Abstract
Saline or alkaline soils present a strong stress on plants that together may be even more deleterious than alone. Australia's soils are old and contain large, sometimes overlapping, areas of high salt and alkalinity. Acacia and other Australian plant lineages have evolved in this stressful soil environment and present an opportunity to understand the evolution of salt and alkalinity tolerance. We investigate this evolution by predicting the average soil salinity and pH for 503 Acacia species and mapping the response onto a maximum-likelihood phylogeny. We find that salinity and alkalinity tolerance have evolved repeatedly and often together over 25 Ma of the Acacia radiation in Australia. Geographically restricted species are often tolerant of extreme conditions. Distantly related species are sympatric in the most extreme soil environments, suggesting lack of niche saturation. There is strong evidence that many Acacia have distributions affected by salinity and alkalinity and that preference is lineage specific.
Keywords: salt-tolerance, soil pH, stress-tolerance
1. Introduction
Most plants are glycophytes that tolerate only low concentrations of salt before they are adversely affected or die. The salt concentration threshold separating glycophytes from salt-tolerant halophytes has been variously set between 80 and 200 mM NaCl [1]; it is often estimated by electrical conductivity (EC) measurements on soil saturated paste extracts. Saline soils have an EC of 4000 μS m−1 or more (approx. 40 mM NaCl) measured on a saturated soil paste extract at 25°C [2]. However, physiological plant responses to salinity start at EC less than 4000 μS m−1 [2] and Australian native plant distributions respond to a salinity gradient starting at low EC [3].
By contrast, there is no precise definition of what characterizes an alkaline-tolerant plant. Most cultivated plants prefer acidic soils (pH 5.5–6.5) [4]. Combined alkaline and salt stresses are more deleterious than salinity alone [5–7]. Even for the naturally alkali-resistant halophyte Chloris virgata, the inhibitory effects of alkali stress on relative growth rate and stored energy are significantly larger than salt stress at neutral pH [6]. Resistance to either or both of these abiotic stresses also may be associated with drought tolerance in arid biome plants [8,9].
Traditionally, research on salt- and/or alkaline-tolerance in plants focused on laboratory experimentation and plant breeding [5–7], but new paradigms have emerged. One uses molecular tools to identify genes involved in stress-tolerance [9,10] and another uses phylogenetics to investigate the evolution of salt-tolerance [1,11,12].
Acacia has radiated into more than 1000 species across Australia over 25 Ma [13] and its distribution is significantly correlated with soil pH and salinity [14]. Acacia species richness and endemicity have been linked to soil chemistry in Western Australia [14] where large localized spatial and seasonal variability in EC and pH [15] create a mosaic of island-like niches. We investigate the evolution of salt- and alkaline-tolerance using a comprehensive Acacia phylogeny. We hypothesize that there is a genetic relationship between salt- and alkaline-tolerance in Acacia. Understanding this evolution may provide new avenues for plant breeding for stress-tolerance and improve prospects for land rehabilitation and agriculture in extreme environments.
2. Material and methods
Locality data were extracted from Australia's Virtual Herbarium (avh.ala.org.au) resulting in 127 259 points with unique geographical coordinates, representing presence data for 1020 Acacia species across the continent [14]. A maximum-likelihood phylogeny of 503 Acacia species was constructed from six DNA regions [16] using RAxML HPC BlackBox tool implemented online in the CIPRES Portal (http://www.phylo.org/).
The National Geochemical Survey of Australia reported the pH and EC on 1 : 5 soil : solution extracts from bulk samples at 1315 georeferenced point measurements across the continent at two depth intervals (top outlet sediment (TOS), 0–10 cm, and bottom outlet sediment (BOS), 60–80 cm) [17]. Salinity in EC1 : 5 extracts is lower than in saturated paste extracts. With geoR, models were constructed to fit variograms and ordinary kriging [18] was used to predict pH and EC for the two depths for all Acacia localities. Average responses of species were used in the phylogenetic analyses.
Phylogenetic signal was tested with Pagel's λ test using Phylosig in R which fits traits on the phylogeny against the null hypothesis (λ = 0) [19]. We tested for character correlation by correcting for non-independence of the observed phylogenetic signal with phylogenetically independent contrasts (PIC) [20,21] which assumes a Brownian motion process of evolution. We divided the geochemistry data into four groups based on quantiles and used parsimony-based ancestral character states reconstruction for visualization.
Relatively few acacias have been tested for salt-tolerance, therefore we defined an Acacia species as salt-tolerant when the average EC value over its range was in the highest 10% (50 species) of 503 species (more than 1979 μS cm−1; electronic supplementary material, table S2; figure 1e). We defined Acacia species as alkaline-tolerant when the average pH value over its range was in the highest 10% of the species (more than 7.61; electronic supplementary material, table S2; figure 1f).
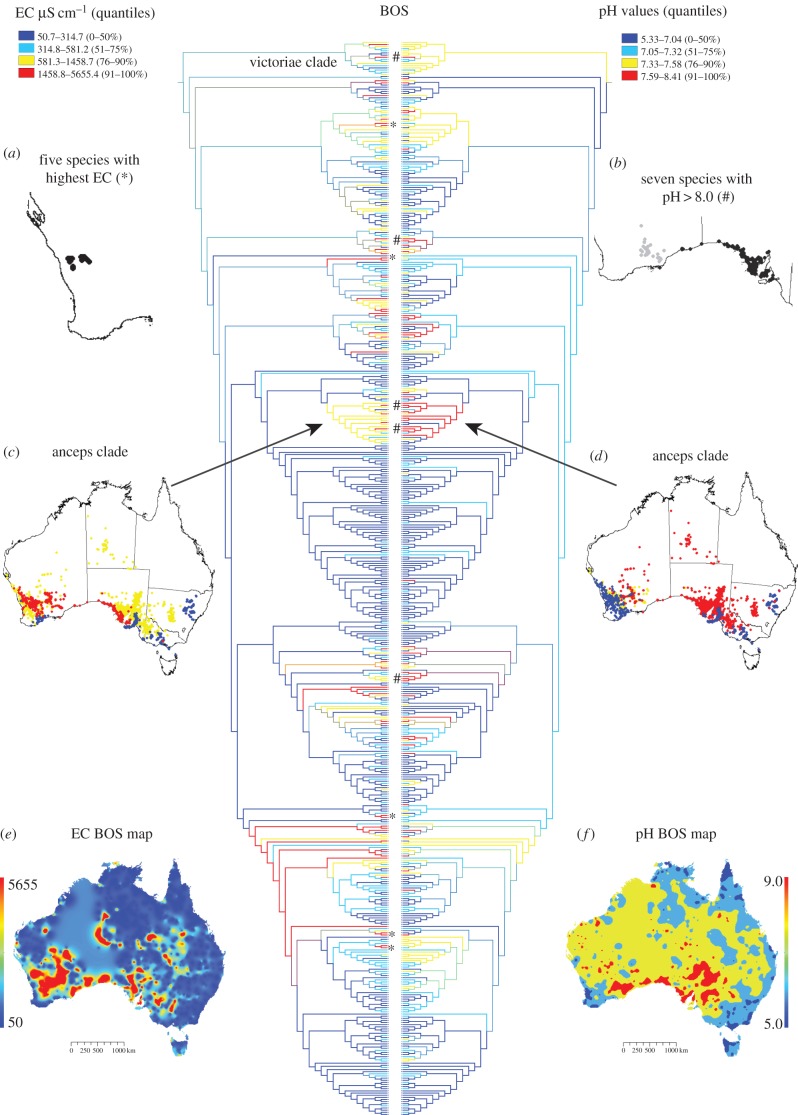
Figure 1.
Phylogenetic relationships of 503 Acacia species with EC_BOS (left) and pH_BOS (right); quantiles mapped and parsimony ancestral state reconstructed. Legend indicates raw values of quantiles. Asterisk (*) indicates species with highest EC_BOS values; symbol # indicates species with pH_BOS values more than 8.0. (a) Distribution map of the five species with highest average EC_BOS values; (b) Distribution map of A. dempsteri (grey), gillii, anceps and other species with subsoil pH > 8.0; (c) A. anceps clade with the species distribution coloured as on the EC phylogeny and (d) as on the pH phylogeny; (e) map of EC_BOS and (f) pH_BOS across Australia.
3. Results
Predicted species averages of EC_BOS values were generally higher than TOS and ranged from 53.5 to 1566 μS cm−1 (TOS) and 50.6–5655 μS cm−1 (BOS). Species averages for pH were slightly higher in the subsoil with values ranging from 5.03 to 7.80 in TOS and 5.33 to 8.41 in BOS (electronic supplementary material, table S2). Because the BOS geochemistry is more extreme and acacias are trees with deep roots [22], we focus on BOS data.
Acacia pterocaulon has the highest average EC_BOS with a value of 5655 μS cm−1, with four other species with values above 4000 μS cm−1; all are endemic to southwestern Western Australia (figure 1a). Twenty-five of the next 27 species with highest EC_BOS values (more than 2000 μS cm−1) co-occur or grow nearby in inland southwestern Western Australia; many are range restricted.
Acacia gillii has the highest average pH_BOS (8.40) and six more species grow in soils where the average pH_BOS is above 8.0. These species, except A. dempsteri, are restricted to coastal regions of South Australia (figure 1b).
Thirteen species have EC and pH values (BOS) in the top 10% of each category and can be considered both salt- and alkaline-tolerant (electronic supplementary material, table S1). Eleven species have EC and pH values (TOS) in the top 10% for each.
Phylogenetic signal analysis indicated that EC and pH average values per species for both TOS and BOS reflect the phylogenetic structure (p < 0.0001). Additionally, PIC analysis determined that both traits are correlated with each other even after taking into account the phylogeny.
Using our definitions of salinity and alkalinity tolerance (50 species), we infer that salinity tolerance evolved 38 times and alkalinity tolerance evolved 31 times. There is more phylogenetic clustering of the alkaline-tolerant than salt-tolerant species. The anceps clade (figure 1) contains 11 alkaline-tolerant species and four salinity-tolerant species, with 12 species in the upper 25% for salinity. The geographical distribution of species in this clade indicates that less tolerant sister species are generally in nearby areas that have soils that are less extreme in EC and pH values (figure 1c,d). Overlap of high EC and pH areas is evident in southern Western Australia and along the Nullarbor Plain into South Australia (figure 1e,f).
4. Discussion
The Australian landscape is old, weathered and has included soils with high levels of salinity and alkalinity for at least the last 2–4 Ma [14,15,23]. Acacia has evolved in this landscape for 25 Ma and has repeatedly adapted to saline and/or alkaline soils, in many lineages and at different times.
Salt-tolerance appears in the victoriae clade, sister to the rest of Acacia (figure 1). Acacia adinophylla is predicted to be associated with relatively high EC soils, while A. victoriae, predicted to be associated with moderate levels of salinity, has been used to rehabilitate salt-affected land [12]. Acacia adinophylla and other highly salt-tolerant species are range restricted and broadly sympatric (figure 1a) in the Transitional Rainfall Province of Western Australia, a locus of species richness and the origin of subsequent radiations [24]; these species are unrelated (asterisk in figure 1) indicating repeated colonization of saline soil. Tolerance to high soil pH also arises in the victoriae clade as all members (figure 1) are predicted to occur on soils with pH_BOS > 7.3.
Many species adapted to high salinity are also adapted to high pH; this is not surprising since both soil properties are associated with aridity [14] and this study cannot determine whether those are independent adaptations to two stresses that happen to exist in the same habitat or examples of cross-tolerance, the induction of adaptive responses to a stress by another. The anceps clade (figure 1c,d) contains four species with average pH_BOS > 8.0 that are also in the upper 25% for salinity. Acacia anceps and several congeners occur across the Nullarbor Plain and Eyre Peninsula. The Nullarbor is an extensive marine limestone formation along the Great Australian Bight, exposed since approximately 14 Ma, and the derived overlying soils are uniformly high in pedogenic carbonates [25]. Colonization of the Nullarbor is associated with the evolution of a new clade adapted to calcareous substrate.
The sympatry of multiple Acacia lineages on saline and alkaline soils could be the result of alternative mechanisms. Firstly, the Acacia ancestral condition could be tolerance to these soils, however ancestral state reconstruction does not support this hypothesis (EC_BOS = 897, pH_BOS = 7.1). Alternatively, the results suggest that there have been multiple colonizations of the highly saline and alkaline areas by multiple lineages at different time-points spanning 25 Ma. This means that even in areas where the soil conditions are providing strong selective pressures through high EC and/or pH, the environmental niche is not saturated by the first colonizing Acacia lineage, i.e. new species are able to invade communities where ecologically similar species are already present [26].
A few apparently salt-tolerant species occur on acid soils; this may point to independent adaptations to stress. There are also large radiations of Acacia associated with soils with both lower salinity and alkalinity, especially in eastern Australia where most of the Acacia species with bipinnate leaves occur [13].
5. Conclusion
We have found in the Australian Acacia that phylogenetic, geographical and soil chemistry relationships provide foundation to investigate how stress tolerance traits have evolved. In particular, southwestern Australia had many species from multiple Acacia lineages that appear adapted to both high salinity and alkalinity. Phylogenetic methods can be powerful tools to identify taxa suitable for generating new genetic combinations by plant breeding which are salt- and/or alkaline-tolerant. However, combined spatial and phylogenetic data cannot establish the role of soil chemistry on niche width. Manipulative experiments are required to test the effect of salt chemistry and pH and other abiotic and biotic factors in determining niche preferences.
Supplementary Material
Supplementary Material
References
- 1.Flowers TJ, Galal HK, Bromham L. 2010. Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct. Plant Biol. 37, 604–612. ( 10.1071/FP09269) [DOI] [Google Scholar]
- 2.United States Salinity Laboratory Staff. 1969. Diagnosis and improvement of saline and alkali soils. Agriculture Handbook No. 60, Rev. ed Washington, DC: United States Department of Agriculture. [Google Scholar]
- 3.Bui EN, Henderson BL. 2003. Vegetation indicators of soil salinity in north Queensland. Austral Ecol. 28, 539–552. ( 10.1046/j.1442-9993.2003.01311.x) [DOI] [Google Scholar]
- 4.Islam AKMS, Edwards DG, Asher CJ. 1980. pH optima for crop growth. Plant Soil 54, 339–357. ( 10.1007/BF02181830) [DOI] [Google Scholar]
- 5.Li C, Fang B, Yang C, Shi D, Wang D. 2009. Effects of various salt–alkaline mixed stresses on the state of mineral elements in nutrient solutions and the growth of alkali resistant halophyte Chloris virgata. J. Plant Nutr. 32, 1137–1147. ( 10.1080/01904160902943163) [DOI] [Google Scholar]
- 6.Yang CW, Jianaer A, Li CY, Shi DC, Wang DL. 2008. Comparison of the effects of salt-stress and alkali-stress on photosynthesis and energy storage of an alkali-resistant halophyte Chloris virgata. Photosynthetica 46, 273–278. ( 10.1007/s11099-008-0047-3) [DOI] [Google Scholar]
- 7.Javid M, Ford R, Nicolas ME. 2012. Tolerance responses of Brassica juncea to salinity, alkalinity and alkaline salinity. Funct. Plant Biol. 39, 699–707. ( 10.1071/FP12109) [DOI] [PubMed] [Google Scholar]
- 8.Bromham L, Saslis-Lagoudakis CH, Bennett TH, Flowers TJ. 2013. Soil alkalinity and salt tolerance: adapting to multiple stresses. Biol. Lett. 9, 20130642 ( 10.1098/rsbl.2013.0642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamirisa S, Vudem DR, Khareedu VR. 2014. Overexpression of pigeonpea stress-induced cold and drought regulatory gene (CcCDR) confers drought, salt, and cold tolerance in Arabidopsis. J. Exp. Bot. ( 10.1093/jxb/eru224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munns R. 2005. Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. ( 10.1111/j.1469-8137.2005.01487.x) [DOI] [PubMed] [Google Scholar]
- 11.Bennett TH, Flowers TJ, Bromham L. 2013. Repeated evolution of salt-tolerance in grasses. Biol. Lett. 9, 20130029 ( 10.1098/rsbl.2013.0029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph S, Bhave M, Miller JT, Murphy DJ. 2013. Rapid identification of Acacia species with potential salt tolerance by using nuclear ribosomal DNA markers. Sust. Agric. Res. 2, 77–86. [Google Scholar]
- 13.Miller JT, Murphy D, Ho SYW, Cantrill DJ, Seigler D. 2013. Comparative dating of Acacia: combining fossils and multiple phylogenies to infer ages of clades with poor fossil records. Austral J. Bot. 61, 436–445. ( 10.1071/BT13149) [DOI] [Google Scholar]
- 14.Bui EN, González-Orozco CE, Miller JT. 2014. Acacia, climate, and geochemistry in Australia. Plant Soil 381, 161–175. ( 10.1007/s11104-014-2113-x) [DOI] [Google Scholar]
- 15.Bowen BB, Benison KC. 2009. Geochemical characteristics of naturally acid and alkaline saline lakes in southern Western Australia. Appl. Geochem. 24, 268–284. ( 10.1016/j.apgeochem.2008.11.013) [DOI] [Google Scholar]
- 16.Miller JT, Murphy D, Brown GK, Richardson DM, González-Orozco CE. 2011. The evolution and phylogenetic placement of invasive Acacia species. Div. Distrib. 17, 848–860. ( 10.1111/j.1472-4642.2011.00780.x) [DOI] [Google Scholar]
- 17.de Caritat P, Cooper M. 2011. National geochemical survey of Australia: the geochemical atlas of Australia. Geoscience Australia: Canberra, ACT, Australia. [Google Scholar]
- 18.Diggle PJ, Ribeiro PJ. 2007. Model-based geostatistics. Springer: New York, NY, USA. [Google Scholar]
- 19.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 21.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 22.Canadell J, Jackson RB, Ehleringer JB, Mooney HA, Sala OE, Schulze ED. 1996. Maximum rooting depth of vegetation types at the global scale. Oecologia 108, 583–595. ( 10.1007/BF00329030) [DOI] [PubMed] [Google Scholar]
- 23.Hopper SD. 2009. OCBIL theory: towards an integrated understanding of the evolution, ecology and conservation of biodiversity on old climatically buffered, infertile landscapes. Plant Soil 322, 49–86. ( 10.1007/s11104-009-0068-0) [DOI] [Google Scholar]
- 24.Hopper SD, Gioia P. 2004. The southwest Australian floristic region: evolution and conservation of a global hot spot of biodiversity. Annu. Rev. Ecol. Evol. Syst. 35, 623–650. ( 10.1146/annurev.ecolsys.35.112202.130201) [DOI] [Google Scholar]
- 25.Webb A, James JM. 2006. Karst evolution of the Nullarbor Plain, Australia. In Perspectives on karst geomorphology, hydrology and geochemistry, vol. 404 (eds Harmon RS, Wicks CM.), pp. 65–78. Geological Society of America: Boulder, CO, USA. [Google Scholar]
- 26.Wiens JJ. 2011. The niche, biogeography and species interactions. Phil. Trans. R. Soc. B 366, 2336–2350. ( 10.1098/rstb.2011.0059) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.