Abstract
The AU-rich element (ARE) was discovered in 1986 as a conserved mRNA sequence found in the 3′ untranslated region of the TNF-α transcript and other transcripts encoding cytokines and inflammatory mediators. Shortly thereafter, the ARE was shown to function as a regulator of mRNA degradation, and AREs were later shown to regulate other posttranscriptional mechanisms such as translation and mRNA localization. AREs coordinately regulate networks of chemokine, cytokine, and growth regulatory transcripts involved in cellular activation, proliferation, and inflammation. ARE-mediated regulation is carried out by a host of ARE-binding proteins, whose activity is regulated in a cell type and activation-dependent manner. The last 25 years of ARE research has offered insight into the mechanisms and regulation of ARE-mediated mRNA decay, and has provided a road map for the discovery of additional mRNA regulatory motifs. The future of ARE research will transition from a discovery phase to a phase focused on translating basic biological findings into novel therapeutic targets. Our understanding of ARE-mediated gene regulation and posttranscriptional control has implications for many fields of study including developmental biology, neuroscience, immunobiology, and cancer biology.
INTRODUCTION
In 1986, Caput et al. described a 33 nucleotide sequence composed of entirely T and A in the 3′ untranslated region (UTR) of the TNF-α cDNA that was completely conserved between mice and humans.1 Furthermore, the investigators found that AU-rich sequences, termed AU-rich elements (AREs), were statistically enriched in the 3′ UTRs of several other inflammatory mediators, suggesting they served a regulatory function.1 This regulatory function was confirmed shortly thereafter when Shaw and Kamen showed that a 51 nucleotide AU-rich sequence from the 3′ UTR of the GM-CSF transcript caused rapid decay when inserted into the 3′ UTR of an otherwise stable β-globin transcript.2 These studies launched the field of ARE-mediated mRNA decay, and opened the door for subsequent insights into posttranscriptional regulatory networks in diverse fields including immunology, cancer biology, development, and neuroscience.
ARE-containing transcripts comprise approximately 5–8% of the transcriptome,3 and this mode of regulation provides the cell with a rapid and precise mechanism to alter gene expression patterns in response to extracellular stimuli.4 While the field of ARE-mediated mRNA regulation has matured greatly, the early pioneers in ARE research introduced important themes that remain evident in the literature today. For example, a major theme which still looms large in ARE research is that regulation of gene expression in inflammatory cells is highly regulated at posttranscriptional levels and precise mRNA decay regulation is crucial for normal immune responses.5,6 Models of immune cell activation have provided fruitful avenues for understanding posttranscriptional regulation, and the field continues to work toward understanding with how networks of posttranscriptional gene regulation control the outcome of immune responses. A second important theme that has emerged from ARE research is that many advances in our understanding of posttranscriptional regulation have been, and continue to be, through bioinformatics approaches. Through the use of genome-wide technologies and the computational analysis of large data sets, we have been able to identify networks of coordinated ARE-mediated mRNA decay that would have been impossible through analysis of individual transcripts.7,8 These approaches will become even more important as the field grapples with more complex questions about how these networks of ARE-containing transcripts are regulated. This review summarizes how our understanding of the ARE has influenced the field of mRNA decay research over the past 25 years as well as the future challenges.
CLASSES OF AREs
As researchers began dissecting the minimum sequence elements necessary for ARE-mediated mRNA decay, it was recognized that not all AREs conferred the same functional outcomes,9,10 leading to the classification of AREs into subgroups.11 In this classification system, class I AREs contain 1–3 copies of scattered AUUUA motifs with a nearby U-rich region and caused synchronous deadenylation. Class II AREs are composed of at least two overlapping copies of the UUAUUUA(U/A)(U/A) nonamer in a U-rich region and caused asynchronous deadenylation. Finally, class III AREs were said to contain a U-rich region and other ‘poorly defined features’ and caused synchronous deadenylation. Certain Class III AREs are similar to GU-rich elements (GREs), discussed later, since the class III AREs, such as the ARE in the c-jun proto-oncogene, can function as binding sites for the GRE-binding protein, CELF1. The organization of AREs into these three classes has proved to be useful in understanding the observed heterogeneity in the behavior of ARE-containing transcripts.
A second classification scheme was introduced by the developers of the ARE-database, an online database of ARE-containing transcripts from mouse, rat, and man.3 This system classified AREs into five groups based on the number of overlapping AUUUA pentamers found in the 3′UTR of a transcript. In this classification scheme, group 1 through group 5 AREs have five to one overlapping AUUUA pentamers, respectively. The developers of this classification system found that group 1 AREs were enriched in secreted proteins involved in the growth of hematopoietic and immune cells, while the other ARE groups are found in a diverse set of transcripts and exhibit no gene ontology enrichment.
While these classification systems capture some important aspects of ARE-mediated decay, they do not sufficiently describe the spectrum of function of AREs and cannot predict which AREs are functional in specific contexts. It is likely that several motif characteristics including surrounding sequences, secondary RNA structure and number of overlapping AUUUA pentamers fine tune the efficacy and function of AREs. These sequence and structural characteristics likely define ARE subgroups by influencing the recruitment of specific ARE-binding proteins. In the future, the most precise ARE classification scheme will likely be constructed based on the defined interactions between individual AREs and the various ARE-binding proteins as well as other components of functional ARE-binding complexes.
Given their heterogeneity, it is not surprising that AREs can promote a variety of outcomes to harboring transcripts. The first discovered function of AREs was they promote the rapid deadenylation and subsequent degradation of a transcript.2,9,12 As the field moved forward, it became clear that AREs can function to increase or decrease the translational efficiency of a transcript.13,14 For example, the ARE-binding proteins, HuR/ELAVL1 and TIA-1, exert opposing translational effects on the cytochrome c transcript.15 An additional function of the ARE is to influence the cytoplasmic location of its harboring transcript. Processing bodies (P-bodies) are discrete cytoplasmic granules that contain much of the mRNA degradation machinery necessary for 5′ → 3′ mRNA decay.16 The decay promoting ARE-binding proteins, TTP and BRF-1/2, are found in P-bodies, suggesting that they may function to recruit ARE-containing transcripts to P-bodies for degradation.17 Stress granules are a separate type of cytoplasmic granule that can be induced under conditions of cellular stress and are thought to function to temporarily and reversibly store transcripts in a state of stalled translational initiation. Depending on the duration and severity of the stress, transcripts harbored in stress granules can either return to the translationally active pool of cytoplasmic mRNA, or be shuttled to P-bodies for decay.18 The ARE-binding proteins TIA-1 and TIAR, among others, are intimately involved in the nucleation and structure of stress granules, and recruit their target transcripts to stress granules in an ARE-dependent manner.18
ARE-BINDING PROTEINS
In 1989, the first RNA-binding protein with specificity for the ARE was identified in cytoplasmic extracts from a lymphocyte cell line.19 Subsequently, two distinct ARE-binding proteins were identified in primary human T lymphocytes: one of these proteins was expressed constitutively and the other was induced upon T cell stimulation.20,21 The constitutive ARE-binding protein was later shown to be HuR (also known as HuA) and the induced ARE-binding protein was shown to be tristetraprolin (TTP),22 both of which are discussed in more detail below. Over the next several years, scientists identified numerous other ARE-binding proteins in a variety of cell types,23,24 and at least 20 ARE-binding proteins with a variety of structural characteristics have been described.25 Despite their heterogeneity in protein structure, many ARE-binding proteins are involved in regulation of transcript half-life. A large group of ARE-binding proteins promote transcript destabilization through binding to the ARE and subsequently recruiting cellular deadenylases and enzymes involved in both 5′ → 3′ and 3′ → 5′ mRNA decay.26 The best-characterized ARE-binding proteins that function to destabilize mRNA are tristetraprolin (TTP), BRF-1, BRF-2, KSRP, and AUF1.25 Within this group of destabilizing proteins, TTP and BRF-1/2 show remarkable similarity in their structure and binding specificity, but exhibit different tissue distributions suggesting that they may perform their roles in a tissue and context specific manner.27,28 A smaller group of ARE-binding proteins, such as HuR and HuD, function to stabilize target transcripts.25 In addition to regulating mRNA stability, ARE-binding proteins regulate almost all aspects of RNA metabolism including alternative splicing,29,30 nucleocytoplasmic export,31 cytoplasmic localization,32 and translational efficiency.33–35 A summary of the biochemical functions and binding specificities associated with several ARE-binding proteins is shown in Table 1. This table only includes ARE-binding proteins whose binding sites have been evaluated through high throughput methods. The intricate network of ARE-binding proteins and their target transcripts is made more complex due to the regulation of these proteins through posttranslational modifications, particularly phosphorylation.36 Phosphorylation of ARE-binding proteins can impact their function by altering subcellular localization31 or cofactor recruitment37 and their function can change over time over the course of cellular activation or immune responses.
TABLE 1.
Binding Specificities and Biochemical Functions Associated with ARE-binding Proteins that have been Investigated by High-throughput Experimental Methods
| RNA-Binding Protein | Binding Motif | Function | Reference |
|---|---|---|---|
| HuR/ELAVL1 |

|
mRNA stabilization, translational regulation | 38 |
| AUF1 |

|
mRNA degradation | 39 |
| TIAR |

|
Translational inhibition (via stress granule formation) | 40 |
| TIA-1 |
|
Translational inhibition | 41 |
| TTP | UUAUUUAUU | mRNA destabilization | 42 |
| HuD |
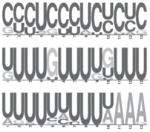
|
mRNA stabilization in neurons | 43 |
| CUGBP1/CELF1 |

|
mRNA destabilization, alternative splicing, translational control | 44 |
Studies of immune responses and inflammation have provided important insight into the function of AREs and ARE-binding proteins. A major breakthrough in the mechanisms of ARE-mediated mRNA decay occurred through evaluation of mice that were genetically deficient in the zinc finger ARE-binding protein TTP.45 Shortly after birth, TTP deficient mice developed a syndrome of cachexia, arthritis, and autoimmunity due to the overproduction of TNF-α by macrophages due to stabilization of TNF-α mRNA.45 This study highlighted the crucial role that ARE-mediated mRNA decay plays in regulating the dynamics of inflammatory and immune responses. While TTP knockout mice were useful in demonstrating that TTP functions as a mediator of mRNA decay, additional biochemical experiments have elucidated the mechanisms by which TTP functions to degrade mRNA. Co-immunprecipitation, gel-shift, and mass spectrometry techniques have suggested a model whereby TTP promotes mRNA decay by recruiting components of the mRNA degradation machinery to ARE-containing transcripts (Figure 1(b)). Upon binding to an ARE, TTP recruits and activates deadenylases including poly A-ribonuclease (PARN)46 and CAF1.47 In addition, TTP recruits enzymes involved in both 5′ → 3′ mRNA decay (Dcp1, Dcp2, and Xrn1) and 3′ → 5′mRNA decay (exosome subunits).48,49 The ability of TTP to bind to and recruit components of the cellular RNA decay machinery to ARE-containing transcripts was shown to be regulated by phosphorylation in a mitogen dependent manner.37,48,50–52 For example, TTP-mediated decay is regulated by phosphorylation of TTP by p38 MAPK-activated protein kinase 2.37 LPS stimulation of macrophages activates this kinase, causing TTP phosphorylation. TTP phosphorylation in turn prevents TTP from recruiting a deadenylase to the bound transcript by promoting TTP’s association with 14-3-3 proteins, while maintaining TTP’s ability to bind to the ARE.37 Through this mechanism, TTP target transcripts such as IL-1, IL-6, and COX-2 are stabilized and expressed at higher levels, allowing for an effective immune response.53 Eventually TTP phosphorylation is reversed by the phosphatase PP2A, allowing for TTP to return to its baseline function.54,55 Dysregulation of this balance between kinase and phosphatase activity on TTP function can lead to abnormal target transcript expression and ultimately result in disease states such as autoimmunity or cancer.56,57
FIGURE 1.
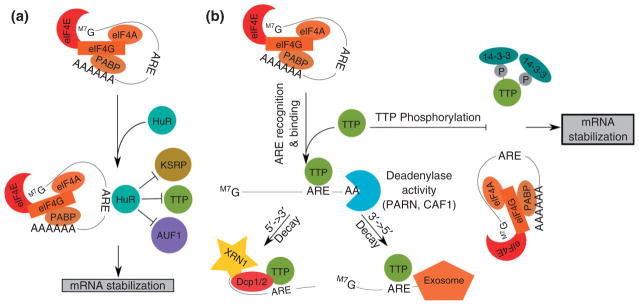
The ARE-binding proteins TTP and HuR exert opposite effects on ARE-containing transcripts. (a) HuR binds to the ARE to stabilize the transcript, likely through competitive inhibition of destabilizing ARE-binding proteins. (b) TTP binds to the ARE and recruits deadenylases as well as proteins involved in both the 5′ → 3′ and 3′ → 5′ decay pathways. The ability of TTP to recruit components of the mRNA decay machinery is blocked by phosphorylation and recruitment of the 14-3-3 adaptor protein.
In addition to phosphorylation of individual ARE-binding proteins, such as TTP, following cellular activation, the dynamic interaction between different ARE-binding proteins is also regulated over the course of immune activation. For example, throughout the process of T cell activation, ARE-containing transcripts are regulated through a dynamic process by alterations of their decay rates.58 ARE-containing transcripts can be either stabilized or destabilized during T cell activation, as a result of the functional relationships between various ARE-binding proteins.59 The ARE-binding proteins, HuR and TTP, compete with each other for certain ARE-containing transcripts expressed in T cells and promote opposing effects of stabilization and destabilization, respectively. A model of the dynamic interplay between HuR and TTP in mRNA decay regulation following T cell activation is illustrated in Figure 1(a). As early as 1 h following T cell activation, cytoplasmic HuR levels increase above those seen in resting T cells and transiently stabilizes a network of ARE-containing transcripts to allow their increased expression.22 At 3–6 h following activation, TTP is induced and displaces HuR on ARE-containing transcripts and mediates their rapid decay.22 This model provides a mechanism for coordinate regulation of the transient expression and subsequent degradation of a network of transcripts involved in T cell activation including proto-oncogene transcripts involved in cell growth and proliferation as well as transcripts that encode cytokines and other mediators of immune responses.
IDENTIFICATION OF NETWORKS OF ARE-CONTAINING TRANSCRIPTS
Experiments performed to evaluate individual-ARE containing transcripts have provided mechanistic insights into ARE-binding protein function and regulation, but are unable to illuminate the coordinate regulation of networks of ARE-containing transcripts. A major advance in our understanding of ARE-mediated mRNA decay was made possible by a technique that utilized immunoprecipitation of RNA-binding proteins followed by microarray analysis of the associated transcripts, allowing for the unbiased, genome-wide identification of RNA-binding protein target transcripts.60 This technique involves immunoprecipitation of an RNA-binding protein of interest from cellular lysates under conditions that maintain protein–RNA binding. After RNA-binding protein immunopurification, the co-purified RNA is then eluted and interrogated using genome-wide approaches such as microarray analysis. The first application of this technique led to the identification of EIF4E, HuB, and PolyA-binding protein (PABP) target transcripts.60 The target transcripts of numerous ARE-binding proteins have subsequently been determined, including HuR,7,61,62 TTP,63 AUF1,39 TIAR,40 and Pum1.64 This approach has also been applied to identifying targets of CELF1, a protein that binds to GREs and Class III AREs.44,65–67 The identification of these target transcripts has allowed for the computational identification of specific recognition motifs of individual ARE-binding proteins, as well as a better understanding of the combinatorial regulation of subnetworks of ARE-binding protein targets. The results of these experiments have led to a network model of RNA regulation, whereby a small number of RNA-binding proteins regulate functionally related, overlapping networks of transcripts.6 The expression of these networks can be altered quickly and precisely by changing the function of a small number of proteins through posttranslational modification, allowing for a rapid cellular response to environmental cues.
Recently a new technique, termed PAR-CLIP, utilizing photoactivatable nucleoside analogs that form reversible protein–RNA crosslinks has allowed for high resolution mapping of RNA-binding protein binding sites in a genome-wide fashion.68 The PAR-CLIP technique is powerful because it is able to identify individual binding sites, allowing for much more accurate inference of binding specificity as well as combinatorial regulation. This technique has been used to determine the target transcripts and binding sites of a few RNA-binding proteins, including HuR.69 Comparison of HuR target transcripts identified through the traditional RNA-immunoprecipitation methods to those identified through the PAR-CLIP method revealed that a much larger number of HuR target transcripts was identified by PAR-CLIP (~7500 transcripts identified by PAR-CLIP vs ~1900 transcripts identified by traditional RNA-immunoprecipitation), and relatively modest overlap (65% of targets identified by RNA-IP were also identified by PAR-CLIP). These results raise questions about whether all targets identified by PAR-CLIP are biologically relevant, or whether this very sensitive technique is identifying a large number of transient protein–RNA interactions that are not biologically meaningful. More work is needed to understand the networks of ARE-binding proteins, and techniques such as RNA-immunoprecipitation and PAR-CLIP will be useful tools for furthering our understanding of these networks.
THE ARE IS A PARADIGM FOR CONSERVED CIS-REGULATORY NETWORKS
The discovery of the ARE is an example of how bioinformatics analysis of genomic data led to the identification of novel regulatory sequences and laid a pathway for the discovery of novel sequences involved in the coordinate regulation of networks of mRNA. The value of a bioinformatic approach is also highlighted by the discovery of the Iron Response Element (IRE) as a conserved sequence in 1987, just a year after the discovery of the ARE.70 A comparison of the sequences in the 5′ UTR of the heavy and light ferritin subunits from rat and man led to the identification of a conserved 28 bp sequence which was postulated to control the translation of these genes.70 Rapidly this prediction was validated through cloning of the IRE into a reporter construct and showing that translation from the reporter transcripts became iron-regulated.71 The characterization of the IRE was followed by the identification of a cytoplasmic protein (Iron Response Element-Binding Protein) which bound to the IRE and increased the translational efficiency of bound transcripts in an iron-dependent manner.72 It is now known that there are two iron regulatory proteins, and these proteins bind to the stem-loop secondary structure of the IRE which occurs in the 5′ UTRs of a network of transcripts coding for several iron metabolism proteins.73 The discovery of the ARE and the IRE was similar in that the identification of sequence conservation led to the identification and characterization of the biological function of these elements.
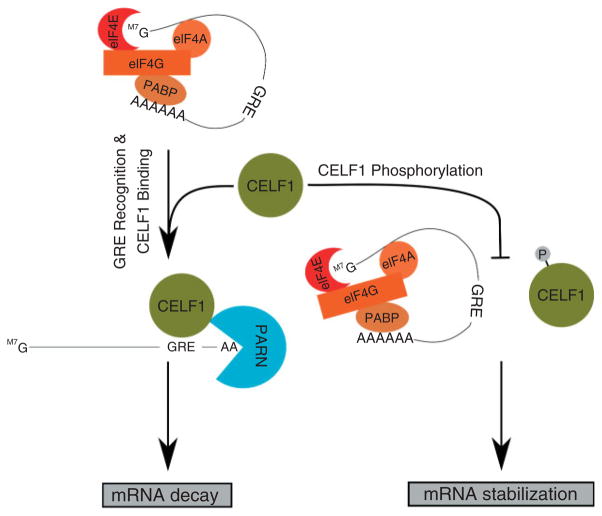
Another example of how the discovery of the ARE as a conserved regulatory element in the 3′ UTR of mRNA informed further discovery of novel regulatory elements is the recent discovery of the GRE, a GU-rich sequence with similarity to certain class III AREs. In order to understand the dynamics of mRNA decay in T cell stimulation, Raghavan et al. used microarray technology to measure the decay rate of thousands of transcripts simultaneously in resting and stimulated primary human T cells.59 The authors found that transcript half-lives were dynamic throughout T cell stimulation, and many transcripts that were upregulated at early time points exhibited rapid decay. In addition, the authors found that while many short-lived transcripts contained an ARE in their 3′UTR, a majority of short-lived transcripts contained no known decay elements. This prompted the authors to perform a bioinformatic search for novel motifs enriched in these short-lived transcripts and led to the discovery of the GU-rich element (GRE) as a conserved 3′ UTR sequence that promoted rapid mRNA decay.8 The GRE was defined as an 11 nucleotide sequence of the form UGU[G/U]UGU[G/U]UGU found to be enriched and conserved in the 3′UTR of short-lived transcripts.44 The GRE resembles class III AREs in that it is U-rich and lacks canonical AUUUA motifs, and certain class III AREs, such as the c-jun ARE, contain the GRE sequence. The GRE serves as a binding site for the RNA-binding protein CUGBP and ETR-3 Like Factor 1 (CELF1; also known as CUGBP1), which functions as a mediator of mRNA decay.74 CELF1 has long been known to be a phosphoprotein,75 and like many ARE-binding proteins, CELF1 function is regulated through phosphorylation.76 Figure 2 depicts a model of CELF1 function in mRNA decay and its regulation by phosphorylation. The CELF1 target transcript, TNF-α, was stabilized under conditions that promote CELF1 phosphorylation,77 and phosphorylation of CELF1 during primary human T cell stimulation led to reduced binding by CELF1 for the GRE, which could contribute to subsequent upregulation of CELF1 target transcripts.67 Overall, the discovery of the GRE and its role in mRNA decay regulation was very similar to the previous discovery of the ARE, in that sequence conservation led to the discovery of a functional element that was found to mediate mRNA degradation through its interaction with an RNA-binding protein.
FIGURE 2.
CELF1 promotes transcript decay by binding to the GRE, followed by the recruitment of deadenylase (i.e., PARN), promoting transcript degradation. CELF1-mediated mRNA decay is regulated by phosphorylation during T cell stimulation which causes decreased ability of CELF1 to bind to target transcripts.
The development of techniques for the genome-wide identification of target transcripts for RNA-binding proteins such as RNA immunoprecipitation and PAR-CLIP have allowed for the determination of subsets of ARE and GRE-containing transcripts targeted by specific RNA-binding proteins. Analysis of the target transcripts of CELF1 for the prevalence of GRE and AUUUA-pentamer-containing transcripts revealed that the number of transcripts containing both motifs is far less than one would expect by chance alone. This finding suggests that mutually exclusive subsets of transcripts can be defined as transcripts regulated by GREs versus transcripts regulated by AUUUA pentamers. These distinct subsets of transcripts may indicate differential regulation by orthogonal mRNA decay pathways. The fact that these elements are conserved throughout evolution suggests that they coevolved under evolutionary pressure which served to separate these two pathways. Clues to the source of this evolutionary divergence comes from functional pathway analysis of the networks of transcripts targeted by GRE- and AUUUA-specific ARE-binding proteins. This analysis revealed some overlapping functions, but transcripts targeted by ARE-binding proteins tend to be involved largely in inflammation and cytokine/chemokine pathways,78 whereas GRE-containing target transcripts tend to be largely involved in cell proliferation and apoptosis related pathways.74 The ability of the cell to regulate these different pathways independently may allow an organism to more precisely respond to environmental changes which could provide an evolutionary advantage.
MICRO-RNAS IN REGULATING ARE-MEDIATED DECAY
In addition to ARE-binding proteins, microRNAs (miRNAs) and the associated molecular machinery may also be involved in regulating ARE-containing transcripts. miRNAs are short (~21 nt) RNA species which bind to complementary sequences in RNA/DNA molecules and can regulate almost all aspects of gene expression including mRNA stability and translational efficiency.79 An RNAi-based screen to identify factors which affect ARE-mediated RNA decay led to the identification of miRNA-16 and proteins involved in miRNA biogenesis and function (Dicer 1, Argonaute 1, and Argonaute 2) as factors required for TTP-mediated degradation of TNF-α mRNA.80 In addition to promoting RNA decay, miRNAs may also negatively regulate TTP-mediated mRNA decay through competitive inhibition as shown in a recent article where miRNA 466I competed with TTP binding to an ARE in the 3′ untranslated region of interleukin-10 mRNA.81 Since decay triggered by miRNA-466I was significantly slower than that triggered by TTP, the result was a functional stabilization of the IL-10 transcript. Another example of regulation through miRNA and ARE interactions is regulation of the CAT-1 transcript by cellular stress in liver hepatocarcinoma cell lines.82 These studies investigated the effects of various cell stresses on CAT-1 regulation and found that upon stress the CAT-1 mRNA was relieved of miRNA-122 mediated repression and translocated from p-bodies into the actively translated pool of cytoplasmic mRNA.82 The investigators further showed that this effect was dependent on cytoplasmic HuR binding to the CAT-1 mRNA.82 It is likely that this effect is mediated by HuR binding adjacent to the miRNA binding site and modulating the function of the miRNA bound Argonaute protein.82,83 The vast diversity of sequences represented by miRNAs, and our inability to accurately predict miRNA target transcripts creates a challenge in understanding fundamental principles regarding ARE mediated decay and its interplay with miRNA regulation. The recent development of techniques to identify both RNA-binding protein and miRNA binding sites on a genome-wide level may provide opportunities to use bioinformatics to begin to decipher the combinatorial code behind these two regulatory mechanisms.
EVOLUTIONARY CONSERVATION OF RNA REGULATORY PATHWAYS
The biologic importance of ARE pathways is highlighted by the fact that AREs and the proteins which recognize them are conserved throughout evolution. AREs have been found to regulate gene expression in all mammalian organisms studied, as well as in evolutionarily distant organisms such as Drosophila and Saccharomyces cerevisiae.84–86 In S. cerevisiae, AREs from the endogenous transcript TIF51A as well as the exogenous transcripts TNF-α and c-fos, placed in the 3′ UTRs of reporter transcripts functioned as mediators of mRNA decay.87,88 Interestingly, ARE-mediated decay in S. cerevisiae was executed through a deadenylation-dependent decapping mechanism similar to that seen in mammalian ARE-mediated mRNA decay.10 The ARE-binding proteins that carry out regulatory functions are also conserved in their structure and function from yeast to humans. For example, the yeast ELAV homolog Pub1p recognizes and binds to the ARE and promotes transcript stabilization.87
GRE-mediated mRNA decay is also highly conserved, extending from worms to humans.74 One highly studied CELF1 homolog is the Xenopus protein, embryo deadenylation element binding protein (EDEN-BP). This protein binds to a UGU-rich element termed the embryo deadenylation element (EDEN) to promote transcript deadenylation.65,89 EDEN-BP is 88% homologous to human CELF1, and CELF1 has been shown to be able to functionally replace EDEN-BP in Xenopus extracts.90 The deadenylation promoting function of CELF1 homologs is conserved through humans, as CELF1 has been shown to recruit deadenylases in order to promote mRNA decay.91 Functionally, EDEN-BP is involved in muscle and bone formation during Xenopus development as evidenced by faulty somatic segregation in EDEN-BP deficient Xenopus embryos.92 The Caenorhabditis elegans protein ETR-1 is also a homolog of human CELF1. The function of ETR-1 in C. elegans development was investigating by inactivating ETR-1 using interfering RNA. This reduction in ETR-1 levels led to a defect in muscle development which caused failure of embryos to elongate and paralysis, which resulted in embryonic lethality.93 This finding that CELF1 homologs are critical for muscle development in Xenopus and Drosophila closely mirrors findings in the human disease, myotonic dystrophy, that show that CELF1 is a key regulator of the disease state in muscle tissue.94 Additionally, CELF1 is a major regulator of mRNA decay in muscle tissue through recognition of the GRE.66 These results suggest that in addition to conservation of the biochemical mechanisms of CELF1-GRE regulation, there may be conservation of the function of CELF1-GRE regulation at the organismal level in regulating specific aspects of developmental programs.
ABBERANT REGULATION OF mRNA DECAY NETWORKS IN CANCER
An important advance in the field of RNA biology in the past decade has been the recognition that altered mRNA metabolism contributes to an oncogenic phenotype.26 Many aspects of RNA metabolism including splicing patterns,95 polyadenylation site usage,96 RNA degradation,97 and translational efficiency98 have been shown to contribute to oncogenesis or malignant transformation. As these stages of the RNA life cycle are controlled by RNA-binding proteins, researchers investigated whether there are altered levels or functions of these RNA binding proteins in cancer relative to healthy tissue.
Increased levels of the ARE-binding protein, HuR, are found in several types of cancer,99–101 and several studies have worked to define mechanisms by which increased HuR may contribute to oncogenesis. HuR has been shown to target the transcript coding for vascular endothelial growth factor (VEGF).102 VEGF is expressed in response to hypoxia and serves as a paracrine factor to promote tissue vascularization. In the setting of cancerous tissue, tumor expression of VEGF promotes vascularization of the tumor allowing for increased delivery of oxygen and other nutrients.103 The interaction between HuR and VEGF serves to stabilize the VEGF transcript and promotes upregulation of the VEGF protein.102 Another HuR target is the cycloxygenase 2 (COX-2) transcript.104 COX-2 is involved in prostanoid synthesis and tends to be upregulated in cancerous tissue.105 Similar to VEGF, the interaction of HuR with the COX-2 enzyme promotes increased COX-2 transcript stability and subsequently increased COX-2 protein.104 Increased HuR expression is correlated with higher tumor grade in colon106 and breast cancer,107 perhaps through stabilization of these and other transcripts that promote tumor development. In addition to upregulation of HuR in the cytoplasm of tumors, it has also been shown that downregulation of TTP is a common finding in a variety of primary cancer tissues.108 TTP promotes the decay of many ARE-containing transcripts, and this downregulation of TTP in cancer promotes the upregulation of target transcripts that may promote cancer. It was further shown that the level of TTP in cancer tissue is inversely proportional to prognosis.108 These and other studies suggest that ARE-binding proteins, in particular HuR, may be promising targets for the design of molecular therapies to combat cancer by inducing ARE-mediated mRNA decay. The development of small molecule inhibitors of HuR might present novel treatment options for cancer patients, sand would provide researchers with useful tools for understanding the basic biology of cancer.
The field of GRE-mediated mRNA decay is in its infancy compared to ARE-mediated mRNA decay, and the relationship between GRE-mediated RNA decay and cancer is not as strong. The identity of GRE-containing target transcripts of the CELF1 protein shows that many of these transcripts encode important regulators of cell growth and apoptosis, suggesting that the dysregulation of the GRE/CELF1 network could promoter oncogenesis. It was recently shown that the ability of cytoplasmic CELF1 to bind to mRNA decreased during early time points of primary T cell stimulation as a result of transient phosphorylation of CELF1.67 This decreased binding by a mediator of mRNA decay allows the networks of transcripts coding for regulators of cell proliferation and apoptosis to rapidly but transiently accumulate in the cell. T cells in the early stages of activation share many characteristics with malignant cells, including reduced responses to apoptotic stimuli and rapid proliferation, and many of the same cell pathways activated at early time points in T cell stimulation are aberrantly regulated in cancer (e.g., PKC and MAPK/ERK pathways).109 We hypothesize that the activation of these signaling pathways in cancer causes phosphorylation and inactivation of CELF1, which contributes to an oncogenic phenotype by preventing the degradation of transcripts that promote cell growth and prevent apoptosis. Investigation of this hypothesis in models of oncogenesis will help to understand the pathways regulating CELF1 in cancer and in normal cell biology.
CONCLUSION
As the ARE turns 25 years old, it is remarkable how far the field has come in a relatively short period of time. The progress made in understanding the function of regulatory networks defined by the ARE and ARE-binding proteins will accelerate with the advent of techniques such as RNA-immunoprecipitation, PAR-CLIP, and next generation sequencing. One challenge to overcome will be to develop more sensitive experimental and bioinformatics approaches to understand the combinatorial regulation of networks of ARE-containing transcripts. Several ARE-binding proteins are expressed simultaneously in the cell and often have opposing functions. Understanding how these subnetworks of ARE-containing transcripts are regulated under various cellular conditions will aid in our ability to leverage this information to develop novel therapies for diseases such as autoimmunity or cancer. In particular, our increased understanding of dysregulated ARE-mediated mRNA decay in cancer will allow a transition from studies focused on biochemical mechanisms to exploiting these findings to develop new anti-cancer therapies that target ARE networks.
References
- 1.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 3.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 6.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 7.Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, Gorospe M, Atasoy U. The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer. BMC Cancer. 2010;10:126. doi: 10.1186/1471-2407-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rat-tenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 10.Chen CY, Shyu AB. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 12.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 13.Aharon T, Schneider RJ. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 15.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38(Pt 1):242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 18.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malter JS. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 20.Bohjanen PR, Petryniak B, June CH, Thompson CB, Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohjanen PR, Petryniak B, June CH, Thompson CB, Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992;267:6302–6309. [PubMed] [Google Scholar]
- 22.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J Biol Chem. 2001;276:47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. Embo J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci: CMLS. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 28.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30(Pt 6):945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 29.Soller M, White K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol Cell Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratti A, Fallini C, Colombrita C, Pascale A, Laforenza U, Quattrone A, Silani V. Post-transcriptional regulation of neuro-oncological ventral antigen 1 by the neuronal RNA-binding proteins ELAV. J Biol Chem. 2008;283:7531–7541. doi: 10.1074/jbc.M706082200. [DOI] [PubMed] [Google Scholar]
- 31.Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cδ coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 2010;30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cok SJ, Morrison AR. The 3′-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J Biol Chem. 2001;276:23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- 34.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36(Pt 3):491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 37.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uren PJ, Burns SC, Ruan J, Singh KK, Smith AD, Penalva LO. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J Biol Chem. 2011;286:37063–37066. doi: 10.1074/jbc.C111.266882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai WS, Carrick DM, Blackshear PJ. Influence of non-americ AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 43.Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 46.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 49.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 50.Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 51.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffer JR, Brooks SA. Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-α mRNA polysome loading. J Immunol. 2012;188:1828–1839. doi: 10.4049/jimmunol.1102837. [DOI] [PubMed] [Google Scholar]
- 53.Tudor C, Marchese FP, Hitti E, Aubareda A, Rawlinson L, Gaestel M, Blackshear PJ, Clark AR, Saklatvala J, Dean JL. The p38 MAPK pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mRNAs in murine macrophages. FEBS Lett. 2009;583:1933–1938. doi: 10.1016/j.febslet.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L, Stoecklin G, Van Way S, Hinkovska-Galcheva V, Guo RF, Anderson P, Shanley TP. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-α mRNA. J Biol Chem. 2007;282:3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- 55.Frasca D, Romero M, Landin AM, Diaz A, Riley RL, Blomberg BB. Protein phosphatase 2A (PP2A) is increased in old murine B cells and mediates p38 MAPK/tristetraprolin dephosphorylation and E47 mRNA instability. Mech Ageing Dev. 2010;131:306–314. doi: 10.1016/j.mad.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carrick DM, Chulada P, Donn R, Fabris M, McNicholl J, Whitworth W, Blackshear PJ. Genetic variations in ZFP36 and their possible relationship to autoimmune diseases. J Autoimmun. 2006;26:182–196. doi: 10.1016/j.jaut.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 57.Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 58.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genom. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen PR. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J Biol Chem. 2012;287:950–960. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leibold EA, Munro HN. Characterization and evolution of the expressed rat ferritin light subunit gene and its pseudogene family. Conservation of sequences within noncoding regions of ferritin genes. J Biol Chem. 1987;262:7335–7341. [PubMed] [Google Scholar]
- 71.Hentze MW, Rouault TA, Caughman SW, Dancis A, Harford JB, Klausner RD. A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc Natl Acad Sci U S A. 1987;84:6730–6734. doi: 10.1073/pnas.84.19.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leibold EA, Munro HN. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci U S A. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cairo G, Recalcati S. Iron-regulatory proteins: molecular biology and pathophysiological implications. Expert Rev Mol Med. 2007;9:1–13. doi: 10.1017/S1462399407000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr Opin Genet Dev. 2011;21:444–451. doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roberts R, Timchenko NA, Miller JW, Reddy S, Caskey CT, Swanson MS, Timchenko LT. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci U S A. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bakheet T, Williams BR, Khabar KS. ARED 3. 0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 80.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 81.Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol. 2010;184:6053–6059. doi: 10.4049/jimmunol.0902308. [DOI] [PubMed] [Google Scholar]
- 82.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 83.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 84.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cairrao F, Halees AS, Khabar KS, Morello D, Vanzo N. AU-rich elements regulate Drosophila gene expression. Mol Cell Biol. 2009;29:2636–2643. doi: 10.1128/MCB.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Apponi LH, Kelly SM, Harreman MT, Lehner AN, Corbett AH, Valentini SR. An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability. Mol Cell Biol. 2007;27:6569–6579. doi: 10.1128/MCB.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- 88.Vasudevan S, Garneau N, Tu Khounh D, Peltz SW. p38 mitogen-activated protein kinase/Hog1p regulates translation of the AU-rich-element-bearing MFA2 transcript. Mol Cell Biol. 2005;25:9753–9763. doi: 10.1128/MCB.25.22.9753-9763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J Biol Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 90.Osborne HB, Gautier-Courteille C, Graindorge A, Barreau C, Audic Y, Thuret R, Pollet N, Paillard L. Post-transcriptional regulation in Xenopus embryos: role and targets of EDEN-BP. Biochem Soc Trans. 2005;33(Pt 6):1541–1543. doi: 10.1042/BST0331541. [DOI] [PubMed] [Google Scholar]
- 91.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- 93.Milne CA, Hodgkin J. ETR-1, a homologue of a protein linked to myotonic dystrophy, is essential for muscle development in Caenorhabditis elegans. Curr Biol. 1999;9:1243–1246. doi: 10.1016/s0960-9822(99)80504-1. [DOI] [PubMed] [Google Scholar]
- 94.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37(Pt 6):1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 96.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vlasova IA, McNabb J, Raghavan A, Reilly C, Williams DA, Bohjanen KA, Bohjanen PR. Coordinate stabilization of growth-regulatory transcripts in T cell malignancies. Genomics. 2005;86:159–171. doi: 10.1016/j.ygeno.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 98.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, Furneaux H, Hla T, Haglund C, Ristimaki A. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 100.Vazquez-Chantada M, Fernandez-Ramos D, Embade N, Martinez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH, et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baou M, Norton JD, Murphy JJ. AU-rich RNA binding proteins in hematopoiesis and leukemogenesis. Blood. 2011;118:5732–5740. doi: 10.1182/blood-2011-07-347237. [DOI] [PubMed] [Google Scholar]
- 102.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 103.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 104.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 106.Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol: Official J U S Canad Acad Pathol. 2006;19:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 107.Denkert C, Weichert W, Winzer KJ, Muller BM, Noske A, Niesporek S, Kristiansen G, Guski H, Dietel M, Hauptmann S. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res: Official J Amer Assoc Cancer Res. 2004;10:5580–5586. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 108.Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]