors in brief. At the end of each section covering a particular nanobiosensor architecture, the reader is pointed to more focused reviews that cover each type of nanobiosensor in more detail.
Sensitivity and Specificity
Sensitivity
One of the many potential advantages of nanobiosensors over more conventional sensing systems is dramatically improved sensitivity. To better compare these sensors, it is important to discuss what is meant by “sensitivity” in this context. In fact, there are several important parameters that relate to how well a sensor can detect an analyte.6,7 The internal sensitivity of a sensor is defined as the ratio of the sensor output signal to a change in a property of the sensor (presumably due to an amount of analyte material bound to the sensor). This parameter can be thought of as a slope defining the ability of the sensor to transduce an input signal (bulk sensor property change) to an output signal. A sensor with a large internal sensitivity value is able to pick out a minute change in a bulk sensor property (due to bound analyte) more easily than one with a small internal sensitivity value.
There is some variation in the literature as to what kind of value the denominator of the sensitivity parameter should be. To take into account the fact that a sensor (often) interacts with an analyte on the sensor surface, the sensitivity may be defined as output signal per (mass/unit sensor area). This method of normalizing away the sensor surface area can often better define the inherent ability of a sensor. While a sensor is often inherently sensitive to an amount of bound analyte, the quantity of interest is typically not a total bound mass (or number of molecules), but a concentration of analyte. This leads one to define a quantity that is expressed as output signal per analyte concentration. The subtlety of this parameter is that it takes into account not only the inherent transducer sensitivity, but also the total exposed area of the sensor, the kinetics of the binding interaction between the sensor and analyte, and other such effects. These factors are particularly important for nanoscale biosensors, as the total exposed sensor surface area may be particularly small, the kinetics of the interaction between the sensor and the analyte may be slow, the affinity of the analyte to the sensor may be poor, etc. Thus, while a sensitivity defined with respect to analyte concentration is in many cases a more useful parameter than one defined with respect to total analyte mass, the effects included in this definition are more complex.
Regardless of the definition of sensitivity used, it can be related to a minimum detectable quantity, called the limit of determination (LOD) or limit of detection or minimum detectable concentration (or mass). This refers to the smallest quantity that can be resolved above a background signal. To define this quantity, the resolution of the sensor must be characterized. If all the various noise processes in the sensor system contribute to a standard deviation in a measurement σ, then the resolution of the sensor is restricted by this standard deviation (often the acceptable spacing between measurements is taken to be 3σ, and thus this is the minimum measurable value above background noise). This leads to a LOD defined by LOD= 3σ/sensitivity. This can be considered the minimum detectable mass or concentration of analyte.6
The term “sensitivity” can also refer to test (or assay or diagnostic) sensitivity, which describes how well a diagnostic test is able to correctly identify a certain population of samples as containing (or not containing) an analyte of interest.8 This definition encompasses several issues, including reproducibility and accuracy of the diagnostic.
Dynamic Range
One parameter rarely discussed in the context of nanobiosensors is the sensor dynamic range. This quantity describes the range over which the sensor is able to accurately produce an output signal indicative of the analyte quantity. The dynamic range is limited on the lower end by the LOD, and on the upper end by saturation of the sensor, breakage of the sensor, or unpredictable changes in the sensor sensitivity. The response curve of the sensor describes the entire range of analyte quantity and corresponding sensor output, and should either be linear or have a corresponding calibration dataset or known scaling law to allow the user to correctly interpret the sensor output. The upper limit of this range may be extended by performing a kinetic (i.e. non-equilibrium) measurement instead of a steady-state measurement; however, this is uncommon because the increased complexity (increased requirement for controlling flow rates, solution properties, temperature, measurement time, etc.) can reduce measurement reproducibility. Depending on the sensor application, dynamic range may or may not be of interest.
Specificity
Besides being able to produce an output signal indicative of the presence of analyte, a sensor must also be able to distinguish between analyte and any “other” material. This quality, called specificity, is what renders a sensor useful in a non-controlled environment containing quantities of unknown material. Unlike sensitivity, specificity can be difficult to measure and confirm, as the number of possible materials that should not produce an output signal is effectively infinite. While several researchers demonstrate specificity by comparing sensor response to the analyte of interest to sensor response to a similar material, the true test of a sensor’s specificity is in the field. The specificity of a sensor becomes particularly important when trying to detect an analyte at low concentration in an environment containing a high concentration of other materials, many of which may bind non-specifically to the sensor and thus produce an anomalous signal.
Biosensors often exploit the complex, specific binding interactions provided by nature, such as antibody-antigen, nucleic acid hybridization, biotin-streptavidin, and enzymatic activity. Several groups have developed artificially synthesized (or discovered) ligands, such as aptamers (artificial nucleic acid ligands that can be selected to specifically bind to a wide range of molecules through an iterative process called systematic evolution of ligands by exponential enrichment (SELEX))9–21 and molecularly imprinted polymers (copolymers made from functional and crosslinking monomers that are polymerized around a template, or “print”, molecule, which is subsequently removed by extraction, leaving a polymer with binding sites in the specific shape and size of the template)22–28, that may have better performance than natural ligands, or may be applicable to a wide range of target molecules. Another method that can be used to obtain some level of specificity is to use an array of non-specific sensors that each react differently to exposed materials; the resulting signals from the sensor array can be used to identify analytes with a known, “signature” response (from a previous calibration experiment). This technique is often used in “electronic nose” or “electronic tongue” sensors, but is currently less commonly used in biosensors.29–32
The techniques used to attach specific capturing molecules to sensor surfaces depend on the sensor surface chemistry. Perhaps the most simplistic method is physical adsorption, where the capture molecules are exposed the sensor surface and allowed to physically adsorb onto the surface (i.e. bind to the surface non-covalently). For gold substrates, it is common to see self-assembled monolayers of molecules with a thiol anchoring group connected (often via an organic chain) to a functional group used for molecular recognition. A description of a multitude of surface functionalization techniques for gold substrates can be found in a review by Shankaran and Miura.33 When silicon surfaces (which, after exposure to atmospheric conditions, contain hydroxyl end groups -Si-OH) are silanized, covalent -Si-O-Si- bonds are formed between the silicon surface and organofunctional alkoxysilanes. The organofunctional alkoxysilanes are used to bind probe molecules using various functional groups, including amines (such as in APTES34), epoxides and thiols. Hydrosilylation of unoxidized silicon (often freshly etched material unexposed to atmosphere, having exposed -Si-H groups on the surface) inserts a carbon-carbon unsaturated bond in the silicon-hydride bond and can create alkyl or alkenyl monolayers which can be further modified to bind probes.35 Magnetic nanoparticles (consisting of iron-based metal oxides) are usually coated with natural or synthetic polymers such as dextran36 and polyethylene glycol (PEG)37 containing desired functional groups. Thus these polymers can interact with functional groups on the probe molecule, such as amines, aldehydes and carboxylic acids, to form covalent bonds and immobilize the probe. A description of many functionalization techniques and polymer coatings for magnetic nanoparticles can be found in a review by McBain et al.38 There are a wide range of processes for functionalizing carbon nanotubes (CNTs) either non-covalently or covalently.39,40 It has been found that proteins and DNA will non-specifically adsorb onto the CNT surface, and aromatic molecules will interact strongly with CNT sidewalls via π-stacking.39 Covalent linking of capture probes to CNTs can be achieved using linking molecules that interact with defects on the CNT (for example, carboxylic acid groups formed by oxidation). The above examples are only a small sample of the available chemistries used to immobilize capture molecules onto sensor surfaces.41
Nanobiosensor Systems
Mechanical Resonators and Static Deflection devices
Sensors exploiting mechanical motion (often called micro/nano-electromechanical systems, or MEMS/NEMS, sensors) to detect analyte can generally be divided into two categories: static deflection devices and resonators (Figure 1). Static devices, also known as deflection based sensors, are typically beams supported at one end by a larger substrate (cantilevers). The top of each cantilever is functionalized with a specific receptor, such as oligonucleotides used for hybridization, aptamers or antibodies. The cantilever deflects up or down because of changes in surface stress when the desired analyte binds to the receptor. The position of the cantilever can then be detected and interpreted.42–45 Detection of cantilever deflection can be accomplished using several different optical and electrical methods, including using a piezoresistive element (an element that will exhibit a change in resistance due to mechanical stress) or reflecting a laser off the cantilever surface and detecting the angle of reflection (typically using a two- or four-quadrant photodiode in a geometry identical to that used in the majority of atomic force microscopes). Static deflection devices employing functionalized cantilevers have been used to detect oligonucleotides (some studies demonstrated the ability to detect a single base-pair mismatch, some studies demonstrated LOD on the ten picomolar range)46–49 and proteins (LOD in the nanomolar range).46,50
Figure 1.
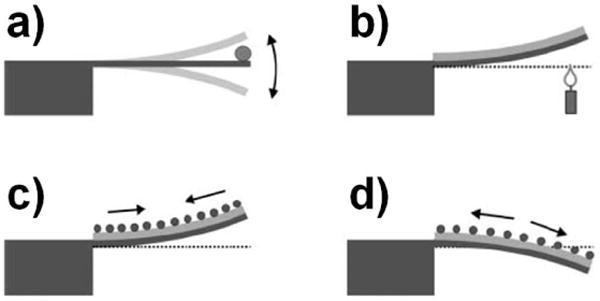
Illustration of several mechanical device configurations. a) A resonating mechanical device (cantilever) that indicates a change in resonator mass (due to bound analyte) by a shift in resonant frequency. b) A static deflection device formed from two materials with different thermal expansion coefficients that bends due to a change in temperature. c,d) Static deflection devices that bend due to analyte binding causing a surface stress.192 Fritz J. Cantilever biosensors. Analyst 2008, 133:855. http://dx.doi.org/10.1039/b718174d Reproduced by permission of The Royal Society of Chemistry
Dynamic, or resonant, devices are oscillators that have a resonant frequency dependent on the resonator mass. When analyte binds to receptors attached to the device, the frequency will shift; this frequency shift can then be interpreted as a mass change (Figure 2). There is significant interest in producing nanoscale mechanical resonator devices because, as the device size decreases, the device mass (m) decreases. Thus the addition of bound analyte molecules (Δm) causes an increased relative change in effective resonator mass (m+Δm )/m, leading to a larger frequency shift. Smaller devices also benefit from an increased surface-to-volume ratio. Moreover, the high quality factors (Q, a parameter that determines the width of the resonance peak; a higher Q corresponds to a more narrow resonance peak and thus allows the discrimination of a smaller frequency shift) of micro- and nano- mechanical devices allow for extremely sensitive mass measurements. These devices have a variety of different geometries such as cantilevers, domes, doubly clamped beams, torsional resonators, trampolines, etc. Methods for driving a dynamic device include thermal excitation, mechanical coupling,51 electrostatic,52 magnetic,53 and piezoelectric.54 Thermal excitation can cause resonance through thermal expansion, causing stress. Response can sometimes be improved using a bilayer of materials with different thermal expansion coefficients. Heat can be introduced to the system by fabricated resistors near the device (to heat via Joule heating), or by a focused, pulsed laser. The simplest actuation methods, piezoelectric and optical, can be external to the resonator and do not require additional fabrication on the resonator. A piezoelectric device (a “buzzer”) can be attached to a chip with a resonator device in order to induce resonance mechanically. To optically excite the resonator, a pulsed focused laser beam can be used to thermally excite oscillation.55 To optically detect resonance, a focused laser beam can be either reflected of the resonator surface at an angle (similar to optical detection of a static deflection device) or used as the illumination source for a Fabry-Pérot interferometer formed by the resonator (as one reflecting surface), the media in which the nanobiosensor operates, and the plane of the chip over which the resonator is suspended (the other reflecting surface).55,56 In this second configuration, the laser is able to probe minute changes in the gap between the reflecting surfaces, even if the resonator width is significantly smaller than the size of the focused laser spot. When the device is optically driven, the detecting laser should be of a different wavelength. Each excitation method and detection method has advantages and disadvantages that depend on the intended use of the device. For example, while excitation and detection methods that employ on-chip circuitry (such as wire-loops, piezoresistors, or capacitive plates) require less complex macroscale experimental apparatus to interrogate the resonator (compared to optically driven and detected systems), they are more complicated to fabricate.
Figure 2.
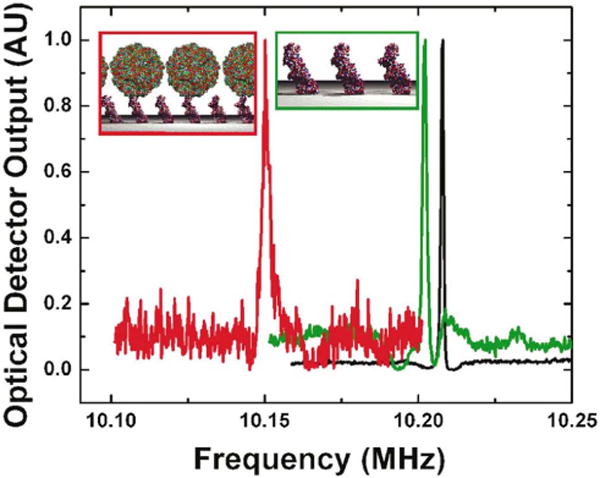
Frequency shifts of a NEMS resonator due to binding of AcV1 antibody (green) and baculovirus particles (red).58 Reprinted with permission Ilic B, Yang Y, Craighead HG. Virus detection using nanoelectromechanical devices. Applied Physics Letters 2004, 85:2604. Copyright 2004, American Institute of Physics.
The Craighead group has used mechanical resonator nanobiosensors to detect a wide variety of biomaterials, including single molecules of DNA (1587 base pairs),57 viruses,58 bacteria,59 prostate-specific antigen (LOD of 1.5 fM using secondary mass labels),60 prion proteins (LOD of 2ng/mL using secondary mass labels),61 other proteins,62 and cells (single-cell detection)63. Some of these results are from multiplexed devices incorporated in fluidic channel systems that are cycled through liquid and vacuum (for detection) cycles.62 Other groups have used mechanical resonators to detect anthrax spores (LOD of 2 spores when detection performed in air, and 50 spores when performed in water),64 viruses (single-virus detection, non-specific),65 and prostate-specific antigen (LOD of 10 pg/mL).66
Both static and dynamic devices are often fabricated in silicon or silicon nitride, though a wide range of other materials, including polymers,67,47,68 have been used. The techniques used in semiconductor manufacturing to make microelectronics, such as photolithography, are also used to fabricate the majority of mechanical biosensor devices. These techniques allow additional circuitry to be added for actuation and detection, as well as the addition of other components needed for integration (fluidics, on-board readout circuitry, etc).
Static devices have an advantage over dynamic devices in that they operate well in both gas and liquid environments, as well as vacuum. However, stress-based detection is limited to the near monolayer regime; single molecule binding typically cannot cause detectable deflection. Dynamic devices have a significantly lower limit of detection compared to static devices. Masses on the attogram and zeptogram scale have been detected using resonant techniques; however these measurements are performed at ultrahigh vacuum and low temperature.69,70 Due to viscous losses and damping, dynamic devices perform poorly in gaseous and aqueous conditions (with quality factors on the order of 10s or 100s), making it difficult to use mechanical resonators for biosensing without the additional steps of drying and evacuating the resonator after its exposure to analyte solutions. These additional steps have, until recently, dramatically inhibited the use of mechanical resonators for real-time sensing in aqueous environments.
Recent work in the area of resonant mechanical nanobiosensors has shown promise in overcoming this hurdle. Manalis and colleagues showed in 2007 that very small masses of cells, proteins and nanoparticles could be resolved using a variation of resonant cantilevers that contained microfluidic channels (Figure 3).71 The novel use of channels inside the resonator allowed the resonator to be operated in a low pressure environment (and thus achieve a high quality factor of 15,000) while still providing a mechanism for liquid solutions to be introduced to a part of the resonator surface (in this case, via an interior volume). Resonator devices containing embedded microfluidic channels have been used to measure the mass of single cells during their growth cycle,72,73 detect cancer biomarkers (LOD of 10ng/mL),74 detect IgG protein (LOD sub-nM),71 and weigh individual bacteria.71 Barton et al. have recently evaluated the performance of doubly clamped beam resonators that contain nanofluidic channels with fluids.75 They expect a minimum detectable mass with this configuration on the femtogram scale. Similar work performed by the Manalis group using resonators with embedded nanofluidic channels demonstrated detection of masses (Au nanoparticles) in fluid at the level of tens of attograms (single nanoparticle sensitivity).76 These “inside-out” mechanical resonators overcome the major limitation of mechanical resonator nanobiosensors: reduced quality factor in liquid and gas environments. As this technology becomes more refined, detection of lower mass entities in solution seems likely. In general, there is still a strong push towards reducing the sizes of mechanical resonator sensors, with carbon nanotubes77,78 and single atomic layers of carbon, called graphene,79 being the ultimate limit. A thorough review on mechanical nanobiosensors can be found in an article by Waggoner and Craighead.80
Figure 3.
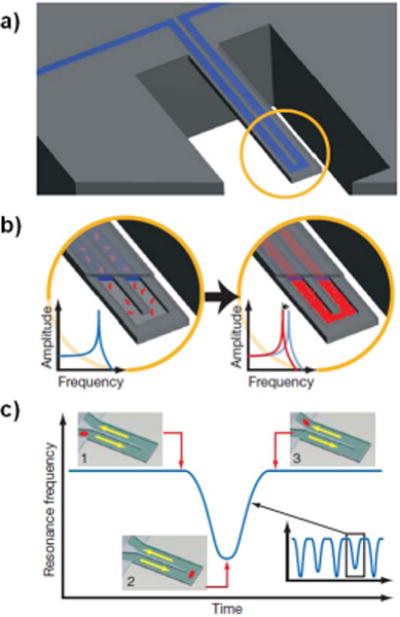
a) Illustration of a mechanical cantilever resonator containing an embedded microfluidic channel. b) Illustration showing a decrease in resonant frequency as the density inside the embedded channel. If molecules that selectively bind to the channel walls are present, they will accumulate in the channel, causing an increased density in the channel that can be detected as a frequency shift. c) If a single particle is cycled through the channel, it will cause a frequency shift that varies with the particle’s position along the cantilever. This allows the measurement of the mass of single, isolated, unbound particles in solution.71 Reprinted by permission from Macmillan Publishers Ltd: Nature. Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 2007, 446:1066–1069. Copyright 2007.
Optical Resonators
Like mechanical resonators, optical resonator sensors rely on a change in resonant frequency to indicate binding of an analyte. While mechanical resonators are based on physical motion, optical resonators are based upon light oscillating within a resonant cavity. Whispering gallery mode (WGM) resonators are a commonly used optical resonator and consist of a circular cavity (such as a microsphere or microdisk) into which light is introduced, often by evanescent coupling of a tapered optical fiber placed next to the cavity.81 The coupled light is confined within the cavity by total internal reflection, and if the wavelength of the light is such that it is able to propagate around the cavity and interfere constructively with itself (in other words, if the cavity circumference is equal to an integer number of wavelengths), then the light will resonate within the cavity. This resonance is measured as a drop in the transmission of light through the nearby optical fiber at the resonant wavelength. Because the resonating light passes multiple times around the cavity, it is able to interact with bound molecules via the evanescent field produced on the cavity surface multiple times before dissipating. The dissipation is inversely related to the quality factor Q of the resonator. The resonant frequencies of a given cavity are ultrasensitive to changes in the effective path length travelled by the resonating light caused by bound analyte at the surface of the cavity. This is illustrated in Figure 4.
Figure 4.
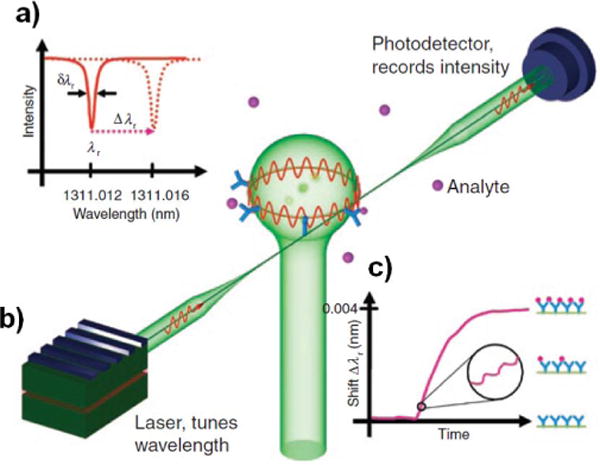
Illustration of a whispering gallery mode biosensor. Light is coupled into a microsphere evanescently via a tapered optical fiber. Certain wavelengths of light will resonate in the cavity, causing dips in transmission through the optical fiber at those wavelengths. Binding of molecules on the cavity surface will cause a shift in the resonant frequency spectrum of the cavity, and thus the resonant frequency dips monitored at the detector will shift. As more analyte binds to the surface, this resonant frequency shift will increase.81 Reprinted by permission from Macmillan Publishers Ltd: Nature Methods. Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat Meth 2008, 5:591–596. Copyright 2008.
Soon after theoretical work describing a WGM resonator biosensor configuration was published (note that this work did not treat the more commonly exploited resonance frequency shifts due to bound molecules as described above, but transmission changes due to nearby absorbing molecules),82 several groups dedicated effort to realizing the exciting potential sensitivity of the WGM resonator architecture. As a first experimental demonstration of a WGM nanobiosensor, Vollmer et al used a 300μm diameter silica glass microparticle as the cavity for a WGM sensor with a Q of 2×106 to detect both non-specific binding of BSA and specific binding of streptavidin to bound BSA-biotin conjugates.83 They concluded that their sensor saturated at BSA concentrations as low as 20nM, defining an upper limit to their dynamic range; this work did not examine the lower concentration limit of detection. Subsequent theoretical work provided some suggestions to improve WGM biosensor system sensitivity.84 Work with a similar experimental system demonstrated multiplexed measurement of DNA hybridization to probes bound on the cavity surface.85 In this system, two microspheres (each with its own “signature” resonance spectrum) were brought near to a tapered optical fiber, one after the other, and thus the resonance dips in the multiplexed transmission spectrum could be uniquely assigned to each cavity. When each microsphere was modified with a different probe oligonucleotide, a resonance dip shift assigned to one of the cavities correctly indicated DNA hybridization upon the introduction of appropriate complementary oligonucleotide into the system. Subsequent introduction of oligonucleotide complementary to the probes on the second microsphere caused a shift in the dip assigned to that cavity, but didn’t cause a detectable shift in the dips assigned to the cavity functionalized with non-complementary probes. The authors estimate the detection limit of this system to be ~6 pg/mm2 nucleic acid. This nanobiosensor system was able to identify a single nucleotide mismatch with high specificity (the cavity functionalized with a matched probe showed a wavelength shift 10× larger than the one functionalized with a mismatched probe, at a final concentration of 1 μM) and a signal-to-noise ratio of 54. The next major step in WGM nanobiosensor technology was achieved by Armani et al86 using a micro-toroid cavity with a Q greater than 108. In this work, the authors demonstrated single-molecule detection sensitivity for interleukin-2 (IL-2) binding to a surface layer of IL-2 antibody on top of Protein G. Not only was this nanobiosensor able to perform real-time single-molecule detection, but it also exhibited a dynamic range of 12 orders of magnitude (working range of 5 aM to 1 μM) with a sigmoidal response curve. The physical mechanism for the improved sensitivity of this measurement system, however, is still under debate in the current literature.87
Optical resonators have been integrated with fluidics in a geometry called a capillary ring resonator (CRR) (sometimes called a liquid core optical ring resonator (LCORR) or opto-fluidic ring resonator (OFRR)). In this sensor configuration, the walls of a glass capillary are used both as the resonator cavity and as a fluid transport system.88 A tapered optical fiber brought near the capillary will couple light into the cavity, and changes in the refractive index near the interior surface of the capillary wall are detected as shifts in the transmission dips corresponding to resonant modes. In this system, liquids flowing through the inside of the capillary may be used to deliver analyte to the interior wall, which may be functionalized to provide sensor specificity (Figure 5). These nanobiosensors have been used to detect BSA (without specificity, LOD below 10pM),89,90 DNA (demonstrated LOD of 10pM),91 and viruses (demonstrated LOD of 2.3×103 pfu/mL of M13 filamentous bacteriophage, dynamic range of 7 orders of magnitude).92
Figure 5.
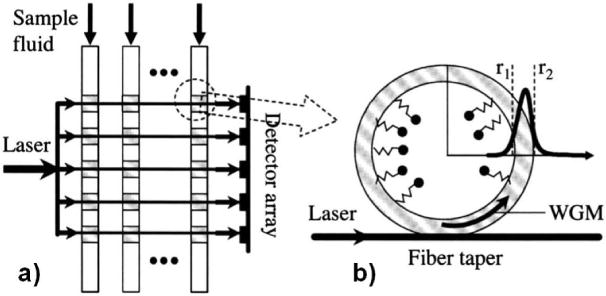
Illustration of a liquid core optical ring resonator sensing system. a) Illustration of a potential method to multiplex multiple liquid core optical ring resonators using a single laser (to excite multiple cavities) and multiple detectors. b) Illustration of a laser coupled to an optical resonator cavity formed by the wall of a glass capillary.88 Reprinted by permission from White IM, Oveys H, Fan X. Liquid-core optical ring-resonator sensors. Optics letters 2006, 31:1319–1321. Copyright 2006.
While WGM nanobiosensors promise single-molecule sensitivity and are easily functionalized to attain high specificity, their inherent complexity has so far prevented their widespread use. They require tunable lasers to inject light into the cavity and high quality optical detectors with sensitivity as the laser wavelength (both of which are costly), careful alignment of the tapered fiber and the cavity, and potential microfabrication processes to form the cavity itself. It is also important to maintain thermal stability in these systems. However, these issues may all be overcome by systems entirely integrated “on-chip”, and thus there is the potential to reduce cost and complexity of these devices by incorporating most or all of the components on a single chip using a scaled-up manufacturing technique. Capillary ring resonators may help overcome scalability problems by integrating the resonant cavity and fluidic transport device into a single system. An in-depth overview of the current state of WGM-based nanobiosensors and the wide range of potential applications may be found in a review by Vollmer and Arnold.81
There are a wide variety of other optical nanobiosensors, including zero mode waveguides,93–95 single metallic nanoparticles,96 nanobiosensors based on surface enhanced Raman scattering (SERS)97,98,9,99–104, Förster (or fluorescence) resonance energy transfer (FRET) between chromophores,105–107 and systems based on the aggregation of nanoparticles upon binding (see below). A review by Erickson et al provides a discussion of a wide range of nanobiosensor configurations, with particular focus on optical nanobiosensors.108
Aggregating Nanoparticles
Nanoparticle-based biosensors are particularly attractive because they can easily be synthesized in bulk using standard chemistry techniques and do not require advanced fabrication approaches (i.e. production is bottom-up instead of top-down). They also offer particularly high surface area due to their extremely small size and the fact that they are typically used suspended in solution (during the time at which they interact with the analyte). Due to their small size, nanoparticles may be taken up by cells,109–111 and thus are promising for in vivo sensing applications. Moreover, nanoparticles can have unique optical or magnetic properties that may exploited. Several sensing platforms have been developed that exploit a change in output signal due to aggregation of suspended nanoparticles caused by the presence of analyte. Typically, nanoparticles are functionalized with a ligand for the desired analyte, and thus upon exposure to analyte in solution the nanoparticles will form a network.
Early work in this field performed in the Mirkin group demonstrated colorimetric detection of oligonucleotides using Au nanoparticles functionalized with probe oligonucleotides.112 Gold nanoparticles suspended in solution exhibit a strong optical absorbance at particular visible optical wavelengths due to plasmon absorption ; this property is dependent on the nanoparticle size.113 Upon hybridization with the target oligonucleotide, the probe molecules attached to the nanoparticles induce the formation of a nanoparticle network, causing a change in solution color from red to blue (Figure 6). This color transition is extremely sharp, and the specificity of this sensing system is good enough to easily detect single base mismatches. The resulting signal upon binding is significant enough to be detected by the naked eye without any artificial equipment to provide amplification or additional transduction. These colorimetric nanoparticle assays work on the principle that aggregation of small metal nanoparticles causes a change in the nanoparticle surface plasmon resonance, and thus the resulting macroscale optical properties of the nanoparticle suspension.114 Similar nanoparticle assays (some of which monitor changes in solution absorbance at a given wavelength due to aggregation) have been used to detect not only nucleic acids,112,115 but metal ions (typical LOD ranging from 1–100μM),116–118 proteins,10,119 adenosine (LOD of 100μM)120 and cocaine (LOD of ~50μM).11 Assays based on nanoparticle aggregation employing amplification by “bio-barcodes”,121 or indicating oligonucleotide strands, have been used to detect both nucleic acids (with sub-attomolar sensitivity)122 and proteins (with attomolar sensitivity),123 and have been combined with colorimetric nanoparticle aggregation detection.124 More information on nanoparticle-based biosensors may be found in a review by Rosi and Mirkin.125
Figure 6.
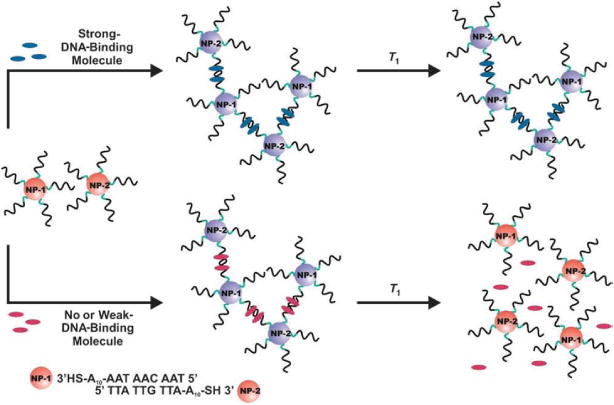
Illustration showing DNA-functionalized Au nanoparticles aggregating upon hybridization with complementary strands115 Han MS, Lytton-Jean AKR, Oh B, Heo J, Mirkin CA. Colorimetric Screening of DNA-Binding Molecules with Gold Nanoparticle Probes. Angew. Chem. Int. Ed. 2006, 45:1807–1810. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.
A similar architecture, developed by Weissleder and co-workers, is based on the aggregation of superparamagnetic nanoparticles and may be used in conjunction with MRI for biosensing purposes. The use of nanoscale particles is necessary because the superparamagentic property depends on the particles each having only a single magnetic domain. Instead of a change in an optical signal, these assays rely on an aggregation-induced change in the spin-spin relaxation time (T2) of water molecules near the nanoparticles, which can be detected using magnetic resonance techniques.126 Thus, this technology is very promising for in vivo sensing applications, as it could be used in conjunction with already established NMR/MRI systems. Similar to the architecture described above, this technology relies on nanoparticles functionalized with ligands that bind the analyte desired, and thus aggregate upon exposure to analyte. This system has been used to detect herpes simplex virus and adenovirus in biological samples (best demonstrated LOD of 5 viral particles in 10μL of liquid),127 influenza,128 nucleic acids,126,129 adenosine,12 enzyme activity,130–133 avadin,134 and other proteins (demonstated LOD often at the nanomolar range).129,135,136 This technology has also been utilized with a Boolean logic analyte binding system, whereby sensor response occurred either in the presence of matrix-metalloproteinase-2 (MMP2) AND matrix-metalloproteinase-7 (MMP7), or in the presence of MMP2 OR MMP7 (where AND and OR refer to the logic functions).137 This provides an attractive opportunity to produce custom, “smart” sensing systems that are carefully tuned to produce an output signal only upon the presence of a particular combination of analytes, and the absence of others. Magnetic nanoparticle-based sensing systems have been integrated into lab-on-a-chip devices, providing a handheld detection system that has been demonstrated for use with bacteria, mammalian cells, and various biomarkers.138 Several reviews on magnetic nanoparticle aggregation biosensors provide a more in-depth discussion of this technology.139,36
Other nanoparticle-based nanobiosensors are based on single metallic nanoparticles,96 mechanical resonators biosensors employing nanoparticle secondary mass labels,60 and nanobiosensors exploiting the SERS effect.99,101–104,140
Nanowires, Nanotubes, and Nanofibers
Several one-dimensional nanostructures have been used as nanobiosensors, typically in an electrical field-effect transistor (FET) configuration with analyte molecules acting as a gate to control the sensing structure’s electrical resistance by causing depletion or accumulation of charge carriers. One or several of the one-dimensional nanostructures are placed between metallic electrodes to allow interfacing to external electronics equipment (Figure 7). These devices offer improved sensitivities due to large surface-to-volume ratios, which enable bound analyte molecules to more significantly affect the bulk electrical properties of the structure. In some cases (e.g. carbon nanotubes), the inherent electrical properties of the device are particularly extraordinary and lend themselves to improved sensor sensitivity.
Figure 7.
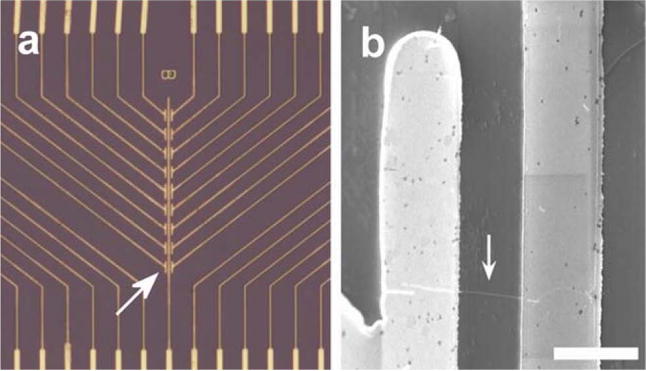
a) Optical microscopy image of a metal electrode array used to interface to nanowire biosensors (field of view is 350×400 μm). b) An individual silicon nanowire bridging two electrodes (scale bar is 2 μm).173 Reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology. Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotech 2005, 23:1294–1301. Copyright 2005.
Carbon nanotubes (CNTs) have received a great deal of attention due to their extremely small size and extraordinary mechanical, optical, and electrical properties. While easily synthesized, they are difficult to work with for several reasons. One major hurdle of working with CNTs is controlling their placement; typical synthesis techniques result in unaligned, “hairy” tangles of CNTs. Recently, techniques have been developed to control the alignment and placement of single and multiple CNTs.141–144 Other improvements have allowed the growth of nanotubes to extremely long lengths, thereby producing structures with extremely high aspect ratios.145,146 Another issue encountered when attempting to produce electrical CNT sensors is that the electrical properties of each CNT depend on its chiral vector, a parameter that describes how the graphene sheet that forms the nanotube is rolled up. CNTs can be metallic or semiconducting (with a wide range of bandgap values), depending on this parameter.147 Typical synthesis processes (laser ablation, chemical vapor deposition, arc-discharge)148 result in a heterogenous distribution of CNTs chiralities and diameters (the two parameters the define the electrical behavior of the CNT). Thus, to achieve production of electrical CNT-based devices with a high level of consistency and reproducibility, techniques have been developed to isolate CNTs with more controlled electrical properties.149,150 CNTs can also be categorized as either single-walled nanotubes (SWNTs) or multi-walled nanotubes (MWNTs), depending on how many concentric cylinders of wrapped graphene form the tube. Several reviews on novel techniques to control alignment, placement, and electrical properties offer more details on CNT synthesis and fabrication advancements.148,147,151
Due to their extreme electrical sensitivity to nearby molecules, CNTs have been incorporated into electrical nanobiosensors by several researchers. A common configuration utilizes semiconducting CNTs as FETs. Dai and coworkers demonstrated CNT-based nanobiosensors based on biofunctionalized SWNT networks used to specifically sense several biomolecules (including several monoclonal antibodies, streptavidin (Figure 8), and other proteins) with nanomolar sensitivity.152,153 They have also discussed in depth the issue of non-specific binding of peptides to CNTs, and techniques to reduce this effect by coating the CNTs with polyethylene oxide (PEO).152,154 Other groups have demonstated CNT-based nanobiosensors able to detect glucose (via functionalization with glucose oxidase),155,156 streptavidin,157,158 thrombin,21 other proteins,157,159 and nucleic acids (with sensitivity in the picomolar range).160,161 CNT networks have also been used as electrodes in electrochemical biosensors, providing improved sensing ability due to both their high surface area and their excellent electrical characteristics.162,163 More discussion of CNT-based biosensors can be found in several review articles written on the subject.125,164
Figure 8.
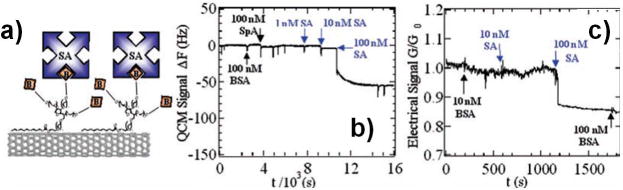
a) Diagram illustrating binding of streptavidin to a biotin-functionalized CNT sensor. b) Frequency shift of a quartz crystal microbalance mass sensor as streptavidin solution is introduced to the functionalized nanobiosensor. Note that the response is specific to the streptavidin solution; negligible response is seen with solutions of other biomolecules. c) Electrical response of the CNT-based nanobiosensor as streptavidin solution is introduced at increasing concentrations. Again, the response is specific to streptavidin.152 Reproduced with permission from Chen RJ, Bangsaruntip S, Drouvalakis KA, Wong Shi Kam N, Shim M, Li Y, Kim W, Utz PJ, Dai H. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proceedings of the National Academy of Sciences of the United States of America 2003, 100:4984. Copyright 2003 National Academy of Sciences, U.S.A.
Nanowires produced from other semiconductors have also been used as nanobiosensors. While their sizes and electrical behavior are typically not as extreme as CNTs, they can typically be produced with more reproducible properties and thus may be more amenable to industrial manufacturing. While they can be patterned lithographically,165–168 nanowires are more often synthesized using a bottom-up chemical growth approach.17,159,169–171 The Lieber group has exploited FETs made using bottom-up synthesis of doped silicon nanowires to specifically sense a wide range of biomaterials, including streptavidin (LOD of roughly 10pM),169 antibodies,169 nucleic acids (LOD of approximately 10fM),170 viruses (at the single virus level),172 and other proteins.173 Some of these studies have incorporated arrays of differently functionalized sensing elements, allowing multiplexed sensing of several analytes on the same chip. Semiconducting polymeric nanowires,174,175 sometimes called nanofibers, can be used in the same configurations as other semiconducting one-dimensional nanostructures.176–178 They are attractive because polymeric materials are in general less expensive than other semiconductors, and offer the possibility of more complex functionalization strategies by choosing appropriate monomers (including biomolecules) to integrate during synthesis. However, they in general exhibit poorer electrical behavior than other semiconducting materials. Further information on nanowire and nanofiber biosensors can be found in several reviews.179,180
Integration and Scalability
The ability to integrate nanobiosensors with the macroscale world is extremely important; without this capability the sensor is (in the majority of cases) effectively useless. Nanobiosensor technology is often paired with micro/nanofluidic devices to deliver analyte to the sensor surface. This allows the usage of small volumes of sample solution. Theoretical treatments of nanobiosensor configurations suggest that sensitivity may in many cases be limited not by the inherent ability of the sensor, but by transport of analyte to the sensor surface within an acceptable timeframe.181–183 Novel fluidic designs based on wicking of solutions in patterned paper,184–188 or on evaporation-driven flow189–191 may help reduce the cost and complexity associated with many fluid delivery systems and have shown tremendous promise in applications in third-world countries. Several nanobiosensor architectures require a high level of integration with external equipment (detectors, pumps, electrical components, lasers, and optics), thereby increasing the cost and complexity (and reducing the portability) of the resulting device.
Nanobiosensors produced using top-down fabrication approaches are in many cases not easily (and inexpensively) scaled up to commercial manufacturing levels (with necessary reproducibility and yield). While one-dimensional nanostructure biosensors are attractive due to their extreme sensitivity, there are several issues regarding scalability and reproducible fabrication (i.e. producing sensing components with reproducible electrical behavior, wiring to external electronics, etc.) that may hinder commercial usage of this architecture. Improved fabrication techniques that allow for multiplexing173 and reproducible device production may aid in the transfer of this technology to commercial diagnostic tools. Diagnostic tools containing several multiplexed highly sensitive nanobiosensors would be invaluable for identifying diseases, and thus the ability to include a large number of sensors specific to different analytes on a single platform is highly attractive. However, only a small fraction of the literature on nanobiosensor technology has demonstrated any multiplexing ability. Nanobiosensors produced using bottom-up synthesis techniques have issues with scalability due to potential difficulty in controllably synthesizing the nanoparticle/nanowire/nanotube with well defined properties (shape, size, or electrical properties). The stability of suspensions of functionalized nanoparticles must also be examined to determine an appropriate “shelf-life”, and they must not be exposed to materials that could cause them to aggregate prematurely.
Conclusion
A wide range of nanobiosensors have been developed in the past two decades, and yet the futuristic goal of low-cost, high throughput, multiplexed clinical diagnostic lab-on-a-chip devices has yet to be truly realized. It is still unclear which nanobiosensor architectures are best matched to which diagnostic tasks. Moreover, nanobiosensors that are functional in the lab may not be of use in the field or clinic for several reasons. Sensitivity and dynamic range should be matched to the analyte and what truly needs to be sensed, and thus it is unclear whether single-molecule sensitivity nanobiosensors will be of clinical use unless they exhibit a dynamic range that extends through a useful concentration range. Preconcentration or pre-dilution of sample could be used to match the analyte concentration to the appropriate concentration range of the nanobiosensor, but this additional step introduces complications and renders the assay inefficient and potentially less accurate. Another consideration when transitioning sensors from the lab to the clinic is their specificity and ability to be fouled by non-specific binding of materials in the sample being interrogated. Finally, the robustness of the sensor architecture, as well as the ease of use, become important parameters when novel diagnostic technology transitions from the development phase to being usable.
Nanobiosensors have generated a great deal of excitement due to their ability to detect a wide range of materials at incredibly small concentrations. As the field (and the multitude of different technologies that it encompasses) matures, it seems highly likely that the diagnostic techniques of today will soon become antiquated, and a new class of low cost, robust, reliable, easy-to-use, and ultra-sensitive diagnostics will become available. This may even spur a dramatic increase in the number of point-of-care diagnostics, as well as diagnostic tools that can be used by patients on their own. Whether a single nanobiosensor architecture will become dominant, or several will transition to commercial devices has yet to be seen. However, it is almost certain that these sensors will allow the detection of pathogens and diseases like never before.
Acknowledgments
The authors acknowledge support from National Institutes of Health Grant EB006365 (to RSL) and F32EB011866 (to LMB) from NIBIB (National Institute of Biomedical Imaging and Bioengineering).
Contributor Information
Leon M Bellan, Email: Lbellan@mit.edu, David H Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA.
Diana Wu, Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA.
Robert S Langer, David H Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA.
References
- 1.Perlmann P, Perlmann H. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 2.Hempen C, Karst U. Labeling strategies for bioassays. Anal Bioanal Chem. 2005;384:572–583. doi: 10.1007/s00216-005-3392-0. [DOI] [PubMed] [Google Scholar]
- 3.Eteshola E, Leckband D. Development and characterization of an ELISA assay in PDMS microfluidic channels. Sensors and Actuators B: Chemical. 2001;72:129–133. [Google Scholar]
- 4.Weiss JB. DNA probes and PCR for diagnosis of parasitic infections. Clinical Microbiology Reviews. 1995;8:113. doi: 10.1128/cmr.8.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto Y. PCR in diagnosis of infection: detection of bacteria in cerebrospinal fluids. Clinical and Vaccine Immunology. 2002;9:508. doi: 10.1128/CDLI.9.3.508-514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper M. Label-free biosensors : techniques and applications. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 7.D’Amico A, Di Natale C. A contribution on some basic definitions of sensors properties. IEEE Sensors J. 2001;1:183–190. [Google Scholar]
- 8.Saah AJ, Hoover DR. “Sensitivity” and “Specificity” Reconsidered: The Meaning of These Terms in Analytical and Diagnostic Settings. Annals of internal medicine. 1997;126:91. doi: 10.7326/0003-4819-126-1-199701010-00026. [DOI] [PubMed] [Google Scholar]
- 9.Huh YS, Erickson D. Aptamer based surface enhanced Raman scattering detection of vasopressin using multilayer nanotube arrays. Biosensors and Bioelectronics. 2010;25:1240–1243. doi: 10.1016/j.bios.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Lu Y. Fast Colorimetric Sensing of Adenosine and Cocaine Based on a General Sensor Design Involving Aptamers and Nanoparticles. Angew Chem Int Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 12.Yigit MV, Mazumdar D, Kim H, Lee JH, Odintsov B, Lu Y. Smart “Turn-on” Magnetic Resonance Contrast Agents Based on Aptamer-Functionalized Superparamagnetic Iron Oxide Nanoparticles. ChemBioChem. 2007;8:1675–1678. doi: 10.1002/cbic.200700323. [DOI] [PubMed] [Google Scholar]
- 13.Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosensors and Bioelectronics. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinical Chemistry. 1999;45:1628. [PubMed] [Google Scholar]
- 15.O’Sullivan C. Aptasensors – the future of biosensing? Analytical and Bioanalytical Chemistry. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 16.Willner I, Zayats M. Electronic Aptamer-Based Sensors. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS, Kim KS, Kim CJ, Hahn SK, Jo MH. Electrical detection of VEGFs for cancer diagnoses using anti-vascular endotherial growth factor aptamer-modified Si nanowire FETs. Biosensors and Bioelectronics. 2009;24:1801–1805. doi: 10.1016/j.bios.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 18.Ikebukuro K, Kiyohara C, Sode K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosensors and Bioelectronics. 2005;20:2168–2172. doi: 10.1016/j.bios.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Yang X, Wang E. Review: Aptamers in microfluidic chips. Analytica Chimica Acta. 2010;683:12–20. doi: 10.1016/j.aca.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 21.So HM, Won K, Kim YH, Kim BK, Ryu BH, Na PS, Kim H, Lee JO. Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J Am Chem Soc. 2005;127:11906–11907. doi: 10.1021/ja053094r. [DOI] [PubMed] [Google Scholar]
- 22.Kriz D, Ramstroem O, Svensson A, Mosbach K. A biomimetic sensor based on a molecularly imprinted polymer as a recognition element combined with fiber-optic detection. Analytical Chemistry. 1995;67:2142–2144. [Google Scholar]
- 23.Kriz D, Kempe M, Mosbach K. Introduction of molecularly imprinted polymers as recognition elements in conductometric chemical sensors. Sensors and Actuators B: Chemical. 1996;33:178–181. [Google Scholar]
- 24.Andersson LI, M\üller R, Vlatakis G, Mosbach K. Mimics of the binding sites of opioid receptors obtained by molecular imprinting of enkephalin and morphine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4788. doi: 10.1073/pnas.92.11.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haupt K, Mosbach K. Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors. Chem Rev. 2000;100:2495–2504. doi: 10.1021/cr990099w. [DOI] [PubMed] [Google Scholar]
- 26.Ansell RJ, Kriz D, Mosbach K. Molecularly imprinted polymers for bioanalysis: chromatography, binding assays and biomimetic sensors. Current Opinion in Biotechnology. 1996;7:89–94. doi: 10.1016/s0958-1669(96)80101-7. [DOI] [PubMed] [Google Scholar]
- 27.Sellergren B. Molecularly imprinted polymers: man-made mimics of antibodies and their applications in analytical chemistry. Elsevier; 2001. [Google Scholar]
- 28.Hedborg E, Winquist F, Lundström I, Andersson LI, Mosbach K. Some studies of molecularly-imprinted polymer membranes in combination with field-effect devices. Sensors and Actuators A: Physical. :37–38. 796–799. [Google Scholar]
- 29.Freund MS, Lewis NS. A chemically diverse conducting polymer-based “electronic nose”. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2652–2656. doi: 10.1073/pnas.92.7.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. Cross-Reactive Chemical Sensor Arrays. Chem Rev. 2000;100:2595–2626. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 31.Vlasov Y, Legin A, Rudnitskaya A, Di Natale C, D’Amico A. Nonspecific sensor arrays (“electronic tongue”) for chemical analysis of liquids (IUPAC Technical Report) Pure Appl Chem. 2005;77:1965–1983. [Google Scholar]
- 32.Legin A, Smirnova A, Rudnitskaya A, Lvova L, Suglobova E, Vlasov Y. Chemical sensor array for multicomponent analysis of biological liquids. Analytica Chimica Acta. 1999;385:131–135. [Google Scholar]
- 33.Shankaran DR, Miura N. Trends in interfacial design for surface plasmon resonance based immunoassays. Journal of Physics D: Applied Physics. 2007;40:7187. [Google Scholar]
- 34.Howarter JA, Youngblood JP. Optimization of Silica Silanization by 3-Aminopropyltriethoxysilane. Langmuir. 2006;22:11142–11147. doi: 10.1021/la061240g. [DOI] [PubMed] [Google Scholar]
- 35.Buriak JM. Organometallic chemistry on silicon surfaces: formation of functional monolayers bound through Si–C bonds. Chemical Communications. 1999;1999:1051–1060. [Google Scholar]
- 36.Haun JB, Yoon T, Lee H, Weissleder R. Magnetic nanoparticle biosensors. WIREs Nanmed Nanobiotech. 2010 doi: 10.1002/wnan.84. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 37.Nitin N, LaConte L, Zurkiya O, Hu X, Bao G. Functionalization and peptide-based delivery of magnetic nanoparticles as an intracellular MRI contrast agent. J Biol Inorg Chem. 2004;9 doi: 10.1007/s00775-004-0560-1. [DOI] [PubMed] [Google Scholar]
- 38.McBain SC, Yiu HH, Dobson J. Magnetic nanoparticles for gene and drug delivery. Int J Nanomedicine. 2008;3:169–180. doi: 10.2147/ijn.s1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirsch A, Vostrowsky O. Functionalization of carbon nanotubes. Functional molecular nanostructures. 2005:193–237. [Google Scholar]
- 40.Veetil JV, Ye K. Development of Immunosensors Using Carbon Nanotubes. Biotechnol Progress. 2008;23:517–531. doi: 10.1021/bp0602395. [DOI] [PubMed] [Google Scholar]
- 41.Hermanson GT. Bioconjugate techniques. Academic Press; 2008. [Google Scholar]
- 42.Datar R, Kim S, Jeon S, Hesketh P, Manalis S, Boisen A, Thundat T. Cantilever Sensors. MRS BULLETIN. 2009;34 [Google Scholar]
- 43.Boisen A, Thundat T. Design & fabrication of cantilever array biosensors. Materials Today. 2009;12:32–38. [Google Scholar]
- 44.Ziegler C. Cantilever-based biosensors. Anal Bioanal Chem. 2004;379 doi: 10.1007/s00216-004-2694-y. [DOI] [PubMed] [Google Scholar]
- 45.Lavrik NV, Sepaniak MJ, Datskos PG. Cantilever transducers as a platform for chemical and biological sensors. Rev Sci Instrum. 2004;75:2229. [Google Scholar]
- 46.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, Meyer E, Güntherodt H, Gerber C, Gimzewski J. Translating Biomolecular Recognition into Nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 47.Lechugaa LM, Boisenb A. Highly sensitive polymer-based cantilever-sensors for DNA detection. Ultramicroscopy. 2005;105:215–222. doi: 10.1016/j.ultramic.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 48.Lechuga LM, Tamayo J, Alvarez M, Carrascosa LG, Yufera A, Doldán R, Peralias E, Rueda A, Plaza JA, Zinoviev K, et al. A highly sensitive microsystem based on nanomechanical biosensors for genomics applications. Sensors and Actuators B: Chemical. 2006;118:2–10. [Google Scholar]
- 49.Zhang J, Lang HP, Huber F, Bietsch A, Grange W, Certa U, Mckendry R, Güntherodt H, Hegner M, Gerber C. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nature Nanotech. 2006;1:214–220. doi: 10.1038/nnano.2006.134. [DOI] [PubMed] [Google Scholar]
- 50.Backmann N, Zahnd C, Huber F, Bietsch A, Pl\ückthun A, Lang HP, G\üntherodt HJ, Hegner M, Gerber C. A label-free immunosensor array using single-chain antibody fragments. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14587. doi: 10.1073/pnas.0504917102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verbridge SS, Parpia JM, Reichenbach RB, Bellan LM, Craighead HG. High quality factor resonance at room temperature with nanostrings under high tensile stress. J Appl Phys. 2006;99:124304. [Google Scholar]
- 52.Evoy S, Carr DW, Sekaric L, Olkhovets A, Parpia JM, Craighead HG. Nanofabrication and electrostatic operation of single-crystal silicon paddle oscillators. J Appl Phys. 1999;86:6072. [Google Scholar]
- 53.Cleland AN, Roukes ML. Fabrication of high frequency nanometer scale mechanical resonators from bulk Si crystals. Applied Physics Letters. 1996;69:2653. [Google Scholar]
- 54.Masmanidis SC, Karabalin RB, De Vlaminck I, Borghs G, Freeman MR, Roukes ML. Multifunctional Nanomechanical Systems via Tunably Coupled Piezoelectric Actuation. Science. 2007;317:780–783. doi: 10.1126/science.1144793. [DOI] [PubMed] [Google Scholar]
- 55.Ilic B, Krylov S, Aubin K, Reichenbach R, Craighead HG. Optical excitation of nanoelectromechanical oscillators. Appl Phys Lett. 2005;86:193114. [Google Scholar]
- 56.Carr D, Craighead H. Fabrication of nanoelectromechanical systems in single crystal silicon using silicon on insulator substrates and electron beam lithography. JOURNAL OF VACUUM SCIENCE & TECHNOLOGY B. 1997;15:2760–2763. [Google Scholar]
- 57.Ilic B, Yang Y, Aubin K, Reichenbach R, Krylov S, Craighead HG. Enumeration of DNA Molecules Bound to a Nanomechanical Oscillator. Nano Lett. 2005;5:925–929. doi: 10.1021/nl050456k. [DOI] [PubMed] [Google Scholar]
- 58.Ilic B, Yang Y, Craighead HG. Virus detection using nanoelectromechanical devices. Applied Physics Letters. 2004;85:2604. [Google Scholar]
- 59.Ilic B, Czaplewski D, Craighead HG, Neuzil P, Campagnolo C, Batt C. Mechanical resonant immunospecific biological detector. Appl Phys Lett. 2000;77:450. [Google Scholar]
- 60.Waggoner PS, Varshney M, Craighead HG. Detection of prostate specific antigen with nanomechanical resonators. Lab Chip. 2009;9:3095. doi: 10.1039/b907309b. [DOI] [PubMed] [Google Scholar]
- 61.Varshney M, Waggoner PS, Tan CP, Aubin K, Montagna RA, Craighead HG. Prion Protein Detection Using Nanomechanical Resonator Arrays and Secondary Mass Labeling. Anal Chem. 2008;80:2141–2148. doi: 10.1021/ac702153p. [DOI] [PubMed] [Google Scholar]
- 62.Waggoner PS, Tan CP, Craighead HG. Microfluidic integration of nanomechanical resonators for protein analysis in serum. Sensors and Actuators B: Chemical. 2010 [Google Scholar]
- 63.Ilic B, Czaplewski D, Zalalutdinov M, Craighead HG, Neuzil P, Campagnolo C, Batt C. Single cell detection with micromechanical oscillators. J Vac Sci Technol B. 2001;19:2825. [Google Scholar]
- 64.Davila AP, Jang J, Gupta AK, Walter T, Aronson A, Bashir R. Microresonator mass sensors for detection of Bacillus anthracis Sterne spores in air and water. Biosensors and Bioelectronics. 2007;22:3028–3035. doi: 10.1016/j.bios.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 65.Gupta A, Akin D, Bashir R. Single virus particle mass detection using microresonators with nanoscale thickness. Appl Phys Lett. 2004;84:1976. [Google Scholar]
- 66.Lee JH, Hwang KS, Park J, Yoon KH, Yoon DS, Kim TS. Immunoassay of prostate-specific antigen (PSA) using resonant frequency shift of piezoelectric nanomechanical microcantilever. Biosensors and Bioelectronics. 2005;20:2157–2162. doi: 10.1016/j.bios.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 67.Zhang XR, Xu X. Development of a biosensor based on laser-fabricated polymer microcantilevers. Appl Phys Lett. 2004;85:2423. [Google Scholar]
- 68.Calleja M, Tamayo J, Nordstroöm M, Boisen A. Low-noise polymeric nanomechanical biosensors. Appl Phys Lett. 2006;88:113901. [Google Scholar]
- 69.Ekinci KL, Huang XMH, Roukes ML. Ultrasensitive nanoelectromechanical mass detection. Appl Phys Lett. 2004;84:4469. [Google Scholar]
- 70.Yang YT, Callegari C, Feng XL, Ekinci KL, Roukes ML. Zeptogram-Scale Nanomechanical Mass Sensing. Nano Lett. 2006;6:583–586. doi: 10.1021/nl052134m. [DOI] [PubMed] [Google Scholar]
- 71.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 72.Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, Jorgensen P, Payer K, Grossman AD, Kirschner MW, et al. Using buoyant mass to measure the growth of single cells. Nat Meth. 2010;7:387–390. doi: 10.1038/nmeth.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryan AK, Goranov A, Amon A, Manalis SR. Measurement of mass, density, and volume during the cell cycle of yeast. Proceedings of the National Academy of Sciences. 2010;107:999. doi: 10.1073/pnas.0901851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Muhlen MG, Brault ND, Knudsen SM, Jiang S, Manalis SR. Label-Free Biomarker Sensing in Undiluted Serum with Suspended Microchannel Resonators. Anal Chem. 2010;82:1905–1910. doi: 10.1021/ac9027356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barton RA, Ilic B, Verbridge SS, Cipriany BR, Parpia JM, Craighead HG. Fabrication of a Nanomechanical Mass Sensor Containing a Nanofluidic Channel. Nano Letters. 2010;10:2058–2063. doi: 10.1021/nl100193g. [DOI] [PubMed] [Google Scholar]
- 76.Lee J, Shen W, Payer K, Burg TP, Manalis SR. Toward Attogram Mass Measurements in Solution with Suspended Nanochannel Resonators. Nano letters. 2010:1976–1978. doi: 10.1021/nl101107u. [DOI] [PubMed] [Google Scholar]
- 77.Sazonova V, Yaish Y, \Üst\ünel H, Roundy D, Arias TA, McEuen PL. A tunable carbon nanotube electromechanical oscillator. Nature. 2004;431:284–287. doi: 10.1038/nature02905. [DOI] [PubMed] [Google Scholar]
- 78.Chiu H, Hung P, Postma HWC, Bockrath M. Atomic-Scale Mass Sensing Using Carbon Nanotube Resonators. Nano Lett. 2008;8:4342–4346. doi: 10.1021/nl802181c. [DOI] [PubMed] [Google Scholar]
- 79.Bunch JS, van der Zande AM, Verbridge SS, Frank IW, Tanenbaum DM, Parpia JM, Craighead HG, McEuen PL. Electromechanical Resonators from Graphene Sheets. Science. 2007;315:490–493. doi: 10.1126/science.1136836. [DOI] [PubMed] [Google Scholar]
- 80.Waggoner PS, Craighead HG. Micro- and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip. 2007;7:1238. doi: 10.1039/b707401h. [DOI] [PubMed] [Google Scholar]
- 81.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat Meth. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 82.Boyd RW, Heebner JE. Sensitive disk resonator photonic biosensor. Applied Optics. 2001;40:5742–5747. doi: 10.1364/ao.40.005742. [DOI] [PubMed] [Google Scholar]
- 83.Vollmer F, Braun D, Libchaber A, Khoshsima M, Teraoka I, Arnold S. Protein detection by optical shift of a resonant microcavity. Appl Phys Lett. 2002;80:4057. [Google Scholar]
- 84.Arnold S, Khoshsima M, Teraoka I, Holler S, Vollmer F. Shift of whispering-gallery modes in microspheres by protein adsorption. Optics letters. 2003;28:272–274. doi: 10.1364/ol.28.000272. [DOI] [PubMed] [Google Scholar]
- 85.Vollmer F, Arnold S, Braun D, Teraoka I, Libchaber A. Multiplexed DNA Quantification by Spectroscopic Shift of Two Microsphere Cavities. Biophys J. 2003;85:1974–1979. doi: 10.1016/S0006-3495(03)74625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Label-free, single-molecule detection with optical microcavities. Science. 2007;317:783. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 87.Arnold S, Shopova SI, Holler S. Whispering gallery mode bio-sensor for label-free detection of single molecules: thermo-optic vs. reactive mechanism. Optics Express. 2010;18:281–287. doi: 10.1364/OE.18.000281. [DOI] [PubMed] [Google Scholar]
- 88.White IM, Oveys H, Fan X. Liquid-core optical ring-resonator sensors. Optics letters. 2006;31:1319–1321. doi: 10.1364/ol.31.001319. [DOI] [PubMed] [Google Scholar]
- 89.Zhu H, White IM, Suter JD, Dale PS, Fan X. Analysis of biomolecule detection with optofluidic ring resonator sensors. Opt Lett. 1995;20:654–656. doi: 10.1364/oe.15.009139. [DOI] [PubMed] [Google Scholar]
- 90.White IM, Oveys H, Fan X, Smith TL, Zhang J. Integrated multiplexed biosensors based on liquid core optical ring resonators and antiresonant reflecting optical waveguides. Appl Phys Lett. 2006;89:191106. [Google Scholar]
- 91.Suter JD, White IM, Zhu H, Shi H, Caldwell CW, Fan X. Label-free quantitative DNA detection using the liquid core optical ring resonator. Biosensors and Bioelectronics. 2008;23:1003–1009. doi: 10.1016/j.bios.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 92.Zhu H, White IM, Suter JD, Zourob M, Fan X. Opto-fluidic micro-ring resonator for sensitive label-free viral detection. Analyst. 2008;133:356. doi: 10.1039/b716834a. [DOI] [PubMed] [Google Scholar]
- 93.Samiee KT, Moran-Mirabal JM, Cheung YK, Craighead HG. Zero mode waveguides for single-molecule spectroscopy on lipid membranes. Biophysical journal. 2006;90:3288–3299. doi: 10.1529/biophysj.105.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levene MJ, Korlach J, Turner S, Foquet M, Craighead HG, Webb W. Zero-Mode Waveguides for Single-Molecule Analysis at High Concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 95.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 96.McFarland AD, Van Duyne RP. Single Silver Nanoparticles as Real-Time Optical Sensors with Zeptomole Sensitivity. Nano Lett. 2003;3:1057–1062. [Google Scholar]
- 97.Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. Rapid and Sensitive Detection of Respiratory Virus Molecular Signatures Using a Silver Nanorod Array SERS Substrate. Nano Lett. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 98.Vo-Dinh T. SERS chemical sensors and biosensors: new tools for environmental and biological analysis* 1. Sensors and Actuators B: Chemical. 1995;29:183–189. [Google Scholar]
- 99.Cao YC, Jin R, Nam JM, Thaxton CS, Mirkin CA. Raman dye-labeled nanoparticle probes for proteins. J Am Chem Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 100.Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Ultrasensitive Chemical Analysis by Raman Spectroscopy. Chem Rev. 1999;99:2957–2976. doi: 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- 101.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 102.Lutz B, Dentinger C, Sun L, Nguyen L, Zhang J, Chmura AJ, Allen A, Chan S, Knudsen B. Raman nanoparticle probes for antibody-based protein detection in tissues. Journal of Histochemistry and Cytochemistry. 2008;56:371. doi: 10.1369/jhc.7A7313.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su X, Zhang J, Sun L, Koo T, Chan S, Sundararajan N, Yamakawa M, Berlin AA. Composite Organic–Inorganic Nanoparticles (COINs) with Chemically Encoded Optical Signatures. Nano Lett. 2005;5:49–54. doi: 10.1021/nl0484088. [DOI] [PubMed] [Google Scholar]
- 104.Sun L, Sung K, Dentinger C, Lutz B, Nguyen L, Zhang J, Qin H, Yamakawa M, Cao M, Lu Y, et al. Composite Organic–Inorganic Nanoparticles as Raman Labels for Tissue Analysis. Nano Lett. 2007;7:351–356. doi: 10.1021/nl062453t. [DOI] [PubMed] [Google Scholar]
- 105.Jareserijman E, Jovin T. Imaging molecular interactions in living cells by FRET microscopy. Current Opinion in Chemical Biology. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 106.Ai H, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Meth. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 107.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 108.Erickson D, Mandal S, Yang AHJ, Cordovez B. Nanobiosensors: optofluidic, electrical and mechanical approaches to biomolecular detection at the nanoscale. Microfluid Nanofluid. 2007;4:33–52. doi: 10.1007/s10404-007-0198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chithrani BD, Ghazani AA, Chan WCW. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Zhang J, Ekman JM, Kenis PJ, Lu Y. DNA-Mediated Control of Metal Nanoparticle Shape: One-Pot Synthesis and Cellular Uptake of Highly Stable and Functional Gold Nanoflowers. Nano letters. 2010;10:1886–1891. doi: 10.1021/nl100675p. [DOI] [PubMed] [Google Scholar]
- 111.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 113.Link S, El-Sayed MA. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J Phys Chem B. 1999;103:4212–4217. [Google Scholar]
- 114.Kreibig U, Genzel L. Optical absorption of small metallic particles. Surface Science. 1985;156:678–700. [Google Scholar]
- 115.Han MS, Lytton-Jean AKR, Oh B, Heo J, Mirkin CA. Colorimetric Screening of DNA-Binding Molecules with Gold Nanoparticle Probes. Angew Chem Int Ed. 2006;45:1807–1810. doi: 10.1002/anie.200504277. [DOI] [PubMed] [Google Scholar]
- 116.Liu J, Lu Y. Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J Am Chem Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- 117.Kim Y, Johnson RC, Hupp JT. Gold Nanoparticle-Based Sensing of “Spectroscopically Silent” Heavy Metal Ions. Nano Lett. 2001;1:165–167. [Google Scholar]
- 118.Xue X, Wang F, Liu X. One-Step, Room Temperature, Colorimetric Detection of Mercury (Hg2+) Using DNA/Nanoparticle Conjugates. Journal of the American Chemical Society. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 119.Thanh NT, Rosenzweig Z. Development of an aggregation-based immunoassay for anti-protein A using gold nanoparticles. Anal Chem. 2002;74:1624–1628. doi: 10.1021/ac011127p. [DOI] [PubMed] [Google Scholar]
- 120.Liu J, Lu Y. Adenosine-Dependent Assembly of Aptazyme-Functionalized Gold Nanoparticles and Its Application as a Colorimetric Biosensor. Anal Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 121.Nam JM, Park SJ, Mirkin CA. Bio-barcodes based on oligonucleotide-modified nanoparticles. J Am Chem Soc. 2002;124:3820–3821. doi: 10.1021/ja0178766. [DOI] [PubMed] [Google Scholar]
- 122.Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 123.Nam J, Thaxton C, Mirkin CA. Nanoparticle-Based Bio-Bar Codes for the Ultrasensitive Detection of Proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 124.Nam J, Jang K, Groves JT. Detection of proteins using a colorimetric bio-barcode assay. Nat Protoc. 2007;2:1438–1444. doi: 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]
- 125.Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 126.Josephson L, Perez JM, Weissleder R. Magnetic nanosensors for the detection of oligonucleotide sequences. Angewandte Chemie. 2001;113:3304–3306. doi: 10.1002/1521-3773(20010903)40:17<3204::AID-ANIE3204>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 127.Perez JM, Simeone FJ, Saeki Y, Josephson L, Weissleder R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J Am Chem Soc. 2003;125:10192–10193. doi: 10.1021/ja036409g. [DOI] [PubMed] [Google Scholar]
- 128.Koh I, Hong R, Weissleder R, Josephson L. Sensitive NMR Sensors Detect Antibodies to Influenza. Angew Chem Int Ed. 2008;47:4119–4121. doi: 10.1002/anie.200800069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perez JM, Josephson L, O’Loughlin T, Hogemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotech. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 130.von Maltzahn G, Min D, Zhang Y, Park J, Harris T, Sailor M, Bhatia S. Nanoparticle Self-Assembly Directed by Antagonistic Kinase and Phosphatase Activities. Adv Mater. 2007;19:3579–3583. [Google Scholar]
- 131.Perez JM, O’Loughin T, Simeone FJ, Weissleder R, Josephson L. DNA-based magnetic nanoparticle assembly acts as a magnetic relaxation nanoswitch allowing screening of DNA-cleaving agents. J Am Chem Soc. 2002;124:2856–2857. doi: 10.1021/ja017773n. [DOI] [PubMed] [Google Scholar]
- 132.Zhao M, Josephson L, Tang Y, Weissleder R. Magnetic sensors for protease assays. Angewandte Chemie. 2003;115:1413–1416. doi: 10.1002/anie.200390352. [DOI] [PubMed] [Google Scholar]
- 133.Perez JM, Simeone FJ, Tsourkas A, Josephson L, Weissleder R. Peroxidase Substrate Nanosensors for MR Imaging. Nano Lett. 2004;4:119–122. [Google Scholar]
- 134.Taktak S, Sosnovik D, Cima MJ, Weissleder R, Josephson L. Multiparameter Magnetic Relaxation Switch Assays. Anal Chem. 2007;79:8863–8869. doi: 10.1021/ac701976p. [DOI] [PubMed] [Google Scholar]
- 135.Kim GY, Josephson L, Langer R, Cima MJ. Magnetic Relaxation Switch Detection of Human Chorionic Gonadotrophin. Bioconjugate Chem. 2007;18:2024–2028. doi: 10.1021/bc070110w. [DOI] [PubMed] [Google Scholar]
- 136.Daniel KD, Kim GY, Vassiliou CC, Jalali-Yazdi F, Langer R, Cima MJ. Multi-reservoir device for detecting a soluble cancer biomarker. Lab Chip. 2007;7:1288. doi: 10.1039/b705143c. [DOI] [PubMed] [Google Scholar]
- 137.von Maltzahn G, Harris TJ, Park JH, Min DH, Schmidt AJ, Sailor MJ, Bhatia SN. Nanoparticle self-assembly gated by logical proteolytic triggers. J Am Chem Soc. 2007;129:6064–6065. doi: 10.1021/ja070461l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee H, Sun E, Ham D, Weissleder R. Chip–NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perez JM, Josephson L, Weissleder R. Use of magnetic nanoparticles as nanosensors to probe for molecular interactions. Chembiochem. 2004;5:261–264. doi: 10.1002/cbic.200300730. [DOI] [PubMed] [Google Scholar]
- 140.Chou I, Benford M, Beier HT, Coteé GL, Wang M, Jing N, Kameoka J, Good TA. Nanofluidic Biosensing for β-Amyloid Detection Using Surface Enhanced Raman Spectroscopy. Nano Lett. 2008;8:1729–1735. doi: 10.1021/nl0808132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang XMH, Caldwell R, Huang L, Jun SC, Huang M, Sfeir MY, O’Brien SP, Hone J. Controlled Placement of Individual Carbon Nanotubes. Nano Lett. 2005;5:1515–1518. doi: 10.1021/nl050886a. [DOI] [PubMed] [Google Scholar]
- 142.Nagahara LA, Amlani I, Lewenstein J, Tsui RK. Directed placement of suspended carbon nanotubes for nanometer-scale assembly. Appl Phys Lett. 2002;80:3826. [Google Scholar]
- 143.Lewenstein JC, Burgin TP, Ribayrol A, Nagahara LA, Tsui RK. High-Yield Selective Placement of Carbon Nanotubes on Pre-Patterned Electrodes. Nano Lett. 2002;2:443–446. [Google Scholar]
- 144.Cassell AM, Franklin NR, Tombler TW, Chan EM, Han J, Dai H. Directed Growth of Free-StandingSingle-Walled Carbon Nanotubes. J Am Chem Soc. 1999;121:7975–7976. [Google Scholar]
- 145.Zheng LX, O’Connell MJ, Doorn SK, Liao XZ, Zhao YH, Akhadov EA, Hoffbauer MA, Roop BJ, Jia QX, Dye RC, et al. Ultralong single-wall carbon nanotubes. Nat Mater. 2004;3:673–676. doi: 10.1038/nmat1216. [DOI] [PubMed] [Google Scholar]
- 146.Hong BH, Lee JY, Beetz T, Zhu Y, Kim P, Kim KS. Quasi-continuous growth of ultralong carbon nanotube arrays. J Am Chem Soc. 2005;127:15336–15337. doi: 10.1021/ja054454d. [DOI] [PubMed] [Google Scholar]
- 147.Dai H. Carbon Nanotubes: Synthesis, Integration, and Properties. Acc Chem Res. 2002;35:1035–1044. doi: 10.1021/ar0101640. [DOI] [PubMed] [Google Scholar]
- 148.Dai H. Nanotube growth and characterization. Carbon Nanotubes. 2001:29–53. [Google Scholar]
- 149.Wang B, Poa CH, Wei L, Li LJ, Yang Y, Chen Y. (n, m) Selectivity of Single-Walled Carbon Nanotubes by Different Carbon Precursors on Co- Mo Catalysts. J Am Chem Soc. 2007;129:9014–9019. doi: 10.1021/ja070808k. [DOI] [PubMed] [Google Scholar]
- 150.Bandow S, Asaka S, Saito Y, Rao AM, Grigorian L, Richter E, Eklund PC. Effect of the growth temperature on the diameter distribution and chirality of single-wall carbon nanotubes. Physical Review Letters. 1998;80:3779–3782. [Google Scholar]
- 151.Dai H. Controlling nanotube growth. Physics World. 2000;13:43–47. [Google Scholar]
- 152.Chen RJ, Bangsaruntip S, Drouvalakis KA, Wong Shi Kam N, Shim M, Li Y, Kim W, Utz PJ, Dai H. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4984. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Drouvalakis KA, Bangsaruntip S, Hueber W, Kozar LG, Utz PJ, Dai H. Peptide-coated nanotube-based biosensor for the detection of disease-specific autoantibodies in human serum. Biosensors and Bioelectronics. 2008;23:1413–1421. doi: 10.1016/j.bios.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen RJ, Choi HC, Bangsaruntip S, Yenilmez E, Tang X, Wang Q, Chang Y, Dai H. An Investigation of the Mechanisms of Electronic Sensing of Protein Adsorption on Carbon Nanotube Devices. J Am Chem Soc. 2004;126:1563–1568. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- 155.Sotiropoulou S, Chaniotakis NA. Carbon nanotube array-based biosensor. Analytical and Bioanalytical Chemistry. 2003;375:103–105. doi: 10.1007/s00216-002-1617-z. [DOI] [PubMed] [Google Scholar]
- 156.Besteman K, Lee J, Wiertz FGM, Heering HA, Dekker C. Enzyme-Coated Carbon Nanotubes as Single-Molecule Biosensors. Nano Lett. 2003;3:727–730. [Google Scholar]
- 157.Bradley K, Briman M, Star A, Grüner G. Charge Transfer from Adsorbed Proteins. Nano Lett. 2004;4:253–256. [Google Scholar]
- 158.Hu P, Fasoli A, Park J, Choi Y, Estrela P, Maeng SL, Milne WI, Ferrari AC. Self-assembled nanotube field-effect transistors for label-free protein biosensors. J Appl Phys. 2008;104:074310. [Google Scholar]
- 159.Li C, Curreli M, Lin H, Lei B, Ishikawa FN, Datar R, Cote RJ, Thompson ME, Zhou C. Complementary detection of prostate-specific antigen using In2O3 nanowires and carbon nanotubes. J Am Chem Soc. 2005;127:12484–12485. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 160.Star A, Tu E, Niemann J, Gabriel JC, Joiner CS, Valcke C. Label-free detection of DNA hybridization using carbon nanotube network field-effect transistors. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:921. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gui EL, Li L, Zhang K, Xu Y, Dong X, Ho X, Lee PS, Kasim J, Shen ZX, Rogers JA, et al. DNA Sensing by Field-Effect Transistors Based on Networks of Carbon Nanotubes. J Am Chem Soc. 2007;129:14427–14432. doi: 10.1021/ja075176g. [DOI] [PubMed] [Google Scholar]
- 162.Li J, Ng HT, Cassell A, Fan W, Chen H, Ye Q, Koehne J, Han J, Meyyappan M. Carbon Nanotube Nanoelectrode Array for Ultrasensitive DNA Detection. Nano Lett. 2003;3:597–602. [Google Scholar]
- 163.Wang J. Carbon-nanotube based electrochemical biosensors: a review. Electroanalysis. 2005;17:7–14. [Google Scholar]
- 164.Allen B, Kichambare P, Star A. Carbon Nanotube Field-Effect-Transistor-Based Biosensors. Adv Mater. 2007;19:1439–1451. [Google Scholar]
- 165.Zhang GJ, Chua JH, Chee RE, Agarwal A, Wong SM. Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosensors and Bioelectronics. 2009;24:2504–2508. doi: 10.1016/j.bios.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 166.Zhang GJ, Chua JH, Chee RE, Agarwal A, Wong SM, Buddharaju KD, Balasubramanian N. Highly sensitive measurements of PNA-DNA hybridization using oxide-etched silicon nanowire biosensors. Biosensors and Bioelectronics. 2008;23:1701–1707. doi: 10.1016/j.bios.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 167.Zhang G, Zhang G, Chua JH, Chee R, Wong EH, Agarwal A, Buddharaju KD, Singh N, Gao Z, Balasubramanian N. DNA Sensing by Silicon Nanowire: Charge Layer Distance Dependence. Nano Lett. 2008;8:1066–1070. doi: 10.1021/nl072991l. [DOI] [PubMed] [Google Scholar]
- 168.Gao Z, Agarwal A, Trigg AD, Singh N, Fang C, Tung C, Fan Y, Buddharaju KD, Kong J. Silicon Nanowire Arrays for Label-Free Detection of DNA. Anal Chem. 2007;79:3291–3297. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 169.Cui Y, Wei Q, Park H, Lieber CM. Nanowire Nanosensors for Highly Sensitive and Selective Detection of Biological and Chemical Species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 170.Hahm J, Lieber CM. Direct Ultrasensitive Electrical Detection of DNA and DNA Sequence Variations Using Nanowire Nanosensors. Nano Lett. 2004;4:51–54. [Google Scholar]
- 171.Heo K, Cho E, Yang J, Kim M, Lee M, Lee BY, Kwon SG, Lee M, Jo M, Choi H, et al. Large-Scale Assembly of Silicon Nanowire Network-Based Devices Using Conventional Microfabrication Facilities. Nano Lett. 2008;8:4523–4527. doi: 10.1021/nl802570m. [DOI] [PubMed] [Google Scholar]
- 172.Patolsky F, Zheng G, Hayden O, Lakadamyali M, Zhuang X, Lieber CM. Electrical detection of single viruses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14017. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotech. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 174.Malhotra BD, Chaubey A, Singh SP. Prospects of conducting polymers in biosensors. Analytica chimica acta. 2006;578:59–74. doi: 10.1016/j.aca.2006.04.055. [DOI] [PubMed] [Google Scholar]
- 175.Bangar MA, Chen W, Myung NV, Mulchandani A. Conducting polymer 1-dimensional nanostructures for FET sensors. Thin Solid Films. 2010;519:964–973. [Google Scholar]
- 176.Ramanathan K, Bangar MA, Yun M, Chen W, Myung NV, Mulchandani A. Bioaffinity sensing using biologically functionalized conducting-polymer nanowire. J Am Chem Soc. 2005;127:496–497. doi: 10.1021/ja044486l. [DOI] [PubMed] [Google Scholar]
- 177.Zhu N, Chang Z, He P, Fang Y. Electrochemically fabricated polyaniline nanowire-modified electrode for voltammetric detection of DNA hybridization. Electrochimica Acta. 2006;51:3758–3762. [Google Scholar]
- 178.Bangar MA, Shirale DJ, Chen W, Myung NV, Mulchandani A. Single Conducting Polymer Nanowire Chemiresistive Label-Free Immunosensor for Cancer Biomarker. Analytical Chemistry. 2009;81:2168–2175. doi: 10.1021/ac802319f. [DOI] [PubMed] [Google Scholar]
- 179.He B, Morrow TJ, Keating CD. Nanowire sensors for multiplexed detection of biomolecules. Current Opinion in Chemical Biology. 2008;12:522–528. doi: 10.1016/j.cbpa.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Patolsky F, Zheng G, Lieber CM. Nanowire-based biosensors. Analytical Chemistry. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 181.Sheehan PE, Whitman LJ. Detection Limits for Nanoscale Biosensors. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 182.Nair PR, Alam MA. Performance limits of nanobiosensors. Appl Phys Lett. 2006;88:233120. [Google Scholar]
- 183.Nair PR, Alam MA. Screening-Limited Response of NanoBiosensors. Nano Letters. 2008;8:1281–1285. doi: 10.1021/nl072593i. [DOI] [PubMed] [Google Scholar]
- 184.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 185.Martinez AW, Phillips ST, Wiley BJ, Gupta M, Whitesides GM. FLASH: A rapid method for prototyping paper-based microfluidic devices. Lab Chip. 2008;8:2146. doi: 10.1039/b811135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martinez AW, Phillips ST, Carrilho E, Thomas SW, Sindi H, Whitesides GM. Simple Telemedicine for Developing Regions: Camera Phones and Paper-Based Microfluidic Devices for Real-Time, Off-Site Diagnosis. Anal Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Martinez AW, Phillips ST, Whitesides GM. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proceedings of the National Academy of Sciences. 2008;105:19606. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Paper-Based Bioassays Using Gold Nanoparticle Colorimetric Probes. Anal Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 189.Zimmermann M, Bentley S, Schmid H, Hunziker P, Delamarche E. Continuous flow in open microfluidics using controlled evaporation. Lab Chip. 2005;5:1355. doi: 10.1039/b510044e. [DOI] [PubMed] [Google Scholar]
- 190.Goedecke N, Eijkel J, Manz A. Evaporation driven pumping for chromatography application. Lab Chip. 2002;2:219. doi: 10.1039/b208031c. [DOI] [PubMed] [Google Scholar]
- 191.Lynn NS, Dandy DS. Passive microfluidic pumping using coupled capillary/evaporation effects. Lab Chip. 2009;9:3422. doi: 10.1039/b912213c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fritz J. Cantilever biosensors. Analyst. 2008;133:855. doi: 10.1039/b718174d. [DOI] [PubMed] [Google Scholar]


