Abstract
We report on the reversible detection of CaptAvidin, a tyrosine modified avidin, with single-walled carbon nanotube (SWNT) field-effect transistors (FETs) noncovalently functionalized with biotin moieties using 1-pyrenebutyric acid as a linker. Binding affinities at different pH values were quantified, and the sensor’s response at various ionic strengths was analyzed. Furthermore, protein “fingerprints” of NeutrAvidin and streptavidin were obtained by monitoring their adsorption at several pH values. Moreover, gold nanoparticle decorated SWNT FETs were functionalized with biotin using 1-pyrenebutyric acid as a linker for the CNT surface and (±)-α-lipoic acid linkers for the gold surface, and reversible CaptAvidin binding is shown, paving the way for potential dual mode measurements with the addition of surface enhanced Raman spectroscopy (SERS).
Introduction
Medical diagnostics stands to benefit from the development of cost-effective and reliable protein detection methods. Detection of protein–protein or protein–ligand interactions, which are vital processes in living organisms, deepens the understanding of associated biological processes and provides invaluable tools to diagnose many diseases. In this context, the suitability of field-effect transistors (FETs) based on one-dimensional nanomaterials, such as nanowires and single-walled carbon nanotubes (SWNTs), as versatile and label-free electronic biosensors has been thoroughly studied.1 Their ease of integration into next-generation electronic circuits, coupled with dimensions on the size order of individual molecules, results in promising potential applications in diagnostics.1,2 When testing a biosensor’s feasibility, well-known biosystems, such as the hybridization of single-stranded nucleic acids3 or the binding of streptavidin or avidin to biotin4 play an important role. Usually, one of the system’s components is immobilized onto the sensitive area of the sensor as a probe, while the other is used as a target analyte. Both of these testing systems utilize properties that are exceptional in nature, taking advantage of DNA’s high charge and the high binding affinity between streptavidin and avidin to biotin (KD is in the order of 10–14 M).5 These strong interactions have helped immensely in understanding the underlying sensing mechanisms and restrictions of FET sensors in detail. The dominant sensing mechanism for functionalized SWNT FETs is commonly accepted as electrostatic gating due to the presence of charged moieties in the proximity of the nanotube’s surface, leading to a shift of the threshold voltage of the transistor.6 While this effect is largely responsible for the ability to perform label-free sensing, devices are limited in terms of the electric double layer forming a so-called Debye screening length, rendering sensing in physiological conditions challenging.7−9
In this Article, we address the need to develop alternative biosystems closer to real demands of FET sensors, which can be used to optimize the sensor for real-life applications, such as point-of-care (POC) testing. For this purpose, we used the CaptAvidin–biotin system, which offers a pH-tunable binding affinity with strong affinity occurring at pH 4 and complete dissociation of the protein–ligand complex occurring at pH 10.10 Hence, these biotinylated devices can be regenerated by washing with pH 10 buffers, rendering this system interesting for engineering biosensors capable of selective protein detection.11,12 Additionally, the strong interactions of the biotin-(strept)avidin system, widely used in biotechnology,13 were utilized with our devices to create protein “fingerprint” sensors by studying SWNT FET response under varying pH values. Finally, CaptAvidin experiments were also performed with gold nanoparticle (AuNP) decorated devices, offering the possibility for complementary surface enhanced Raman spectroscopy (SERS) measurements.
Experimental Section
SWNT FET devices were fabricated as previously reported.14 Briefly, interdigitated electrodes (with channel length of 10 μm) were patterned via a standard photolithography procedure on silicon substrates (200 nm SiO2, 30 nm Ti, 100 nm Au). P2-SWNTs from Carbon Solutions, Inc. (purity >90%) were dispersed in N,N-dimethylformamide (DMF) and deposited as the active layer between the predefined electrodes via alternating current dielectrophoresis.15 The chips were placed onto a ceramic dual in-line package fitted with a liquid compartment affixed by polydimethylsiloxane (PDMS). Electrical characterization in this work was conducted by recording transfer curves at a fixed source-drain bias of 50 mV and sweeping the gate potential from −0.6 V to +0.6 V. The transistors were operated in electrolyte-gate mode with a Ag/AgCl electrode immersed in the buffer solution (Figure 1a). All electrical measurements were conducted using two Keithley 2400 sourcemeters.
Figure 1.
(a) Schematic illustration of an electrolyte-gated single-walled carbon nanotube field-effect transistor (SWNT FET). (b) Typical transfer curves recorded for the electrical detection of CaptAvidin. Transfer curve 1 was taken before exposure to the protein, 2 after incubation with 140 nM CaptAvidin, and 3 after 15 min exposure to pH 10 buffer, which is known to reverse the biotin/CaptAvidin binding. (c) Normalized sensor response (I0–IP)/gm at different buffer pH as a function of CaptAvidin concentration. (d) Effect of Debye screening length on sensor response to 140 nM CaptAvidin in pH 4 buffer with varying concentrations of KCl.
AuNP-SWNT hybrid devices were fabricated on chip via bulk electrolysis in a three-electrode configuration. The nucleation of gold nanoparticles of varying size from chloroauric acid (1 mM AuCl3 in 0.1 M HCl) onto the SWNTs was realized with a nucleation potential fixed at −0.4 V and varying pulse durations (10–40 s, further details are supplied in the Supporting Information).
For the detection of biotin-binding proteins, the SWNTs were functionalized with pyrene-biotin (P–B) by incubating the devices in a 50 μM P–B solution in anhydrous methanol overnight. Pyrene is known to adhere onto carbon nanotube sidewalls via π–π stacking interactions and is well established for functionalizing carbon nanotubes in biosensors.16−20 CaptAvidin experiments with AuNP decorated FETs were conducted after incubation with 50 μM P–B and 50 μM (±)-α-lipoic acid–biotin (LA-B) for 42 h in methanol. The functionalized devices allow for reproducible results even after extended storage (see Supporting Information).
For protein experiments, a universal buffer (Britton-Robinson) was used and adjusted to the desired pH value by KOH titration.21,22 The compositions of the buffers were adjusted to the requirements of each experiment as detailed in the Supporting Information. Biotin-binding proteins CaptAvidin, NeutrAvidin, and streptavidin were purchased from Life Technologies.
Further information regarding the device fabrication, functionalization, characterization, additional SEM images of AuNP decorated FETs, as well as details on the synthesis of the functionalization molecules are provided in the Supporting Information.
Results and Discussion
CaptAvidin is a commercially available, tyrosine modified avidin, which offers the advantage of a pH-dependent binding affinity toward biotin with its highest association constant of >109 M–1 at pH 4 and complete dissociation occurring at pH 10.10 This system has been already successfully exploited using surface plasmon resonance biosensors by García-Aljaro et al.11 Here, we demonstrate that the biotinylated SWNT FET surface can be regenerated after a pH 10 washing step by monitoring the transfer characteristics before and after protein incubation as well as after pH 10 device regeneration. As previously demonstrated, protein sensing may suffer from parasitic signals that can eventually occur from protein attachment onto the gate electrode.23 Thus, we avoided any contact of the electrode with solutions containing the protein by removing the electrode from the fluid chamber during protein incubation.
Figure 1b shows a typical CaptAvidin experiment. A transfer curve (ID vs VG) of the device was taken in pH 4 buffer (1) before incubation in 140 nM CaptAvidin for 15 min. After thoroughly rinsing the device with pH 4 buffer to ensure the removal of all excess protein from the solution, a second ID vs VG transfer characteristic was recorded (2). Bound protein was then removed with a pH 10 washing step (detailed in Supporting Information) and then transfer curve 3 was recorded.
After protein incubation, a threshold voltage shift of about 20 mV toward negative voltages was observed. This shift was entirely reversed after the pH 10 washing step, indicating a complete dissociation of the biotin-CaptAvidin complex. This procedure involving the reversible immobilization of CaptAvidin on P–B functionalized SWNT FETs is repeatable multiple times and was successfully used for various experiments. Control experiments were performed to verify that pristine, unfunctionalized devices do not show this recovery after washing (Supporting Information Figure S4c).
Using this biosystem, further experiments were performed to gain information about binding strength and the effect of Debye screening length on the sensor’s response. To probe the pH-dependent binding affinity of the biotin-CaptAvidin complex, data obtained from four different transistors were analyzed and the results are shown in Figure 1c. In the graph, normalized sensing response ((|Io – IP|)/gm)|VD is plotted as a function of CaptAvidin concentration, allowing comparison between devices by compensating for device-to-device variations. I0, IP, and gm are extracted from transfer curves, where I0 is the drain current at VG = −0.6 V before protein incubation, IP is the drain current at the same potential after protein incubation, and gm is the maximum of the transconductance ∂ID/∂VG. Using previously published analytical models to determine the dissociation constant of CaptAvidin at pH 4 and pH 5, data were fitted according to eq 1.24,25
| 1 |
In this equation, [B]max is the maximum surface density of binding sites on the surface of the carbon nanotubes, C0 is the analyte/channel capacitance, and qp is the charge from the protein.
Our sensing protocol differs significantly from previous work utilizing streptavidin and other proteins with extremely high binding affinities, wherein sensors cannot be reused after a protein binding experiment.4,18 Here, CaptAvidin was released from the sensor’s surface after incubation with a certain protein concentration by a pH 10 washing step and devices could be reused to obtain data for subsequent protein concentrations. As SWNT FETs are intrinsically sensitive toward changes of the electrolyte’s pH, all transfer curves were taken at pH 4, however the 15 min protein incubations were performed at pH 4 or pH 5.26,27 At pH 4 and pH 5, dissociation constants of 43 ± 7 and 308 ± 170 nM were obtained, respectively.
As previously mentioned, experiments addressing the effect of ionic strength on protein sensing can also be performed using the CaptAvidin–biotin system. By consecutively sensing the response upon incubation of 140 nM CaptAvidin in pH 4 buffer with varying KCl concentrations ranging from 0.25 mM to 50 mM, the response of four transistors was analyzed and the results are depicted in Figure 1d. The graph shows the normalized response as a function of the Debye screening length (LD), which was calculated by solving eq 2 for monovalent ions.
| 2 |
Here, LB is the Bjerrum length (0.7 nm) and ρi and zi the density and the valence of ion species, i. These results are in agreement with the work of Sorgenfrei et al. on FET biosensors, in that the sensing response depends exponentially on the ionic strength of the electrolyte.8 Furthermore, we find that the response drops significantly in a LD regime on the order of the hydrodynamic diameter of avidin at a distance from the nanotubes surface, which is given by the length of the functionalization molecule.28 The P–B length in the lowest-energy conformation estimated using molecular modeling (Spartan 10 software) was 2.7 nm.
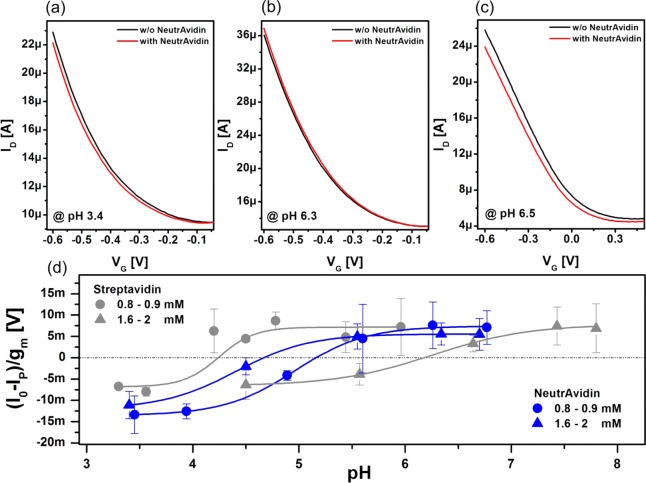
Additionally, pH-dependent measurements with two other biotin-binding proteins, NeutrAvidin and streptavidin, were performed. For each pH value, transfer curves were recorded before and after protein incubation. All protein incubations were conducted in 10 mM PBS buffer (pH 7) for 15 min. In contrast to measurements with CaptAvidin, however, sensors cannot be regenerated after binding to biotin; thus, each measurement requires a new transistor. Typical transfer curves are shown in Figure 2. The curves in panels a and b were collected for biotinylated devices at buffer pH’s of 3.4 and 6.3, respectively. We observed that the threshold voltage shift after NeutrAvidin incubation depends on the buffer pH (experiments with streptavidin reveal the same trend, thus transfer characteristics are not shown). For the experiment conducted at pH 3.4, the threshold voltage shifts toward more negative gate voltages and for pH 6.3 the threshold voltage shifts in the opposite direction. This phenomenon does not occur for experiments with unfunctionalized SWNT FETs as shown in Figure 2c. The ID–VG measurements in Figure 2c show an overall decrease in device conductance and a slight tilt of the curve after protein adsorption, indicating that the mobility of the device is affected upon protein adsorption, a phenomenon which does not appear if a spacer provided by the functionalization prevents unspecific protein adsorption onto the sidewalls of the carbon nanotubes.
Figure 2.
Transistor characteristics of a biotinylated SWNT FET before (black) and after (red) NeutrAvidin adsorption, measured at pH 3.4 (a) and at pH 6.3 (b). (c) Transfer characteristics of an unfunctionalized device before (black) and after (red) NeutrAvidin adsorption measured at pH 6.5. (d) Normalized sensor response (I0 – IP)/gm of pyrene-biotin-functionalized SWNT FET toward NeutrAvidin (blue) and streptavidin (gray) under different buffer concentrations as a function of buffer pH. The protein response was measured in 1.6–2 mM buffer (triangles) and 0.8–0.9 mM buffer (circles) concentrations. Error bars result from averaging responses of several devices. The solid lines represent sigmoidal Boltzmann fits.
To further investigate this phenomenon, we conducted the same experiments for a broad range of pH values for NeutrAvidin and streptavidin; the results thereof are presented in Figure 2d. The data points of this graph are calculated by taking the normalized currents at a gate potential of −0.6 V from the transfer curves before and after protein binding. The solid curves represent sigmoidal Boltzmann fits. The experiments for both proteins were conducted at two different buffer dilutions which resulted in buffer concentrations of 1.6–2 mM (triangles) and 0.8–0.9 mM (circles). Details regarding the buffers are presented in the Supporting Information. Irrespective of the parameters applied, a transition from a negative to a positive transistor response can be observed, which is best described by the intercept of the fitted curves with the horizontal, which is plotted as dashed-dotted gray line. The intercept will be referred to as point of zero response (PZR). The measurements at the higher buffer concentration yielded a PZR at pH 4.7 for NeutrAvidin (blue) and a PZR at pH 6.2 for streptavidin (gray). At the lower buffer concentration, a PZR at pH 5.1 for NeutrAvidin and a PZR at pH 4.2 for streptavidin were obtained. In general, we suspect that the transition from negative to positive response is associated with the zeta potential of the proteins, which is the potential at the hydrodynamic shear boundary. In particular, for proteins, surface charges arise due to pH-dependent ionization of the acidic or basic side groups of amino acids located at the surface. In this regard, the zeta potential, which can be positive or negative depending on the pH value, gives information on a protein’s electric landscape in solution.29 At the transition point from positive to negative charge, there is a point of net neutral charge, or the isoelectric point (IEP). The zeta potential and the IEP are usually determined by measuring the particle motion under an applied electric field in solution for various buffer pH. Similarly, a pH dependent transition from a positive to a negative transistor response, including a point of zero response, should be also detectable with a field-effect transistor.30 Namely, if a positively charged particle approaches the vicinity of the surface of a SWNT FET, the net charge of this particle will cause a threshold voltage shift toward the positive x-axis, which results in I0 – IP < 0 and vice versa for negatively charged particles. NeutrAvidin is an engineered protein with an almost neutral IEP of 6.3 as stated by the provider (Invitrogen). Various IEPs for streptavidin are reported throughout the literature and range from roughly 5 to 7.5.5 However, those values were obtained in solutions with higher ionic strength than those used in this work. Hence, to compare our data with previous results, experiments with NeutrAvidin and streptavidin under different pH values were conducted using a zeta potential analyzer (details and corresponding data are provided in the Supporting Information). NeutrAvidin revealed an IEP of 4.8 for both buffer concentrations and streptavidin yielded an IEP of 5.1 in 1.6 mM PBS and of 4.8 in 0.8 mM PBS (Supporting Information Figure S6). For NeutrAvidin, the values obtained from the two independent experiments are in good agreement. However, the discrepancy between the zeta analyzer and the FET experiments is significant for streptavidin. In both experiments, the IEP and PZR depend on the electrolytic conditions wherein a shift toward more acidic pH for lower buffer concentrations is observed. However, the shift is much greater for the FET measurements, possibly because of changes in salt concentration having an effect on the equilibrium of charges in basic and acidic amino acids, which can modify the protein’s IEP. However, predicting the PZR obtained with an FET for a specific protein and at a certain ionic strength requires exact knowledge of the surrounding potential and charge distribution of the protein. Although the observed behavior for our two model proteins cannot be considered universal because of well-known batch-to-batch variations (a broad spectrum of isoelectric points is reported for streptavidin),5 we believe that such zeta-potential-like SWNT FET measurements can provide a new tool to differentiate between proteins and thus give rise to new cost-effective methods applicable in fields such as protein engineering.
There is an increasing interest in surface enhanced Raman spectroscopy (SERS) for applications in chemical and biological sensing, due to its unique sensitivity with regards to molecular fingerprinting.31−33 Surface enhanced Raman spectroscopy can be realized by decorating carbon nanotubes with gold nanoparticles directly on chip by electrodeposition from a AuCl3 solution.34,35 To assess the feasibility of this approach for solution processed SWNT FETs, we utilized AuNP-SWNT hybrid devices, the fabrication of which is detailed in the Experimental Section. In Figure 3a, a scanning electron microscope image of a gold decorated device (deposition parameters −0.4 V for 10 s) reveals a distribution of particle sizes ranging from 50 to 200 nm.
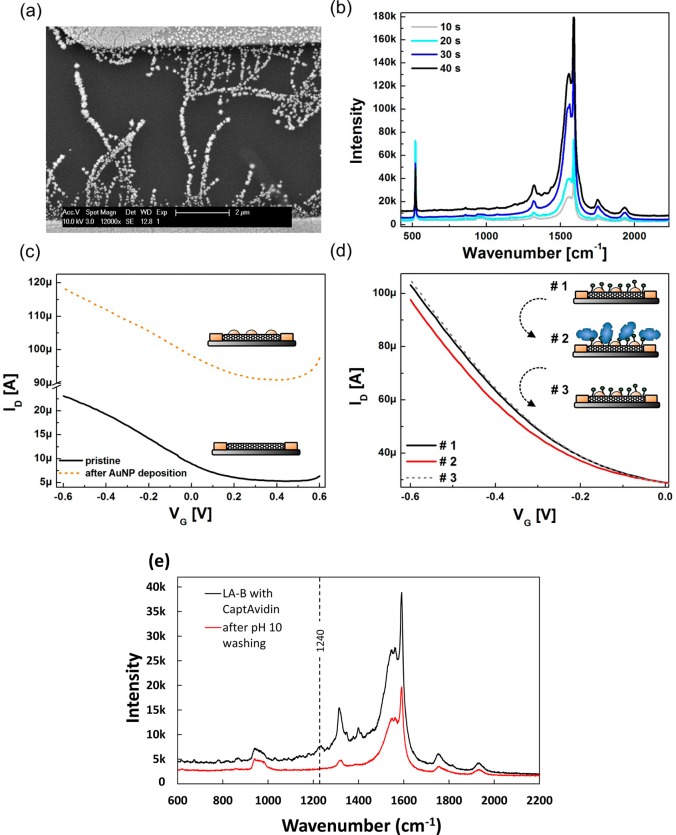
Figure 3.
(a) SEM image of gold nanoparticle decorated SWNTs. The particle sizes obtained from the applied parameters for this device are 50 to 200 nm. (b) Raman spectra of gold nanoparticle decorated SWNTs using varying deposition times of bulk electrolysis (deposition voltage −0.4 V). (c) Transfer characteristics of a SWNT FET recorded before and after functionalization with gold nanoparticles. (d) Transfer characteristics recorded for CaptAvidin detection with a LA-B functionalized device. (1) Transfer curve was taken before exposure to CaptAvidin, (2) after incubation with 140 nM CaptAvidin, and (3) after 15 min exposure to pH 10 buffer. (e) Raman spectra of gold nanoparticle-decorated SWNTs functionalized with LA-B after incubation with CaptAvidin and after pH 10 washing. Peaks unique to the protein can be discerned after incubation which disappear upon rinsing.
Figure 3b shows the SERS spectra for different gold nanoparticle deposition times. The enhancement of the SWNT SERS signal is proportional to both the size of the nanoparticles as well as the variation in interparticle distances, and thus on the duration of the deposition pulse.36 We do not observe a shift in the frequency of the enhanced modes, from which we conclude that electromagnetic enhancement plays the predominant role of SERS here. Electrical characterization of an SWNT FET before and after gold deposition is shown in Figure 3c. The rise in conductance of the devices after gold deposition can be attributed to additional metallic pathways between the source and drain electrodes as well as the direct charge transfer between AuNPs and SWNTs.37
For protein sensing, (±)-α-lipoic acid−biotin (LA-B) molecules were synthesized to enable covalent biotinylation of the gold particles. Nanotube sidewalls were biotinylated as previously with P–B to prevent signals from unspecific protein adsorption onto the pristine nanotubes.38 In Figure 3d, transfer curves upon CaptAvidin binding and dissociation are shown. Similar to the experiments without gold nanoparticles, we observe a 20 mV shift of the threshold voltage. In addition to electrostatic gating, the threshold voltage shift may be explainable by a modulation of AuNP-SWNT Schottky barriers due to the presence of the protein.39−41
Analysis of Raman spectra agrees well with the FET results. After attachment of CaptAvidin, spectra show clear protein signals, specifically the 1240 cm–1 peak, indicative of β-sheet protein structures,42 which disappear after pH 10 washing (Figure 3e). For SERS to occur the analyte species must be in close proximity to the SERS substrate surface, in this case the AuNPs. This Raman signal enhancement is generally aided by specific binding such as DNA hybridization,43 protein-antibody interactions,44 and avidin–biotin coupling.45 Advantages of using SERS in combination with FET sensors include its ability to perform dry measurements, whereas the ionic strength of electrolyte solution and background signal drift may influence the FET device; SERS provides molecular fingerprints32 allowing for facile differentiation of proteins; last it is a nonelectrical method, which is immune to parasitic signals from analyte-electrode interactions, and complements FET characterization. This type of dual mode measurement could be extremely useful for development of future biomedical applications.
Conclusion
In conclusion, we successfully detected reversible protein–ligand binding with pyrene-biotin functionalized SWNT FETs. Hereby, we introduced CaptAvidin, a tyrosine modified avidin, as a useful protein to engineer and optimize FET biosensors due to its reversible binding to biotin. Using this biosystem, we were able to probe the dissociation constant of CaptAvidin at two different pH values and to demonstrate that the sensor signal depends on the Debye screening length. Furthermore, we were able to differentiate between two different biotin-binding molecules, streptavidin and NeutrAvidin, by their pH-dependent sensor response. Additionally, SWNT FETs decorated with gold nanoparticles were shown to be a promising platform providing a supplemental surface enhanced Raman spectroscopy substrate useful for the detection of proteins.
Acknowledgments
This work (A. M. M.; K. M.; G. S.) was supported by the International Graduate School for Science and Engineering (IGSSE) at the Technische Universität München. The authors would like to thank Prof. Dr. Martin Zacharias for valuable comments and discussion. This work was partially supported by NIEHS Award R01ES019304.
Supporting Information Available
Synthesis of functional biotin moieties, device fabrication, experimental protocols, and additional imaging and experiments are available in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Feigel I. M.; Vedala H.; Star A. Biosensors Based on One-Dimensional Nanostructures. J. Mater. Chem. 2011, 21, 8940–8945. [Google Scholar]
- Zhang Y.; Guo Y.; Xianyu Y.; Chen W.; Zhao Y.; Jiang X. Nanomaterials for Ultrasensitive Protein Detection. Adv. Mater. 2013, 25, 3802–3819. [DOI] [PubMed] [Google Scholar]
- Star A.; Tu E.; Niemann J.; Gabriel J.-C. P.; Joiner C. S.; Valcke C. Label-Free Detection of DNA Hybridization Using Carbon Nanotube Network Field-Effect Transistors. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star A.; Gabriel J.-C. P.; Bradley K.; Grüner G. Electronic Detection of Specific Protein Binding Using Nanotube FET Devices. Nano Lett. 2003, 3, 459–463. [Google Scholar]
- Green M. N.[5] Avidin and Streptavidin. In Methods Enzymology; Wilchek M., Bayer E. A., Eds.; Academic Press: San Diego, CA, 1990; Vol. 184, 51–67. [DOI] [PubMed] [Google Scholar]
- Heller I.; Janssens A. M.; Männik J.; Minot E. D.; Lemay S. G.; Dekker C. Identifying the Mechanism of Biosensing with Carbon Nanotube Transistors. Nano Lett. 2008, 8, 591–595. [DOI] [PubMed] [Google Scholar]
- Stern E.; Wagner R.; Sigworth F. J.; Breaker R.; Fahmy T. M.; Reed M. A. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007, 7, 3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgenfrei S.; Chiu C.-y.; Johnston M.; Nuckolls C.; Shepard K. L. Debye Screening in Single-Molecule Carbon Nanotube Field-Effect Sensors. Nano Lett. 2011, 11, 3739–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni G. S.; Zhong Z. Detection Beyond the Debye Screening Length in a High-Frequency Nanoelectronic Biosensor. Nano Lett. 2012, 12, 719–723. [DOI] [PubMed] [Google Scholar]
- Morag E.; Bayer E. A.; Wilchek M. Reversibility of Biotin-Binding by Selective Modification of Tyrosine in Avidin. Biochem. J. 1996, 316, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Aljaro C.; Munoz F. X.; Baldrich E. Captavidin: A New Regenerable Biocomponent for Biosensing?. Analyst 2009, 134, 2338–2343. [DOI] [PubMed] [Google Scholar]
- Campbell G. A.; Mutharasan R. Detection and Quantification of Proteins Using Self-Excited PZT-Glass Millimeter-Sized Cantilever. Biosens. Bioelectron. 2005, 21, 597–607. [DOI] [PubMed] [Google Scholar]
- Diamandis E. P.; Christopoulos T. K. The Biotin–(Strept)avidin System: Principles and Applications in Biotechnology. Clin. Chem. 1991, 37, 625–636. [PubMed] [Google Scholar]
- a Vedala H.; Chen Y.; Cecioni S.; Imberty A.; Vidal S.; Star A. Nanoelectronic Detection of Lectin–Carbohydrate Interactions Using Carbon Nanotubes. Nano Lett. 2011, 11, 170–175. [DOI] [PubMed] [Google Scholar]; b Chen Y.; Vedala H.; Kotchey G. P.; Audfray A.; Cecioni S.; Imberty A.; Vidal S.; Star A. Electronic Detection of Lectins Using Carbohydrate-Functionalized Nanostructures: Graphene versus Carbon Nanotubes. ACS Nano 2012, 6, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-B.; Liu X.-J.; Campbell E. E. B.; Zhang S.-L. Alternating Current Dielectrophoresis of Carbon Nanotubes. J. Appl. Phys. 2005, 98, 056103. [Google Scholar]
- Chen R. J.; Zhang Y.; Wang D.; Dai H. Noncovalent Sidewall Functionalization of Single-Walled Carbon Nanotubes for Protein Immobilization. J. Am. Chem. Soc. 2001, 123, 3838–3839. [DOI] [PubMed] [Google Scholar]
- Choi Y.; Olsen T. J.; Sims P. C.; Moody I. S.; Corso B. L.; Dang M. N.; Weiss G. A.; Collins P. G. Dissecting Single-Molecule Signal Transduction in Carbon Nanotube Circuits with Protein Engineering. Nano Lett. 2013, 13, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios M.; Martin-Fernandez I.; Borrise X.; del Valle M.; Bartroli J.; Lora-Tamayo E.; Godignon P.; Perez-Murano F.; Esplandiu M. J. Real Time Protein Recognition in a Liquid-Gated Carbon Nanotube Field-Effect Transistor Modified with Aptamers. Nanoscale 2012, 4, 5917–5923. [DOI] [PubMed] [Google Scholar]
- Maehashi K.; Katsura T.; Kerman K.; Takamura Y.; Matsumoto K.; Tamiya E. Label-Free Protein Biosensor Based on Aptamer-Modified Carbon Nanotube Field-Effect Transistors. Anal. Chem. 2006, 79, 782–787. [DOI] [PubMed] [Google Scholar]
- Besteman K.; Lee J.-O.; Wiertz F. G. M.; Heering H. A.; Dekker C. Enzyme-Coated Carbon Nanotubes as Single-Molecule Biosensors. Nano Lett. 2003, 3, 727–730. [Google Scholar]
- Britton H. T. S.; Robinson R. A. CXCVIII—Universal Buffer Solutions and the Dissociation Constant of Veronal. J. Chem. Soc. 1931, 1456–1462. [Google Scholar]
- Fernández C. M.; Martin V. C. Preparation d’un Tampon Universel de Force Ionique 0,3 M. Talanta 1977, 24, 747–748. [DOI] [PubMed] [Google Scholar]
- Minot E. D.; Janssens A. M.; Heller I.; Heering H. A.; Dekker C.; Lemay S. G. Carbon Nanotube Biosensors: The Critical Role of the Reference Electrode. Appl. Phys. Lett. 2007, 91, 093507. [Google Scholar]
- Duan X.; Li Y.; Rajan N. K.; Routenberg D. A.; Modis Y.; Reed M. A. Quantification of the Affinities and Kinetics of Protein Interactions Using Silicon Nanowire Biosensors. Nat. Nanotechnol. 2012, 7, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. Y.; Sung M. G.; Lee J.; Baik K. Y.; Kwon Y.-K.; Lee M.-S.; Hong S. Universal Parameters for Carbon Nanotube Network-Based Sensors: Can Nanotube Sensors Be Reproducible?. ACS Nano 2011, 5, 4373–4379. [DOI] [PubMed] [Google Scholar]
- Haeberle T.; Münzer A. M.; Buth F.; Garrido J. A.; Abdellah A.; Fabel B.; Lugli P.; Scarpa G. Solution Processable Carbon Nanotube Network Thin-Film Transistors Operated in Electrolytic Solutions at Various pH. Appl. Phys. Lett. 2012, 101, 223101–223105. [Google Scholar]
- Münzer A. M.; Heimgreiter M.; Melzer K.; Weise A.; Fabel B.; Abdellah A.; Lugli P.; Scarpa G. Back-Gated Spray-Deposited Carbon Nanotube Thin Film Transistors Operated in Electrolytic Solutions: an Assessment Towards Future Biosensing Applications. J. Mater. Chem. B 2013, 1, 3797–3802. [DOI] [PubMed] [Google Scholar]
- Firnkes M.; Pedone D.; Knezevic J.; Döblinger M.; Rant U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett. 2010, 10, 2162–2167. [DOI] [PubMed] [Google Scholar]
- Bowen W. R.; Hall N. J.; Pan L.-C.; Sharif A. O.; Williams P. M. The Relevance of Particle Size and Zeta-Potential in Protein Processing. Nat. Biotechnol. 1998, 16, 785–787. [DOI] [PubMed] [Google Scholar]
- Koch S.; Woias P.; Meixner L. K.; Drost S.; Wolf H. Protein Detection with a Novel ISFET-Based Zeta Potential Analyzer. Biosens. Bioelectron. 1999, 14, 413–421. [DOI] [PubMed] [Google Scholar]
- Vo-Dinh T.; Yan F.; Wabuyele M. B. Surface-Enhanced Raman Scattering for Medical Diagnostics and Biological Imaging. J. Raman Spectrosc. 2005, 36, 640–647. [Google Scholar]
- Huh Y.; Chung A.; Erickson D. Surface Enhanced Raman Spectroscopy and Its Application to Molecular and Cellular Analysis. Microfluid. Nanofluid. 2009, 6, 285–297. [Google Scholar]
- Li T.; Guo L.; Wang Z. Gold Nanoparticle-Based Surface Enhanced Raman Scattering Spectroscopic Assay for the Detection of Protein-Protein Interactions. Anal. Sci. 2008, 24, 907–910. [DOI] [PubMed] [Google Scholar]
- Singhal R.; Orynbayeva Z.; Kalyana Sundaram R. V.; Niu J. J.; Bhattacharyya S.; Vitol E. A.; Schrlau M. G.; Papazoglou E. S.; Friedman G.; Gogotsi Y. Multifunctional Carbon-Nanotube Cellular Endoscopes. Nat. Nanotechnol. 2011, 6, 57–64. [DOI] [PubMed] [Google Scholar]
- Quinn B. M.; Dekker C.; Lemay S. G. Electrodeposition of Noble Metal Nanoparticles on Carbon Nanotubes. J. Am. Chem. Soc. 2005, 127, 6146–6147. [DOI] [PubMed] [Google Scholar]
- a Sharma H.; Agarwal D. C.; Shukla A. K.; Avasthi D. K.; Vankar V. D. Surface-Enhanced Raman Scattering and Fluorescence Emission of Gold Nanoparticle–Multiwalled Carbon Nanotube Hybrids. J. Raman Spectrosc. 2013, 44, 12–20. [Google Scholar]; b Thomas R.; Swathi R. S. Organization of Metal Nanoparticles for Surface-Enhanced Spectroscopy: A Difference in Size Matters. J. Phys. Chem. C 2012, 116, 21982–21991. [Google Scholar]
- Ding M.; Tang Y.; Star A. Understanding Interfaces in Metal–Graphitic Hybrid Nanostructures. J. Phys. Chem. Lett. 2013, 4, 147–160. [DOI] [PubMed] [Google Scholar]
- Bradley K.; Briman M.; Star A.; Grüner G. Charge Transfer from Adsorbed Proteins. Nano Lett. 2004, 4, 253–256. [Google Scholar]
- Pachauri V.; Kern K.; Balasubramanian K. Field-Effect-Based Chemical Sensing Using Nanowire-Nanoparticle Hybrids: The Ion-Sensitive Metal-Semiconductor Field-Effect Transistor. Appl. Phys. Lett. 2013, 102, 023501. [Google Scholar]
- Kim B.; Lee J.; Namgung S.; Kim J.; Park J. Y.; Lee M.-S.; Hong S. DNA Sensors Based on CNT-FET with Floating Electrodes. Sens. Actuators B 2012, 169, 182–187. [Google Scholar]
- a Kauffman D. R.; Sorescu D. C.; Schofield D. P.; Allen B. L.; Jordan K. D.; Star A. Understanding the Sensor Response of Metal-Decorated Carbon Nanotubes. Nano Lett. 2010, 10, 958–963. [DOI] [PubMed] [Google Scholar]; b Kauffman D. R.; Star A. Chemically Induced Potential Barriers at the Carbon Nanotube–Metal Nanoparticle Interface. Nano Lett. 2007, 7, 1863–1868. [DOI] [PubMed] [Google Scholar]; c Star A.; Joshi V.; Skarupo S.; Thomas D.; Gabriel J.-C. P. Gas Sensor Array Based on Metal-Decorated Carbon Nanotubes. J. Phys. Chem. B 2006, 110, 21014–21020. [DOI] [PubMed] [Google Scholar]
- Rygula A.; Majzner K.; Marzec K. M.; Kaczor A.; Pilarczyka M.; Baranska M. Raman Spectroscopy of Proteins: A Review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar]
- Huh Y. S.; Chung A. J.; Cordovez B.; Erickson D. Enhanced On-Chip SERS Based Biomolecular Detection Using Electrokinetically Active Microwells. Lab Chip 2009, 9, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou X.; Yamaguchi Y.; Yamamoto H.; Doi S.; Ozaki Y. NIR SERS Detection of Immune Reaction on Gold Colloid Particles without Bound/Free Antigen Separation. J. Raman Spectrosc. 1998, 29, 739–742. [Google Scholar]
- Li T.; Guo L.; Wang Z. Gold Nanoparticle-Based Surface Enhanded Raman Scattering Spectroscopic Assay for the Detection of Protein–Protein Interactions. Anal. Sci. 2008, 24, 907–910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.