Abstract
Background
YWHAE is a possible susceptibility gene for schizophrenia that encodes 14-3-3epsilon, a Disrupted-in-Schizophrenia 1 (DISC1)-interacting molecule, but the effect of variation in its genotype on brain morphology remains largely unknown.
Methods
In this voxel-based morphometric magnetic resonance imaging study, we conducted whole-brain analyses regarding the effects of YWHAE single-nucleotide polymorphisms (SNPs) (rs28365859, rs11655548, and rs9393) and DISC1 SNP (rs821616) on gray matter volume in a Japanese sample of 72 schizophrenia patients and 86 healthy controls. On the basis of a previous animal study, we also examined the effect of rs28365859 genotype specifically on hippocampal volume.
Results
Whole-brain analyses showed no significant genotype effect of these SNPs on gray matter volume in all subjects, but we found significant genotype-by-diagnosis interaction for rs28365859 in the left insula and right putamen. The protective C allele carriers of rs28365859 had a significantly larger left insula than the G homozygotes only for schizophrenia patients, while the controls with G allele homozygosity had a significantly larger right putamen than the C allele carriers. The C allele carriers had a larger right hippocampus than the G allele homozygotes in schizophrenia patients, but not in healthy controls. No significant interaction was found between rs28365859 and DISC1 SNP on gray matter volume.
Conclusions
These different effects of the YWHAE (rs28365859) genotype on brain morphology in schizophrenia and healthy controls suggest that variation in its genotype might be, at least partly, related to the abnormal neurodevelopment, including in the limbic regions, reported in schizophrenia. Our results also suggest its specific role among YWHAE SNPs in the pathophysiology of schizophrenia.
Introduction
Schizophrenia is a heterogeneous psychiatric disorder with a multifactorial etiology in which multiple susceptibility genes interact with environmental factors [1], [2]. Convergent evidence from neuroimaging studies in schizophrenia suggests subtle but widespread gray matter (GM) reductions predominantly in the frontal and temporo–limbic regions (e.g., hippocampus), at least partly as a consequence of early neurodevelopmental insult [3], [4]. These brain morphologic changes in schizophrenia could be useful endophenotypes for unraveling the molecular etiopathology of this complex psychiatric disorder [5], [6].
The Disrupted-in-Schizophrenia 1 (DISC1) gene [7], [8], which is thought to be involved in mechanisms of neurodevelopment and synaptic plasticity in cortical and limbic regions [9]–[13], has been one of the candidate genes for schizophrenia [14], [15]. In addition to the possible effect of DISC1 genotype variation on brain function and structure in the hippocampus [16] and cingulate cortex [17] in healthy subjects, our preliminary magnetic resonance imaging (MRI) study suggested that it might differentially affect GM volume of the neocortical and limbic regions in schizophrenia patients and healthy controls [18]. Several other MRI studies of DISC1 in schizophrenia have yielded inconsistent results [reviewed by Duff et al. [19] and there have also been questions about DISC1 as a genetic risk factor of schizophrenia [20]. However, DISC1 interacts with a complex formed by related molecules [13] and the genetic variation in such DISC1-interacting molecules might have a significant role in the pathophysiology of schizophrenia.
YWHAE is a gene encoding 14-3-3epsilon, one of the DISC1-interacting molecules that is thought to play a crucial role in neuronal development via transport of the NudE-like (NUDEL)/lissencephaly-1 (LIS1) complex [13], [21], and is a possible susceptibility gene for schizophrenia as identified in a Japanese population [22]. Genetic and expression evidence indicated that a functional single-nucleotide polymorphism (SNP) in the 5′ flanking region (rs28365859) was associated with schizophrenia, with subjects with the C allele having a reduced risk of the illness [22]. In addition, animal studies using genetically modified 14-3-3epsilon-deficient mice showed developmental defects of hippocampal neurons [21] as well as working memory deficits [22], which is one of the prominent features of schizophrenia [23]. Despite these observations supporting the significant role of YWHAE in the neurobiology of schizophrenia, the possible association between variation in its genotype and brain morphology in schizophrenia remains largely unknown.
In this MRI study, we used voxel-based morphometry (VBM), which allows automated whole-brain analysis, to explore the effects of a YWHAE SNP (rs28365859) on regional GM volume in a Japanese sample of schizophrenia patients and matched healthy controls. On the basis of the potential role of YWHAE in neuronal development as well as previous MRI findings in schizophrenia [3], [4], we predicted significant diagnosis-by-genotype interaction predominantly in frontal and temporo–limbic regions, with patients with the protective C allele having a larger GM volume. As previous animal studies suggested the impact of YWHAE on the hippocampus [21], we also examined the effect of its genotype specifically on hippocampal volume using small volume correction (SVC) of VBM analyses, with the hypothesis that subjects with the C allele would have a larger hippocampal volume, especially in schizophrenia patients.
To investigate the specificity of the effect of rs28365859 on brain morphology, we also examined two putative non-risk SNPs in YWHAE (rs11655548 that was associated with schizophrenia but located in the intron region and rs9393, a functional SNP with no difference in genotype distribution between schizophrenia and controls) [22]. Possible interaction effect between rs28365859 and DISC1 Ser704Cys SNP (rs821616) on brain morphology was also examined.
Methods
Ethics statement
This protocol was approved by Committee on Medical Ethics of Toyama University and Nagoya University Graduate School of Medicine. After a complete and detail description of the study was given, subjects provided written informed consent. Clinical staff explained the nature of the study to the subjects, the risks and benefits, and the option not to participate in this research. If the mental status of a subject was impaired to the point where s/he could not understand these issues, the subject was not asked to participate in this research. If there was a possibility that the capacity of a participant to consent was compromised, an additional consent form was obtained from the next of kin, care takers, or guardians of such subjects.
Subjects
Seventy-two patients with schizophrenia (39 males and 33 females; mean age = 27.5 years, SD = 6.0) who met the ICD-10 research criteria [24] were recruited from inpatient and outpatient clinics of the Department of Neuropsychiatry of Toyama University Hospital. The patients were diagnosed following a structured clinical interview by psychiatrists using the Comprehensive Assessment of Symptoms and History (CASH) [25]. Clinical symptoms were rated at the time of scanning using the Scale for the Assessment of Negative Symptoms (SANS) [26] and the Scale for the Assessment of Positive Symptoms (SAPS) [27]. Sixty-eight patients were right-handed and four patients were mixed-handed.
The control subjects consisted of 86 right-handed healthy volunteers (45 males and 41 females; mean age = 26.4 years, SD = 6.6) recruited from members of the local community, hospital staff, and university students. They were asked to complete a questionnaire consisting of 15 items concerning their personal (13 items; including a history of obstetric complications, substantial head injury, seizures, neurological or psychiatric disease, impaired thyroid function, hypertension, diabetes, and substance abuse) and family (2 items) histories of illness. Subjects with any personal or family history of psychiatric illness among their first-degree relatives were excluded.
All subjects were Japanese and physically healthy at the time of the study. None had a lifetime history of serious head trauma, neurological illness, serious medical or surgical illness, or substance abuse. All participants were also screened for gross brain abnormalities by neuroradiologists. The subject overlap with our previous publication included 30/72 schizophrenia patients and 28/86 controls, where we reported the effect of DISC1 Ser704Cys polymorphism (rs821616) on brain morphology [18].
SNP genotyping
Genomic DNA was extracted from EDTA-containing venous blood samples according to standard procedures. The genotyping of SNPs in YWHAE (rs28365859, rs11655548, and rs9393) and DISC1 (rs821616) was performed by TaqMan assays (Applied Biosystems, Foster City, CA). TaqMan SNP Genotyping Assay and Universal PCR Master Mix were obtained from Applied Biosystems. Allelic-specific fluorescence was measured using the ABI PRISM 7900 Sequence Detector System (Applied Biosystems).
MRI procedures
MR images were obtained using 1.5 T Magnetom Vision (Siemens Medical System, Inc., Erlangen, Germany) with a three-dimensional gradient-echo sequence FLASH (fast low-angle shots) yielding 160–180 contiguous T1-weighted slices of 1.0 mm thickness in the sagittal plane. The imaging parameters were as follows: repetition time = 24 ms; echo time = 5 ms; flip angle = 40°; field of view = 256 mm; and matrix size = 256×256 pixels. The voxel size was 1.0×1.0×1.0 mm. The scanner was calibrated weekly with the same phantom to ensure measurement stability.
T1-weighted MR images were processed using Statistical Parametric Mapping 8 (SPM8, Wellcome Institute of Neurology, University College London, UK, http://www.fil.icon.ucl.ac.uk/spm) running under MATLAB R2012b (The MathWorks Inc., USA). The images were preprocessed using the VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm/), which is an extension of the unified segmentation model consisting of spatial normalization, bias field correction, and tissue segmentation [28]. Registration to the stereotactic space of the Montreal Neurological Institute (MNI) consisted of linear affine transformation and nonlinear deformation using high-dimensional Diffeomorphic Anatomical Registration through Exponential Lie Algebra (DARTEL) normalization [29]. Estimation options were set as follows: extremely light bias regulation; bias cut-off full width at half maximum (FWHM) = 30 mm; affine regulation = International Consortium for Brain Mapping (ICBM) space template of East Asian brains; and the others were defaults. The normalized and segmented images were modulated by applying a nonlinear deformation, which allows comparison of absolute amounts of tissue corrected for individual differences in brain size. The bias-corrected, modulated, and warped tissue maps were then written with an isotopic voxel resolution of 1.5×1.5×1.5 mm and smoothed with an 8-mm FWHM Gaussian kernel [30], [31].
Exploratory whole-brain analysis of regional GM volume
First, we performed whole-brain analyses using SPM8 to explore the effects of genotype and genotype-by-diagnosis interaction for each of YWHAE (rs28365859, rs11655548, and rs9393) and DISC1 (rs821616) SNPs on GM volume in all subjects. These effects were statistically assessed using a full factorial model for a 2×2 ANOVA, with diagnosis and genotype status as independent variables, and age and sex as covariates of no interest in SPM8. In order to avoid type I error, the significance level was set at p<0.0001 (uncorrected for multiple comparison), and the extent threshold of cluster size was set at k>50. We also explored the gene-gene interaction between rs28365859 and rs821616 on brain morphology using a full factorial model for a 2×2 ANOVA, with genotype status of each SNP as independent variables.
Using the Wake Forest University (WFU) PickAtlas [32], we then performed small volume corrections (SVCs) for each brain region including the clusters with a significant genotype effect or interaction. Each region was defined using the Automated Anatomical Labeling (AAL) atlas [33]. For the regions of interest (ROIs) with significant genotype-by-diagnosis interaction, the genotype effect was examined separately in the patients and controls, with age and sex as covariates of no interest. For these SVC analyses, a family-wise error-corrected (FWE) voxel level threshold of p<0.05 was applied to account for multiple comparisons of the results. Voxel coordinates were given as an indication of location in a standardized brain. Voxels were localized in MNI space and transformed into Talairach and Tournoux coordinates [34] using the WFU PickAtlas [35], [36].
Hypothesis-driven ROI analysis for hippocampus
On the basis of a previous postmortem rat experiment [21], we also examined the effect of rs28365859 on bilateral hippocampi defined by the AAL atlas (FWE, p<0.05). For this hypothesis-driven ROI analysis, we examined the effect of genotype in all subjects as well as in each diagnostic group. Age and sex were used as covariates of no interest in these analyses.
Statistical analysis
Demographic and clinical differences between groups were examined by using chi-square test or one-way analysis of variance (ANOVA) with post hoc Scheffé’s test. Genotypes were tested for Hardy–Weinberg equilibrium (HWE) using the chi-square goodness-of-fit test. Since the number of subjects with C allele homozygosity of rs28365859 was quite small (3 schizophrenia patients and 4 control subjects), and on the basis of a previous report on lymphocytes of healthy control subjects [22], the study participants were categorized into C allele carriers (protective allele group) or G allele homozygotes. For other YWHAE and DISC1 SNPs, on the basis of minor allele frequency [22] and previous report [18], the subjects were divided into G allele carriers vs A allele homozygotes (rs11655548 and rs9393) and T allele homozygotes vs A allele carriers (rs821616), respectively. Statistical significance was defined as p<0.05.
Results
Sample characteristics and genotyping results
Groups were matched for age, sex, height, body weight, and total GM volume, but the controls had attained a higher level of education than the schizophrenia patients (Table 1). In Table1, the different typical and atypical antipsychotic dosages were converted into haloperidol equivalent according to the guidelines by Toru [37]. There was no significant difference in clinical and demographic data between YWHAE (rs28365859) C allele carriers and G allele homozygotes in both schizophrenia and control groups. The genotype frequencies of the SNPs investigated in this study were within the distribution expected according to the HWE. As shown in Table 1, patients with schizophrenia and healthy comparisons did not differ significantly in genotype distributions (chi-square = 1.62, p = 0.204) and allele frequencies (chi-square = 1.00, p = 0.317) of rs28365859.
Table 1. Clinical and YWHAE genotypic description of schizophrenia patients and healthy controls.
| Schizophrenia patients | Controls | Group comparisons | |||
| C allele carriers | G homozygotes | C allele carriers | G homozygotes | ||
| (n = 34) | (n = 38) | (n = 32) | (n = 54) | ||
| Male/female | 14/20 | 25/13 | 19/13 | 26/28 | Chi-square = 3.95, p = 0.27 |
| Age (years) | 27.2±5.9 | 27.9±6.2 | 25.5±6.6 | 27.0±6.6 | F (3,154) = 0.85, p = 0.47 |
| Height (cm) | 162.3±8.7 | 166.4±8.1 | 166.9±9.6 | 164.5±7.4 | F (3,154) = 2.22, p = 0.09 |
| Body weight (kg) | 56.3±9.5 | 62.1±11.6 | 57.9±9.9 | 57.1±9.7 | F (3,154) = 2.48, p = 0.06 |
| Education (years) | 13.9±1.7 | 13.6±2.1 | 16.0±2.2 | 15.9±2.3 | F (3,153 ) = 13.79, p<0.01;Con>Sz |
| Parental education (years) | 13.0±1.8 | 12.4±2.5 | 13.2±2.5 | 13.3±2.4 | F (3,153) = 1.22, p = 0.30 |
| Age of onset (years) | 21.7±4.1 | 23.3±5.1 | - | - | F (1,70) = 2.21, p = 0.14 |
| Duration of illness (years) | 5.4±5.8 | 4.4±4.6 | - | - | F (1,70) = 0.64, p = 0.43 |
| Duration of medication (years) | 2.9±3.9 | 3.2±3.7 | - | - | F (1,70) = 0.11, p = 0.75 |
| Drug dose (haloperidolequivalent, mg/day) | 8.2±7.2 | 9.3±8.3 | - | - | F (1,70) = 0.37, p = 0.55 |
| Total SAPS scorea) | 32.3±26.3 | 28.3±26.6 | - | - | F (1,69) = 0.40, p = 0.53 |
| Total SANS scorea) | 53.1±24.1 | 52.2±20.6 | - | - | F (1,69) = 0.03, p = 0.87 |
| Total gray matter volume (mm3) | 631.3±46.6 | 658.0±64.4 | 655.6±52.3 | 654.5±57.2 | F (3,154) = 1.74, p = 0.16 |
Values represent means ± SDs. Con, controls; SANS, Scale for the Assessment of Negative Sympoms; SAPS, Scale for the Assessment of Positive Symptoms; Sz, schizophrenia.
Data missing for one patient.
For the other SNPs, rs11655548 (3 patients and 3 controls), rs9393 (3 patients and 1 control), and rs821616 (3 patients) were not detected for some participants. There was a group difference in the genotype distribution only for rs9393 (chi-square = 5.65, p = 0.018; less G allele carriers in the patients), but such a difference was not found in a larger sample including the current sample (n = 332) or in a large independent Japanese sample (n = 3157) [22].
Exploratory whole-brain analysis of regional GM volume
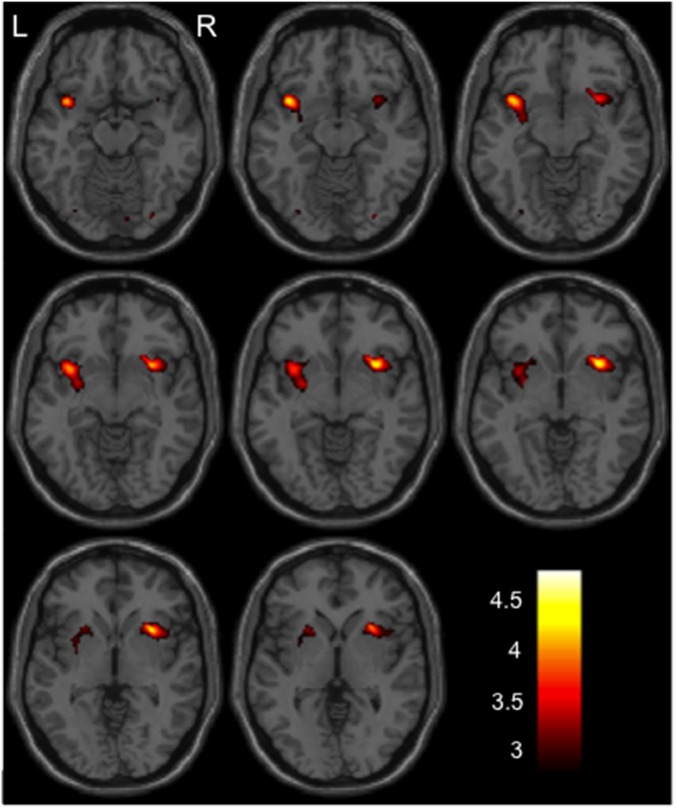
There was no significant genotype effect of YWHAE SNPs or rs821616 on GM volume in all subjects. However, we found significant genotype-by-diagnosis interactions for rs28365859 in the left insula and right putamen GM volume (uncorrected p<0.0001, extent threshold k>50; Table 2 and Fig. 1), which were confirmed by subsequent FWE-corrected SVC analyses (left insula, p = 0.004; right putamen, p = 0.001) (Table 2). Other SNPs (rs11655548, rs9393, and rs821616) had no genotype-by-diagnosis interaction. There was no significant gene-gene interaction on GM volume between rs28365859 and rs821616.
Table 2. Effect of rs28365859 genotype and genotype-by-diagnosis interaction on gray matter volume.
| Brain region | Contrast | Covariates | Talairach coordinate | Cluster size | p | |||
| x | y | z | ||||||
| Interaction on whole brain | ||||||||
| Rt putamen | age, sex | 32 | 13 | −5 | 125 | <0.0001 (uncorrected) | ||
| Lt insula | age, sex | −39 | 10 | −11 | 108 | <0.0001 (uncorrected) | ||
| Interaction on SVC | ||||||||
| Rt putamen | age, sex | 32 | 13 | −5 | 168 | 0.001 (FWE-corrected) | ||
| Lt insula | age, sex | −39 | 10 | −11 | 232 | 0.004 (FWE-corrected) | ||
| Genotype effect on SVCa | ||||||||
| Rt putamen | ConC−>ConC+ | age, sex | 30 | 16 | −1 | 60 | 0.023 (FWE-corrected) | |
| Lt insula | SzC+>SzC− | age, sex | −36 | 8 | −11 | 52 | 0.047 (FWE-corrected) | |
| SzC+>SzC− | age, sex, doi,med | −36 | 8 | −11 | 68 | 0.037 (FWE-corrected) | ||
ConC+, controls with C allele; ConC−, controls without C allele; doi, duration of illness; FWE, family-wise error; Lt, left; med, daily medication dose; Rt, right; SVC, small volume correction; SzC+, schizophrenia patients with C allele; SzC−, schizophrenia without C allele.
There were no suprathreshold clusters for other contrasts.
Figure 1. The YWHAE (rs28365859) genotype-by-diagnosis interaction on gray matter volume.
The regions showing interaction in all subjects are displayed by a hot colormap. The color bar shows t values corresponding to the color in the figure.
On the basis of significant genotype-by-diagnosis interactions of rs28365859, we then separately investigated its genotype effect on GM volume in schizophrenia and control groups. The protective C allele carriers had a significantly larger left insula than G homozygotes only for the schizophrenia patients (FWE-corrected p = 0.047, Fig. 2), while the controls with G allele homozygosity had a significantly larger right putamen than the C allele carriers (FWE-corrected p = 0.023, Fig. 3) (Table 2). The C allele was also related to smaller left insula in controls (FWE-corrected p = 0.144) and larger right putamen in schizophrenia patients (FWE-corrected p = 0.078), although these effects were not statistically significant. The findings reported herein did not change even when we added the illness duration and medication dose as covariates for the SVC analyses for the schizophrenia patients (Table 2).
Figure 2. Impact of the rs28365859 genotype on gray matter volume of left insula in schizophrenia.
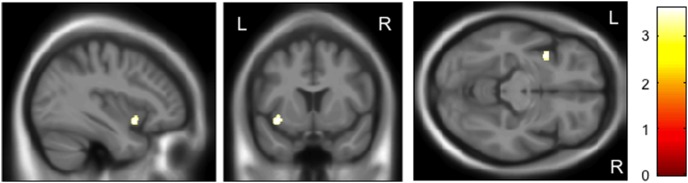
Age, sex, illness duration, and medication dose were used as covariates. The protective C allele carriers had a significantly larger left insula than the G homozygotes. Anatomical localizations are displayed on the normal template MR images in three directions. The color bar shows t values corresponding to the color in the figure.
Figure 3. Impact of the rs28365859 genotype on gray matter volume of the right putamen in healthy controls.
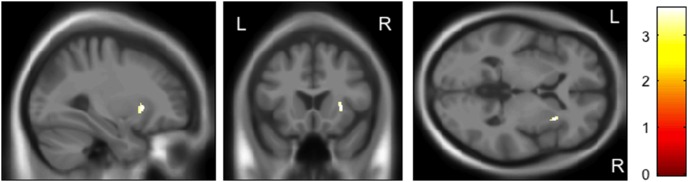
The G allele homozygotes had a significantly larger right putamen than the C allele carriers. Anatomical localizations are displayed on the normal template MR images in three directions. The color bar shows t values corresponding to the color in the figure.
Hypothesis-driven ROI analysis for hippocampus
The protective C allele carriers of rs28365859 had a significantly larger right, but not left, hippocampal volume than the G allele homozygotes (FWE-corrected p = 0.009, Table 3). For the analyses in each diagnostic group, such an effect of YWHAE genotype was significant only in schizophrenia patients (FWE-corrected p = 0.009, Table 3 and Fig. 4). That result in schizophrenia remained the same even when we added illness duration and medication as covariates (Table 3).
Table 3. Effect of rs28365859 genotype on right hippocampal gray matter volume.
| Contrasta | Covariates | Talairach coordinate | Cluster size | FWE p | ||
| x | y | z | ||||
| C+>C− | age, sex | 24 | −35 | 0 | 120 | 0.009 |
| SzC+>SzC− | age, sex | 20 | −33 | 3 | 78 | 0.009 |
| age, sex, doi, med | 20 | −33 | 3 | 120 | 0.002 | |
C+, subjects with C allele; C−, subjects without C allele; doi, duration of illness; FWE, family-wise error; med, daily medication dose; SzC+, schizophrenia patients with C allele; SzC−, schizophrenia patients without C allele.
There were no suprathreshold clusters for other contrasts.
Figure 4. Impact of the rs28365859 genotype on gray matter volume of the right hippocampus in schizophrenia.
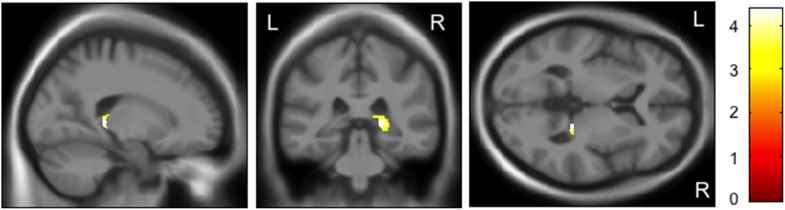
Age, sex, illness duration, and medication dose were used as covariates. The protective C allele carriers had a significantly larger right hippocampus than the G allele homozygotes. Anatomical localizations are displayed on the normal template MR images in three directions. The color bar shows t values corresponding to the color in the figure.
Discussion
This is the first structural MRI study to report the relationship between the functional polymorphism of YWHAE, a gene encoding 14-3-3epsilon, and brain morphology in patients with schizophrenia and healthy controls. While no significant difference was found in clinical and demographic data between the YWHAE (rs28365859) C allele carriers (protective allele group) and G allele homozygotes in both schizophrenia and control groups, the exploratory whole-brain analysis of regional GM volume demonstrated significant genotype-by-diagnosis interaction of rs28365859 on the left insula and right putamen. Subsequent SVC analyses showed that the protective C allele carriers had a significantly larger left insula than G homozygotes only for the schizophrenia patients, while the controls with G allele homozygosity had a significantly larger right putamen than the C allele carriers. Furthermore, the hypothesis-driven ROI analysis revealed that the subjects with the C allele had a larger hippocampal volume, especially for schizophrenia patients. Our report using a Japanese cohort thus suggests that the genotype variation of 14-3-3epsilon, a DISC1-interacting molecule associated with neuronal development [13], [21], may be at least partly related to the abnormalities in brain morphology reported in schizophrenia. Importantly, we found no significant genotype effect of non-risk YWHAE SNPs (rs11655548 and rs9393) on GM volume, supporting the specific role of rs28365859 in the pathophysiology of schizophrenia [22].
Our finding of preserved insula GM volume in schizophrenia patients with protective C allele of rs28365859 is consistent with the literature suggesting a significant role of insula pathology in schizophrenia [38]. GM reduction of the insula, which plays crucial roles in emotional and various cognitive functions as a component of the limbic integration cortex [39], has been repeatedly described in schizophrenia [40], [41]. GM reduction or dysfunction of the insula has also been implicated in the manifestation of psychotic symptoms and cognitive impairments [38]. The exact neurobiological basis for these GM changes of the insula in schizophrenia remains unknown, but the defects in gyrification [42], cytoarchitectural abnormalities [43], [44], and significant volume reduction prior to the illness onset [45], [46] imply early neurodevelopmental abnormalities in this region. A lack of insular GM abnormalities in non-psychotic co-twins within monozygotic twins discordant for schizophrenia [47] suggests that the insular findings in schizophrenia are also attributable to non-genetic factors. In this study, healthy controls with C allele had a non-significantly smaller left insula compared to G homozygotes. The reason for this opposite direction of volume changes related to the same allele between schizophrenia patients and controls is unclear, but our earlier MRI study demonstrated that the DISC1 (rs821616) genotype variation could also differently affect the insula GM volume in schizophrenia patients and healthy comparisons [18]. The current evidence for DISC1 alone as a genetic risk factor of schizophrenia is not strong [20]. Indeed, the present study did not support its effect on brain morphology in schizophrenia. However, considering that DISC1 interacts with a complex formed by related molecules (including 14-3-3epsilon) during processes involved in neuronal development, such as axonal elongation [13], the present results raise the possibility that the genetic variation of DISC1-interacting molecules might have an additive or independent role in alterations of the neural development in schizophrenia, especially regarding the insula pathology [38]. The potential role of genetic variation in DISC1-interacting molecules and its interaction with other genetic/non-genetic factors in the pathophysiology of schizophrenia should be further tested through in vitro and in vivo studies.
We also found significant rs28365859 genotype-by-diagnosis interaction on the right putamen, with the C allele carriers having a smaller putamen volume only for healthy subjects. This finding might have some association with a previous MRI study that demonstrated the relationship between functional DISC1 genotype and striatal volume [48]. Taken together with animal data that the DISC1 gene influences striatal dopamine receptor levels [49], Chakravarty et al. [48] hypothesized that a key risk pathway for schizophrenia might be conferred via DISC1’s effects on the striatum. MRI findings of the putamen in schizophrenia have been highly controversial; smaller [50] or normal [51], [52] volume was reported in first-episode antipsychotic-naïve patients, with both volume expansion [51], [53] and decrease [54] following antipsychotic treatment. We did not find a significant effect of the genetic variation of 14-3-3epsilon, a DISC1-interacting molecule, on the basal ganglia in our sample of chronically medicated schizophrenia patients. However, the possible role of genetic variation of DISC1 and its interacting molecules on brain morphology in schizophrenia should be examined in future, ideally using a larger antipsychotic-naïve sample.
In this study, as hypothesized, we also demonstrated that the subjects with the protective C allele of rs28365859 had a larger hippocampal volume, especially for schizophrenia patients. Hippocampal GM volume is thought to represent an endophenotype associated with the clinical expression of schizophrenia [55]. Brain imaging studies suggest that variants in the DISC1 gene may influence normal neurodevelopment, brain structure, function, and neurochemistry, but the association of the common DISC1 SNPs with hippocampal regions has been inconsistent for both schizophrenia and healthy subjects (reviewed by Duff et al. [19]). However, the expression of DISC1-binding partners such as NUDEL and LIS1, which form a complex with 14-3-3epsilon [13], [21], is reduced in the hippocampus of postmortem schizophrenia brains [56]. More specifically, animal studies using genetically modified 14-3-3epsilon-deficient mice showed developmental defects of hippocampal neurons [21] as well as behavioral changes related to clinical features of schizophrenia (i.e., anxiety-like behavior, working memory deficits) [22]. Schizophrenia is a complex disorder with a variety of pathologies and risk factor genes, and the variation of a single gene could explain only a part of its clinical expression. We found no direct interaction between the YWHAE (rs28365859) and DISC1 (rs821616) SNPs on gray matter volume in schizophrenia in this study. Nevertheless, the present and previous basic studies suggest the possibility that genetically defined impairment of DISC1 and/or 14-3-3epsilon could cause neuronal developmental defects in brain regions including the hippocampus, which result in the increased risk of developing schizophrenia.
There are several confounding factors in the present study. First, in contrast to recent large multinational consortium genome-wide association studies [57], [58], this study examined the effect of the YWHAE genotype only in a relatively small Japanese sample. Our whole-brain analysis found a specific YWHAE genotype effect only on the left insula in schizophrenia, but the current study was potentially underpowered to detect significant genotype effects on other brain regions owing to the small sample size. For example, the relation between the protective C allele of rs28365859 and larger hippocampal volume in all subjects (but more robust in schizophrenia patients) was detectable only by the hypothesis-driven ROI analysis, which is thought to be more sensitive than whole-brain analysis. Furthermore, an animal study by Sekiguchi et al. [59] suggested a relationship between the defect of 14-3-3epsilon and axon elongation abnormality in the prefrontal cortex. As we also found mild diagnosis-by-genotype interaction in frontal regions when we used a significance level of uncorrected p<0.001 in exploratory whole-brain analysis (data not shown), future studies on a larger sample of schizophrenia might detect other YWHAE genotype effects on brain morphology including the frontal regions. Second, we examined schizophrenia patients with an illness duration of approximately 5 years in this study. Illness chronicity [60] and medication with antipsychotics [61], [62] could significantly affect brain morphology. Although there was no difference in these variables between the patients with and without the C allele of rs28365859 (Table 1) and we statistically controlled these factors, the present findings should be replicated using patients at early illness stages. Third, the current study cannot address the disease specificity of our YWHAE findings. There are overlapping GM structural abnormalities in the neurobiology of schizophrenia and bipolar disorder [63] and there are several susceptibility genes (e.g., DISC1) for both of these disorders [19]. Finally, considering that we examined only four selected SNPs in the present study, more comprehensive assessment would be required to clarify the role of genetic variation of DISC1 and its interacting molecules in the pathophysiology of schizophrenia.
In conclusion, we found that the C allele of YWHAE (rs28365859) is related to preserved GM volume of the insula and hippocampus in schizophrenia, major brain regions related to the illness, in a Japanese sample. These findings are likely to provide neurobiological support for previous genetic and expression studies suggesting that this SNP reduces the risk of schizophrenia [22].
Supporting Information
Diagnosis effect on gray matter volume in all subjects analyzed by using the SPM8 full factorial model. Age and sex were used as covariates. Healthy controls had a larger gray matter volume compared with schizophrenia patients predominantly in fronto-temporo-limbic regions (family-wise error-corrected p<0.05). Anatomical localizations are displayed on the normal template MR images in three directions. The color bar shows t values corresponding to the color in the figure.
(TIFF)
Diagnosis effect on gray matter volume in all subjects. Each region was defined using the Automated Anatomical Atlas (AAL) atlas [33].
(DOCX)
Acknowledgments
The authors would like to thank all the participants in this study. We would also like to thank the radiological technologists, especially Mr. Koichi Mori and Mr. Sadanori Ito, who assisted in the MRI data collection at Toyama University Hospital. Thanks are also due to Ms. Hiroko Itoh for her assistance with genomic DNA extraction for all the participants in this study.
Funding Statement
This research was supported in part by Grants-in-Aid for Scientific Research (C) (No. 22591275, 24591699) and Grants-in-Aid for Scientific Research (B) (No. 24390281) from the Japanese Society for the Promotion of Science, Health and Labour Sciences Research Grants (Comprehensive Research on Disability, Health and Welfare, H23-Seishin-Ippan-002 and H23-Seishin-Ippan-009), a Research Grant from the JSPS Asian Core Program, and a Grant-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) from the Ministry of Education, Culture, Sports, Science & Technology of Japan. It was also supported by Grant-in-Aid for “Integrated research on neuropsychiatric disorders” carried out under the Strategic Research Program for Brain Sciences by the Ministry of Education, Culture, Sports, Science and Technology of Japan and Grant-in-Aid for Scientific Research on Innovative Areas, “Glial assembly: a new regulatory machinery of brain function and disorders”. The funding agencies had no further role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
References
- 1. Harrison PJ, Weinberger DR (2005) Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 10: 40–68. [DOI] [PubMed] [Google Scholar]
- 2. Sawa A, Snyder SH (2002) Schizophrenia: diverse approaches to a complex disease. Science 296: 692–695. [DOI] [PubMed] [Google Scholar]
- 3. Shenton ME, Dickey CC, Frumin M, McCarley RW (2001) A review of MRI findings in schizophrenia. Schizophr Res 49: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suzuki M, Nohara S, Hagino H, Kurokawa K, Yotsutsuji T, et al. (2002) Regional changes in brain gray and white matter in patients with schizophrenia demonstrated with voxel-based analysis of MRI. Schizophr Res 55: 41–54. [DOI] [PubMed] [Google Scholar]
- 5. Gottesman II, Gould TD (2003) The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160: 636–645. [DOI] [PubMed] [Google Scholar]
- 6. Keshavan MS, Prasad KM, Pearlson G (2007) Are brain structural abnormalities useful as endophenotypes in schizophrenia? Int Rev Psychiatry 19: 397–406. [DOI] [PubMed] [Google Scholar]
- 7. Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, et al. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9: 1415–1423. [DOI] [PubMed] [Google Scholar]
- 8. St Clair D, Blackwood D, Muir W, Carothers A, Walker M, et al. (1990) Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336: 13–16. [DOI] [PubMed] [Google Scholar]
- 9. James R, Adams RR, Christie S, Buchanan SR, Porteous DJ, et al. (2004) Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol Cell Neurosci 26: 112–122. [DOI] [PubMed] [Google Scholar]
- 10. Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, et al. (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 7: 1167–1178. [DOI] [PubMed] [Google Scholar]
- 11. Kirkpatrick B, Xu L, Cascella N, Ozeki Y, Sawa A, et al. (2006) DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J Comp Neurol 497: 436–450. [DOI] [PubMed] [Google Scholar]
- 12. Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, et al. (2003) Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A 100: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taya S, Shinoda T, Tsuboi D, Asaki J, Nagai K, et al. (2007) DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J Neurosci 27: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishizuka K, Paek M, Kamiya A, Sawa A (2006) A review of Disrupted-in-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol Psychiatry 59: 1189–1197. [DOI] [PubMed] [Google Scholar]
- 15. Roberts RC (2007) Schizophrenia in translation: disrupted in schizophrenia (DISC1): integrating clinical and basic findings. Schizophr Bull 33: 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, et al. (2005) Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A 102: 8627–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, et al. (2006) Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet 15: 3024–3033. [DOI] [PubMed] [Google Scholar]
- 18. Takahashi T, Suzuki M, Tsunoda M, Maeno N, Kawasaki Y, et al. (2009a) The Disrupted-in-Schizophrenia-1 Ser704Cys polymorphism and brain morphology in schizophrenia. Psychiatry Res 172: 128–135. [DOI] [PubMed] [Google Scholar]
- 19. Duff BJ, Macritchie KA, Moorhead TW, Lawrie SM, Blackwood DH (2013) Human brain imaging studies of DISC1 in schizophrenia, bipolar disorder and depression: a systematic review. Schizophr Res 147: 1–13. [DOI] [PubMed] [Google Scholar]
- 20. Sullivan PF (2013) Questions about DISC1 as a genetic risk factor for schizophrenia. Mol Psychiatry 18: 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, et al. (2003) 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet 34: 274–285. [DOI] [PubMed] [Google Scholar]
- 22. Ikeda M, Hikita T, Taya S, Uraguchi-Asaki J, Toyo-oka K, et al. (2008) Identification of YWHAE, a gene encoding 14-3-3epsilon, as a possible susceptibility gene for schizophrenia. Hum Mol Genet 17: 3212–2322. [DOI] [PubMed] [Google Scholar]
- 23. Goldman-Rakic PS (1994) Working memory dysfunction in schizophrenia. J Neuropsychiatry Clin Neurosci 6: 348–357. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization (1993) The ICD-10 classification of mental and behavioural disorders: diagnostic criteria for research. World Health Organization, Geneva.
- 25. Andreasen NC, Flaum M, Arndt S (1992) The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Arch Gen Psychiatry 49: 615–623. [DOI] [PubMed] [Google Scholar]
- 26.Andreasen NC (1984a) Scale for the assessment of negative symptoms (SANS). University of Iowa, Iowa City.
- 27.Andreasen NC (1984b) Scale for the assessment of positive symptoms (SAPS). University of Iowa, Iowa City.
- 28. Ashburner J, Friston KJ (2005) Unified segmentation. NeuroImage 26: 839–851. [DOI] [PubMed] [Google Scholar]
- 29. Ashburner J (2007) A fast diffeomorphic image registration algorithm. NeuroImage 38: 95–113. [DOI] [PubMed] [Google Scholar]
- 30. Jones DK, Symms MR, Cercignani M, Howard RJ (2005) The effect of filter size on VBM analyses of DT-MRI data. Neuroimage 26: 546–554. [DOI] [PubMed] [Google Scholar]
- 31. Salmond CH, Ashburner J, Vargha-Khadem F, Connelly A, Gadian DG, et al. (2002) Distributional assumptions in voxel-based morphometry. Neuroimage 17: 1027–1030. [PubMed] [Google Scholar]
- 32. Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH (2003) An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage 19: 1233–1239. [DOI] [PubMed] [Google Scholar]
- 33. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, et al. (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15: 273–289. [DOI] [PubMed] [Google Scholar]
- 34. Talairach J, Tournoux P (1988) Co-planar stereotaxic atlas of the human brain. 3-Dimensional proportional system: an approach to cerebral imaging. Stuttgart Thieme. [Google Scholar]
- 35. Lancaster JL, Summerln JL, Rainey L, Freitas CS, Fox PT (1997) The Talairach Daemon, a database server for Talairach Atlas Labels. NeuroImage 5: S633. [Google Scholar]
- 36. Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, et al. (2000) Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp 10: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toru M (2008) Psychotropic Manual, Third Edition. Igaku-shoin, Tokyo. (in Japanese).
- 38. Wylie KP, Tregellas JR (2010) The role of the insula in schizophrenia. Schizophr Res 123: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Augustine JR (1996) Circuitry and functional aspects of the insular lobe in primates including humans. Brain Res Brain Res Rev 22: 229–244. [DOI] [PubMed] [Google Scholar]
- 40. Glahn DC, Laird AR, Ellison-Wright I, Thelen SM, Robinson JL, et al. (2008) Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol Psychiatry 64: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shepherd AM, Matheson SL, Laurens KR, Carr VJ, Green MJ (2012) Systematic meta-analysis of insula volume in schizophrenia. Biol Psychiatry 72: 775–784. [DOI] [PubMed] [Google Scholar]
- 42. Palaniyappan L, Liddle PF (2012) Aberrant cortical gyrification in schizophrenia: a surface-based morphometry study. J Psychiatry Neurosci 37: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jakob H, Beckmann H (1986) Prenatal developmental disturbances in the limbic allocortex in schizophrenics. J Neural Transm 65: 303–326. [DOI] [PubMed] [Google Scholar]
- 44. Pennington K, Dicker P, Hudson L, Cotter DR (2008) Evidence for reduced neuronal somal size within the insular cortex in schizophrenia, but not in affective disorders. Schizophr Res 106: 164–171. [DOI] [PubMed] [Google Scholar]
- 45. Borgwardt SJ, Riecher-Rössler A, Dazzan P, Chitnis X, Aston J, et al. (2007) Regional gray matter volume abnormalities in the at risk mental state. Biol Psychiatry 61: 1148–1156. [DOI] [PubMed] [Google Scholar]
- 46. Takahashi T, Wood SJ, Yung AR, Phillips LJ, Soulsby B, et al. (2009b) Insular cortex gray matter changes in individuals at ultra-high-risk of developing psychosis. Schizophr Res 111: 94–102. [DOI] [PubMed] [Google Scholar]
- 47. Borgwardt SJ, Picchioni MM, Ettinger U, Toulopoulou T, Murray R, et al. (2010) Regional gray matter volume in monozygotic twins concordant and discordant for schizophrenia. Biol Psychiatry 67: 956–964. [DOI] [PubMed] [Google Scholar]
- 48. Chakravarty MM, Felsky D, Tampakeras M, Lerch JP, Mulsant BH, et al. (2012) DISC1 and Striatal Volume: A Potential Risk Phenotype For mental Illness. Front Psychiatry 3: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lipina TV, Niwa M, Jaaro-Peled H, Fletcher PJ, Seeman P, et al. (2010) Enhanced dopamine function in DISC1-L100P mutant mice: implications for schizophrenia. Genes Brain Behav 9: 777–789. [DOI] [PubMed] [Google Scholar]
- 50. Ballmaier M, Schlagenhauf F, Toga AW, Gallinat J, Koslowski M, et al. (2008) Regional patterns and clinical correlates of basal ganglia morphology in non-medicated schizophrenia. Schizophr Res 106: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Glenthoj A, Glenthoj BY, Mackeprang T, Pagsberg AK, Hemmingsen RP, et al. (2007) Basal ganglia volumes in drug-naive first-episode schizophrenia patients before and after short-term treatment with either a typical or an atypical antipsychotic drug. Psychiatry Res 154: 199–208. [DOI] [PubMed] [Google Scholar]
- 52. Gunduz H, Wu H, Ashtari M, Bogerts B, Crandall D, et al. (2002) Basal ganglia volumes in first-episode schizophrenia and healthy comparison subjects. Biol Psychiatry 51: 801–808. [DOI] [PubMed] [Google Scholar]
- 53. Li M, Chen Z, Deng W, He Z, Wang Q, et al. (2011) Volume increases in putamen associated with positive symptom reduction in previously drug-naive schizophrenia after 6 weeks antipsychotic treatment. Psychol Med 42: 1475–1483. [DOI] [PubMed] [Google Scholar]
- 54. Ebdrup BH, Skimminge A, Rasmussen H, Aggernaes B, Oranje B, et al. (2011) Progressive striatal and hippocampal volume loss in initially antipsychotic-naive, first-episode schizophrenia patients treated with quetiapine: relationship to dose and symptoms. Int J Neuropsychopharmacol 14: 69–82. [DOI] [PubMed] [Google Scholar]
- 55. Borgwardt S, Smieskova R, Fusar-Poli P (2012) Gray matter pathology of hippocampus - a specific endophenotype for schizophrenia? Psychiatry Res 202: 273–274. [DOI] [PubMed] [Google Scholar]
- 56. Lipska BK, Peters T, Hyde TM, Halim N, Horowitz C, et al. (2006) Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet 15: 1245–1258. [DOI] [PubMed] [Google Scholar]
- 57. Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, et al. (2012) Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet 44: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, et al. (2012) Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sekiguchi H, Iritani S, Habuchi C, Torii Y, Kuroda K, et al. (2011) Impairment of the tyrosine hydroxylase neuronal network in the orbitofrontal cortex of a genetically modified mouse model of schizophrenia. Brain Res 1392: 47–53. [DOI] [PubMed] [Google Scholar]
- 60. Haijma SV, Van Haren N, Cahn W, Koolschijn PC, Hulshoff Pol HE, et al. (2013) Brain volumes in schizophrenia: a meta-analysis in over 18 000 subjects. Schizophr Bull 39: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Andreasen NC, Liu D, Ziebell S, Vora A, Ho BC (2013) Relapse duration, treatment intensity, and brain tissue loss in schizophrenia: a prospective longitudinal MRI study. Am J Psychiatry 170: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lieberman JA, Tollefson GD, Charles C, Zipursky R, Sharma T, et al. (2005) Antipsychotic drug effects on brain morphology in first-episode psychosis. Arch Gen Psychiatry 62: 361–370. [DOI] [PubMed] [Google Scholar]
- 63. Anderson D, Ardekani BA, Burdick KE, Robinson DG, John M, et al. (2013) Overlapping and distinct gray and white matter abnormalities in schizophrenia and bipolar I disorder. Bipolar Disord 15: 680–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagnosis effect on gray matter volume in all subjects analyzed by using the SPM8 full factorial model. Age and sex were used as covariates. Healthy controls had a larger gray matter volume compared with schizophrenia patients predominantly in fronto-temporo-limbic regions (family-wise error-corrected p<0.05). Anatomical localizations are displayed on the normal template MR images in three directions. The color bar shows t values corresponding to the color in the figure.
(TIFF)
Diagnosis effect on gray matter volume in all subjects. Each region was defined using the Automated Anatomical Atlas (AAL) atlas [33].
(DOCX)