Abstract
Bacteriophytochromes (BphPs) constitute a class of photosensory proteins that toggle between Pr and Pfr functional states through absorption of red and far-red light. The photosensory core of BphPs is composed of PAS, GAF, and PHY domains. Here, we apply FTIR spectroscopy to investigate changes in the secondary structure of Rhodopseudomonas palustris BphP2 (RpBphP2) upon Pr to Pfr photoconversion. Our results indicate conversion from a β-sheet to an α-helical element in the so-called tongue region of the PHY domain, consistent with recent X-ray structures of Deinococcus radiodurans DrBphP in dark and light states (Takala H.; et al. Nature 2014, 509, 245−248). A conserved Asp in the GAF domain that noncovalently connects with the PHY domain and a conserved Pro in the tongue region of the PHY domain are essential for the β-sheet-to-α-helix conversion.
Phytochromes are red-light-sensing proteins found in plants, bacteria, cyanobacteria, and fungi that act as photochromic switches activated by distinct wavelengths in the red or far-red regions.2−4 In the dark, most phytochromes adopt a red-absorbing state known as Pr and upon light absorption convert to a far-red-absorbing state known as Pfr. The light activation mechanism involves an isomerization process about the C15=C16 double bond of the linear tetrapyrrole, changing its configuration from 15Z to 15E.5−7 Their light-sensing module is composed of PAS, GAF, and PHY domains3 and covalently binds a linear tetrapyrrole, phycochromobilin (PΦB) in plant phytochromes, phycocyanobilin (PCB) in cyanobacterial phytochromes, and biliverdin (BV) in bacteriophytochromes (BphPs). In BphPs, the N-terminal light-sensing module is covalently linked to an effector module, usually a histidine kinase. BphPs exist as dimers in which interfaces between the monomers are formed both by the N-terminal photosensory core module and the C-terminal kinase domain.8
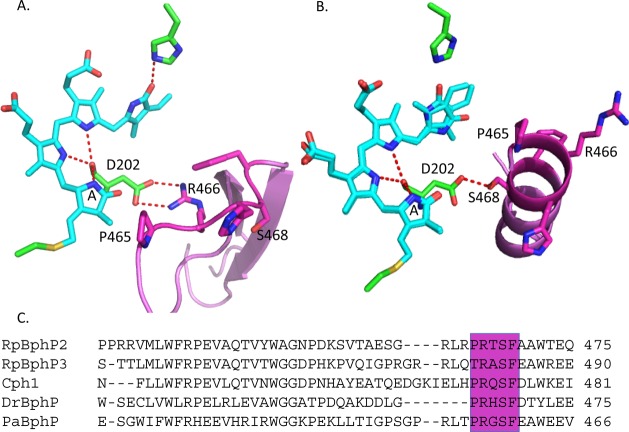
The determination of crystal structures of various BphPs and the cyanobacterial phytochrome Cph1 has explored the light-activated function of phytochromes.5,6,9−12 BphPs bind BV through a covalent thioether linkage to the PAS domain. BV is largely embedded in the GAF domain, which provides most of the hydrogen-bonding, steric, and hydrophobic interactions that secure the chromophore in position.5,9,11,13 The pyrrole rings form an intricate hydrogen-bond network with a conserved Asp of the PASDIP motif in the GAF domain (Figure 1). In addition, a conserved His in the GAF domain and a water molecule were shown to interact with the pyrrole nitrogens in the previously published structures9,10 (the His was omitted in Figure 1 for clarity, while the pyrrole water was not resolved in the DrBphP structures). The PHY domain forms an extension to the chromophore-binding domain of phytochromes that works in tandem with the GAF domain to tune spectral properties and facilitate physiologically functional photochemistry. Crystal structures of the complete photosensory core module comprising the PAS-GAF-PHY domains are available for Synechocystis sp. Cph1 in the Pr state14 and Pseudomonas aeruginosa PaBphP in the Pfr state.5 The latter is a so-called “bathy” phytochrome that assembles as Pr but rapidly thermally converts to its stable Pfr15 form. Deletion of the PHY domain affects the photochemistry and impairs Pfr formation11,16 in all referenced BphPs. The PHY domain engages the chromophore pocket through a salt bridge between this conserved Asp and a conserved Arg and seals off the chromophore from contact with the solvent through a conserved secondary structure element known as the “tongue” (Figure 1).
Figure 1.
DrBphP X-ray structures of BV (cyan)–GAF (green) and tongue in the PHY domain (pink) in Pr (A) and Pfr (B)1 (PDB ID codes 4O0P and 4O01, respectively); partial sequence alignment of previously characterized BphPs and Synechocystis sp. Cph1, highlighting the conserved PRXSF motif of the PHY domain (C).
Recently, X-ray structures were solved of both the Pr and Pfr states of a single species, the DrBphP PAS-GAF-PHY module from Deinococcus radiodurans. The structure of the Pfr state was determined by growing crystals in the light, allowing crystallization to occur from the photoconverted protein.1 One of the striking observations was substantial refolding of the conserved tongue between the Pr and Pfr states. Specifically, in Pr, the tongue region in the PHY domain assumed a loop-two stranded β-sheet conformation, whereas in Pfr, it assumed a loop-α-helix conformation. Questions arise about the significance of this switch in the secondary structure. Does it occur in solution, how is it related to Pfr formation, and which residues are key for the switch to take place? Here, we address these questions through FTIR spectroscopy on a DrBphP homologue, BphP from Rhodopseudomonas palustris (RpBphP2).
Sample preparation, site-directed mutagenesis, and FTIR spectroscopy on BphPs were carried out as described before.11,17 Figure 2 shows the light-minus-dark FTIR spectra of the RpBphP2 PAS-GAF-PHY photosensory core domain (black), the PAS-GAF chromophore-binding domain (red), the PAS-GAF-PHY D202A mutant (blue), and the PAS-GAF-PHY P465T mutant (green). D202 in RpBphP2 represents a well-characterized, conserved Asp from the PASDIP motif of the GAF domain, while P465 represents a conserved Pro from the PRXSF motif of the PHY domain. The spectra were scaled on the highest-wavenumber C1=O bleach near 1738 cm–1. For PAS-GAF-PHY, the spectrum represents the Pfr–Pr spectrum. The FTIR difference spectrum shows a multitude of positive and negative bands, some of which were assigned before in this protein, RpBphP2.17,18 BV-specific bands were identified before on the basis of ultrafast IR experiments on RpBphP2, where optical excitation results in BV-specific signals on femtosecond and picosecond time scales before any signals from the apoprotein arise.17,19,20 Other BV-specific bands were observed and assigned before in Cph1 on the basis of isotope labeling of the chromophore and apoprotein21 and will be discussed below. Here, we focus attention on the highest-amplitude features, the negative band at 1630 cm–1 and positive band at 1653 cm–1. Similar bands were also observed in Cph1 and assigned to amide I.21 Given the amide I wavenumber ranges where distinct secondary structure elements have their main absorption,22 this signal corresponds to a loss of a β-sheet (negative signal at 1630 cm–1) and a gain of an α-helix (positive signal at 1653 cm–1) upon transformation of Pr into Pfr.
Figure 2.
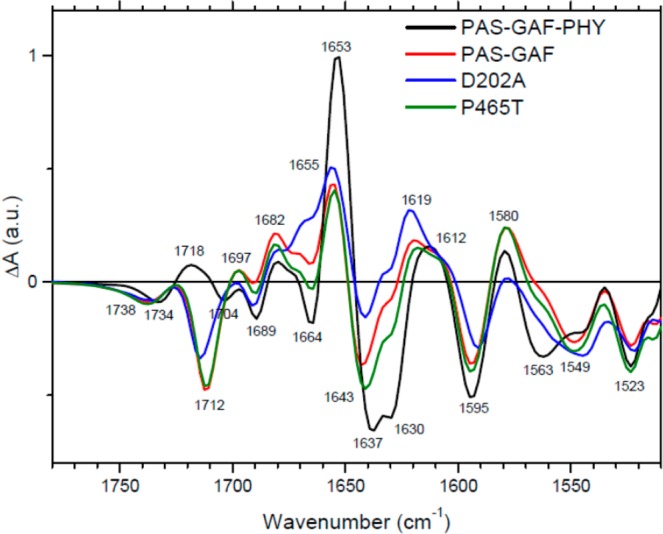
Light-minus-dark FTIR spectra of RpBphP2 PAS-GAF-PHY (black), PAS-GAF (red), PAS-GAF-PHY D202A mutant (blue), and PAS-GAF-PHY P465T mutant (green).
The interpretation of the remaining Pfr–Pr FTIR signals of RpBphP2 PAS-GAF-PHY is made on the basis of earlier results. The 1734 cm–1 band results from BV ring A C1=O, while that at 1704 cm–1 is due to ring D C19=O.17,23,24 The 1689 (−)/1682 (+) cm–1 band is due to amide I (loop/turn) or a propionate COOH, and the 1664 cm–1 band corresponds to loop amide I, as previously assigned in Cph1.21 The 1612 (+) cm–1 band was previously tentatively assigned to amide I (β-sheet structure) in Cph1. The 1595 cm–1 band is due to the BV C–D methine bridge.17,21,25−27 The 1563 cm–1 band corresponds to amide II, as previously assigned for Cph1.21 The 1540 (−) cm–1 band was shown to belong to the BV chromophore17,19,20 but was not interpreted.
To further explore the origin of the prominent 1630 cm–1 (−)/1653 (+) cm–1 signal, we performed FTIR spectroscopy on a shorter construct, RpBphP2 PAS-GAF (also referred to as the chromophore binding domain (CBD)), and on the RpBphP2 PAS-GAF-PHY D202A mutant. Both proteins are deficient in Pfr formation. Upon photoconversion, the D202A mutant becomes arrested in a Meta-R-like state in which the absorption characteristic of the Pr state is bleached and only a small induced absorption is evident, less red-shifted than that in the Pfr state of wild-type11,13 (Figure 3). The PAS-GAF protein converts to a red-shifted state that absorbs at 741 nm11 and fails to form the fully red-shifted Pfr state of wild-type PAS-GAF-PHY, which absorbs at 753 nm (Figure 3).11 The FTIR difference spectra of the PAS-GAF (Figure 2, red line) and the D202A mutant (Figure 2, blue line) proteins are very similar. Strikingly, they both mostly lack the negative band at 1630 cm–1 and the positive band at 1653 cm–1 characteristic of wild-type PAS-GAF-PHY. Evidently, the β-sheet-to-α-helix switch does not occur in these proteins. Given that Asp-202 links the BV chromophore to the PHY domain14 and has proved central in the protonation–deprotonation cycle of BV,13,28 this experimental result strongly suggests that the β-sheet-to-α-helix switch observed in wild-type PAS-GAF-PHY takes place in the PHY domain.
Figure 3.
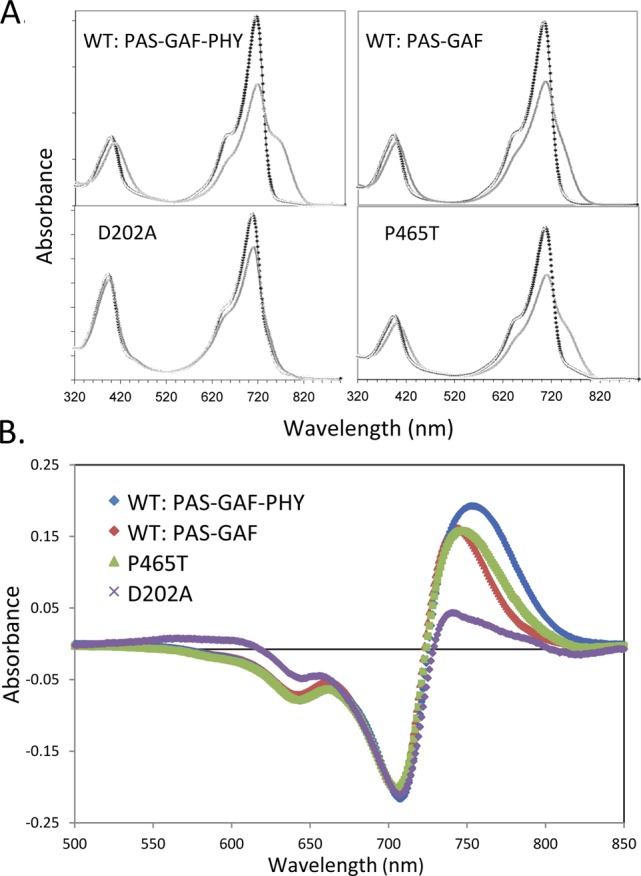
(A) Absorbance spectra for RpBphP2 PAS-GAF-PHY, PAS-GAF, PAS-GAF-PHY D202A, and PAS-GAF-PHY P465T mutants in the Pr state (black line) and photoconverted state (gray line); (B) light-minus-dark difference absorption spectra of RpBphP2 wild-type PAS-GAF-PHY (blue line), PAS-GAF (red line), D202A (magenta), and P465T (green line).
The other bands of RpBphP2 PAS-GAF and the RpBphP2 D202A mutant are similar to those observed in wild-type PAS-GAF-PHY. The 1738 (−) and 1712 (−) cm–1 bands are assigned to BV C1=O and C19=O, respectively. Femtosecond IR experiments showed that in the PAS-GAF domain, the C19=O band is indeed upshifted with respect to PAS-GAF-PHY.17 The remaining bands are conserved with respect to PAS-GAF-PHY. The 1643 (−)/1656 (+) cm–1 feature either corresponds to amide I, where it would correspond to loosening of a helical element in the PAS or GAF domain upon photoconversion, or to C=C stretches of the BV chromophore.17,21 In PAS-GAF-PHY, this feature is present as well and appears additive to the prominent 1630 (−)/1653 (+) cm–1 feature.
Further evidence on the nature of the prominent 1630 (−)/1653 (+) cm–1 signal in wild-type PAS-GAF-PHY comes from the RpBphP2 P465T mutant. Pro-465 is a highly conserved residue located at a key position in the tongue region (Figure 1).5,14 The P465T mutant forms a red-shifted state at 746 nm, similar to the PAS-GAF protein, and is hence deficient in Pfr formation (Figure 3). Its FTIR difference spectrum of the P465T mutant (Figure 2, green line) is almost identical to that of the PAS-GAF protein and lacks the 1630 (−)/1653 cm–1 (+) β-sheet-to-α-helix switch feature.
We conclude that the β-sheet-to-α helix switch observed with FTIR spectroscopy is only present in wild-type RpBphP2 composed of PAS-GAF-PHY domains. It is absent in the protein lacking the PHY domain, the D202A mutant, and the P465T mutant. These observations strongly suggest that the β-sheet-to-α-helix switch as observed in FTIR difference spectroscopy corresponds to the loop-sheet to loop-helix switch in the tongue region of DrBphP recently observed with X-ray crystallography1 and solidifies the argument that this conformational switch is an integral part of phytochrome signaling in vitro and in vivo. Furthermore, our results indicate that refolding of the tongue is required to stabilize the BV chromophore in the fully red-shifted Pfr conformation.
The conserved Asp-202 is instrumental for the sheet–helix switch. It is likely that the β-sheet conformation is stabilized through the salt-bridge interaction with the conserved Arg-466, which is located in the tongue region. This salt bridge is broken in the Pfr state, and Arg-466 is flipped out to the solvent (Figure 1 B).1 Additionally, the conserved Pro-465, part of the tongue and located in the loop adjacent to the double-stranded β-sheet in the Pr state and at the beginning of the newly formed α-helix in the Pfr state, is essential for the sheet-to-helix switch. The proline residue is unique in that its side chain is covalently bonded to the nitrogen atom of the peptide backbone. Consequently, the backbone of Pro cannot form a hydrogen bond, and its N–Cα rotation is rigid. Thus, prolines are frequently found in the first N-terminus turn of an α-helix where the loss of the H-bond to the immino nitrogen does not cause significant effects.29 Proline and glycine are known to destabilize α-helices because they disrupt the regularity of the α-helical backbone conformation. This may be taking place in the Pr conformation of the protein.
Finally, in the unusual bacteriophytochrome RpBphP3, which converts from Pr to a near-red-absorbing state Pnr,18 the amino acid equivalent to P465 in RpBphP2 is actually a threonine (Thr480) (Figure 1 C). To our knowledge, this is the only BphP that lacks a conserved Pro in the PRXSF motif of the PHY domain. In light of the present results, our observations suggest that the signaling mechanism in RpBphP3 might be distinct from canonical BphPs.
Acknowledgments
J.T.M.K. was supported by the Chemical Sciences Council of the NWO (NWO-CW) through a VICI grant. J.T.M.K. and K.C.T. were supported by the Earth and Life Sciences Council of NWO (NWO-ALW) through a VIDI grant. M.T.A.A. was supported by NWO-ALW through the Molecule to Cell programme. K.M. and E.A.S. were supported by NIH Grant GM036452 to K.M.
Author Present Address
⊥ E.A.S.: Department of Biology, Northeastern Illinois University, Chicago, IL.
Author Present Address
# K.C.T.: Mechanobiology Institute, National University of Singapore, Singapore 117411.
Author Present Address
∥ M.B.: Faculty of Applied Sciences, Delft University of Technology, Delft, The Netherlands.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Takala H.; Bjorling A.; Berntsson O.; Lehtivuori H.; Niebling S.; Hoernke M.; Kosheleva I.; Henning R.; Menzel A.; Ihalainen J. A.; Westenhoff S. Signal Amplification and Transduction in Phytochrome Photosensors. Nature 2014, 509, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge M. E.; Forest K. T. Bacterial Phytochromes: More than Meets the Light. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 67–88. [DOI] [PubMed] [Google Scholar]
- Rockwell N. C.; Su Y. S.; Lagarias J. C. Phytochrome Structure and Signaling Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeglich A.; Yang X.; Ayers R. A.; Moffat K. Structure and Function of Plant Photoreceptors. Annu. Rev. Plant Biol. 2010, 61, 21–47. [DOI] [PubMed] [Google Scholar]
- Yang X.; Kuk J.; Moffat K. Crystal Structure of Pseudomonas aeruginosa Bacteriophytochrome: Photoconversion and Signal Transduction. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 14715–14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer T.; Lang C.; Hughes J.; Essen L. O.; Gartner W.; Matysik J. Light-Induced Chromophore Activity and Signal Transduction in Phytochromes Observed by 13C and 15N Magic-angle Spinning NMR. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroginski M. A.; Murgida D. H.; Hildebrandt P. The Chromophore Structural Changes during the Photocycle of Phytochrome: A Combined Resonance Raman and Quantum Chemical Approach. Acc. Chem. Res. 2007, 40, 258–266. [DOI] [PubMed] [Google Scholar]
- Li H.; Zhang J. R.; Vierstra R. D.; Li H. L. Quaternary Organization of a Phytochrome Dimer as Revealed by Cryoelectron Microscopy. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 10872–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. R.; Brunzelle J. S.; Forest K. T.; Vierstra R. D. A Light-Sensing Knot Revealed by the Structure of the Chromophore-Binding Domain of Phytochrome. Nature 2005, 438, 325–331. [DOI] [PubMed] [Google Scholar]
- Wagner J. R.; Zhang J.; Brunzelle J. S.; Vierstra R. D.; Forest K. T. High Resolution Structure of Deinococcus Bacteriophytochrome Yields New Insights into Phytochrome Architecture and Evolution. J. Biol. Chem. 2007, 282, 12298–12309. [DOI] [PubMed] [Google Scholar]
- Yang X.; Stojkovic E. A.; Kuk J.; Moffat K. Crystal Structure of the Chromophore Binding Domain of an Unusual Bacteriophytochrome, RpBphP3, Reveals Residues that Modulate Photoconversion. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12571–12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Ren Z.; Kuk J.; Moffat K. Temperature-Scan Cryocrystallography Reveals Reaction Intermediates in Bacteriophytochrome. Nature 2011, 479, 428–U190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. R.; Zhang J.; von Stetten D.; Gunther M.; Murgida D. H.; Mroginski M. A.; Walker J. M.; Forest K. T.; Hildebrandt P.; Vierstra R. D. Mutational Analysis of Deinococcus radiodurans Bacteriophytochrome Reveals Key Amino Acids Necessary for the Photochromicity and Proton Exchange Cycle of Phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen L. O.; Mailliet J.; Hughes J. The Structure of a Complete Phytochrome Sensory Module in the Pr Ground State. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 14709–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasler R.; Moises T.; Frankenberg-Dinkel N. Biochemical and Spectroscopic Characterization of the Bacterial Phytochrome of Pseudomonas aeruginosa. FEBS J. 2005, 272, 1927–1936. [DOI] [PubMed] [Google Scholar]
- Oka Y.; Matsushita T.; Mochizuki N.; Suzuki T.; Tokutomi S.; Nagatani A. Functional Analysis of a 450-Amino Acid N-terminal Fragment of Phytochrome B in Arabidopsis. Plant Cell 2004, 16, 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K. C.; Stojkovic E. A.; Rupenyan A. B.; van Stokkum I. H. M.; Salumbides M.; Groot M.-L.; Moffat K.; Kennis J. T. M. Primary Reactions of Bacteriophytochrome Observed with Ultrafast Mid-Infrared Spectroscopy. J. Phys. Chem. A 2011, 115, 3778–3786. [DOI] [PubMed] [Google Scholar]
- Giraud E.; Zappa S.; Vuillet L.; Adriano J. M.; Hannibal L.; Fardoux J.; Berthomieu C.; Bouyer P.; Pignol D.; Vermeglio A. A New Type of Bacteriophytochrome Acts in Tandem with a Classical Bacteriophytochrome to Control the Antennae Synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 2005, 280, 32389–32397. [DOI] [PubMed] [Google Scholar]
- van Thor J. J.; Ronayne K. L.; Towrie M. Formation of the Early Photoproduct Lumi-R of Cyanobacterial Phytochrome Cph1 Observed by Ultrafast Mid-Infrared Spectroscopy. J. Am. Chem. Soc. 2007, 129, 126–132. [DOI] [PubMed] [Google Scholar]
- Schumann C.; Gross R.; Michael N.; Lamparter T.; Diller R. Sub-Picosecond Mid-Infrared Spectroscopy of Phytochrome Agp1 from Agrobacterium tumefaciens. ChemPhysChem 2007, 8, 1657–1663. [DOI] [PubMed] [Google Scholar]
- van Thor J. J.; Fisher N.; Rich P. R. Assignments of the Pfr–Pr FTIR Difference Spectrum of Cyanobacterial Phytochrome Cph1 Using 15N and 13C Isotopically Labeled Phycocyanobilin Chromophore. J. Phys. Chem. B 2005, 109, 20597–20604. [DOI] [PubMed] [Google Scholar]
- Krimm S.; Bandekar J. Vibrational Spectroscopy and Conformation of Peptides, Polypeptides and Proteins. Adv. Protein Chem. 1986, 38, 181–364. [DOI] [PubMed] [Google Scholar]
- Smit K.; Matysik J.; Hildebrandt P.; Mark F. Vibrational Analysis of Biliverdin Dimethyl Ester. J. Phys. Chem. 1993, 97, 11887–11900. [Google Scholar]
- Foerstendorf H.; Benda C.; Gartner W.; Storf M.; Scheer H.; Siebert F. FTIR Studies of Phytochrome Photoreactions Reveal the C=O Bands of the Chromophore: Consequences for Its Protonation States, Conformation, and Protein Interaction. Biochemistry 2001, 40, 14952–14959. [DOI] [PubMed] [Google Scholar]
- Margulies L.; Toporowicz M. Resonance Raman Study of Model Compounds of the Phytochrome. 2. Biliverdin Dimethyl Ester. J. Am. Chem. Soc. 1984, 106, 7331–7336. [Google Scholar]
- Andel F.; Lagarias J. C.; Mathies R. A. Resonance Raman Analysis of Chromophore Structure in the Lumi-R Photoproduct of Phytochrome. Biochemistry 1996, 35, 15997–16008. [DOI] [PubMed] [Google Scholar]
- Wagner J. R.; Zhang J.; von Stetten D.; Guenther M.; Murgida D. H.; Mroginski M. A.; Walker J. M.; Forest K. T.; Hildebrandt P.; Vierstra R. D. Mutational Analysis of Deinococcus radiodurans Bacteriophytochrome Reveals Key Amino Acids Necessary for the Photochromicity and Proton Exchange Cycle of Phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stetten D.; Seibeck S.; Michael N.; Scheerer P.; Mroginski M. A.; Murgida D. H.; Krauss N.; Heyn M. P.; Hildebrandt P.; Borucki B.; Lamparter T. Highly Conserved Residues Asp-197 and His-250 in Agp1 Phytochrome Control the Proton Affinity of the Chromophore and Pfr Formation. J. Biol. Chem. 2007, 282, 2116–2123. [DOI] [PubMed] [Google Scholar]
- Kim M. K.; Kang Y. K. Positional Preference of Proline in α-Helices. Protein Sci. 1999, 8, 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]