Abstract
The prognosis of endometrial cancer is strongly associated with stage at diagnosis, suggesting that early detection may reduce mortality. Women who are diagnosed with endometrial carcinoma often have a lengthy history of vaginal bleeding, which offers an opportunity for early diagnosis and curative treatment. We performed DNA methylation profiling on population-based endometrial cancers to identify early detection biomarkers and replicated top candidates in two independent studies. We compared DNA methylation values of 1500 probes representing 807 genes in 148 population-based endometrial carcinoma samples and 23 benign endometrial tissues. Markers were replicated in another set of 69 carcinomas and 40 benign tissues profiled on the same platform. Further replication was conducted in The Cancer Genome Atlas and in prospectively collected endometrial brushings from women with and without endometrial carcinomas. We identified 114 CpG sites showing methylation differences with p-values of ≤10−7 between endometrial carcinoma and normal endometrium. Eight genes (ADCYAP1, ASCL2, HS3ST2, HTR1B, MME, NPY, and SOX1) were selected for further replication. Age-adjusted odds ratios for endometrial cancer ranged from 3.44 (95%-CI: 1.33–8.91) for ASCL2 to 18.61 (95%-CI: 5.50–62.97) for HTR1B. An area under the curve (AUC) of 0.93 was achieved for discriminating carcinoma from benign endometrium. Replication in The Cancer Genome Atlas and in endometrial brushings from an independent study confirmed the candidate markers. This study demonstrates that methylation markers may be used to evaluate women with abnormal vaginal bleeding to distinguish women with endometrial carcinoma from the majority of women without malignancy.
Keywords: endometrial cancer, methylation, biomarker, early detection, vaginal bleeding
Introduction
Endometrial cancer is the most common gynecological malignancy in the United States, with approximately 46,000 newly diagnosed cases and 8,100 related deaths in 2012 (1). The strongest prognostic factor for endometrial cancer is stage at diagnosis. Endometrioid adenocarcinoma, the numerically predominant histological tumor type, typically presents with stage 1 disease, which is an important factor in the overall favorable outcomes for these tumors (2). However, 15% of patients with endometrioid adenocarcinoma present with advanced stage disease, and this subset accounts for 80% of all endometrial cancer deaths (3).
Most endometrial carcinomas appear to develop from precursors termed endometrial hyperplasia that progress over a period of years (4;5). Patients who are ultimately diagnosed with carcinoma often present clinically with complaints of abnormal vaginal bleeding years prior to diagnosis, which offers an opportunity for early diagnosis and curative treatment (6). However, many such patients undergo repeated evaluations (4;6), incurring expense, inconvenience and stress. Conversely, abnormal vaginal bleeding is an extremely common complaint that is unrelated to carcinoma in the substantial majority of women (7). Thus, distinguishing the minority of women who require management for carcinoma precursors or carcinoma from others with benign causes of abnormal bleeding may be challenging. Furthermore, management of postmenopausal bleeding is usually based on pathologic diagnosis of an endometrial biopsy or curettage, the interpretation of which is limited by the adequacy of endometrial sampling, and difficulties related to accurately and reproducibly interpreting small, fragmented specimens (8). Data suggest that community pathologists over-diagnose endometrial hyperplasia compared with experts (8), but 40% of biopsies reported as atypical endometrial hyperplasia, even on review, are associated with prevalent carcinoma at hysterectomy (9;10).
We propose that the development of a molecular test that could aid in the management of abnormal vaginal bleeding could enable triage of patients with carcinoma, while reducing over-treatment of innocuous lesions. Among women with limited health care access, rapid identification of carcinomas may increase chances of cure and reduce the need for more aggressive treatment secondary to disease progression, whereas ruling out high-risk lesions could allow many women to safely opt for conservative management (6;11;12).
Candidate biomarker studies have consistently identified a number of specific molecular alterations in endometrial carcinoma, including mutations, DNA methylation, microsatellite instability, copy number alterations and gene expression patterns (13–16). Among the many characteristic molecular alterations that could provide early detection markers in endometrial carcinoma, DNA methylation is notable because of its early occurrence in carcinogenesis, stability and detectability using highly sensitive and specific assays. However, efforts to discover and validate DNA methylation markers using population-based samples as a means of identifying an early detection marker panel are limited. Accordingly, we describe results of DNA methylation profiling of endometrial carcinoma and benign tissues for biomarker discovery and the validation of promising candidate markers in independent samples by pyrosequencing.
Materials and Methods
Sample sources for biomarker discovery
We performed DNA methylation profiling for marker discovery in three studies: the Polish Endometrial Cancer Study (PECS), the Endometrial Hyperplasia Study (EH), and the Benign Reproductive Tissue Evaluation Study (BRTE) (Supplementary Table 1). Briefly, PECS was a population-based endometrial cancer case-control study conducted from 2001 to 2003 that included 551 histologically confirmed endometrial cancer cases (17) of which 148 with available fixed tissue cores were included in this analysis. The EH study was a nested case-control study evaluating endometrial hyperplasia progression in the Kaiser Permanente Northwest health plan from 1970 to 2003 (4;5). This investigation included 138 histologically confirmed endometrial cancer cases that developed a year or more after a diagnosis of endometrial hyperplasia or disordered proliferative endometrium, of which 69 are included in this study. In the BRTE protocol, normal endometrial tissue was collected from 142 women undergoing hysterectomy for benign uterine conditions, of which 63 were included in the current profiling analysis as a comparison group for the carcinomas.
Data and sample sources for candidate biomarker validation
We evaluated methylation of candidate markers in 444 endometrial carcinomas and 43 normal endometrial tissues from TCGA that were evaluated for methylation using high density arrays (14). For each candidate gene, we used average methylation values for all CpGs in a 200bp window around the sites that were included in the discovery effort. To further validate candidate methylation markers discovered through profiling analyses, we collected specimens from 38 women with endometrial carcinoma and 37 women undergoing hysterectomy for benign reasons at the Mayo Clinic Departments of Gynecology and Gynecologic Oncology (Supplementary table 1). Specifically, on the day of hysterectomy, women underwent endometrial sampling with a Tao brush (Cook Medical, Bloomington IN) (18) which was rinsed in PreservCyt (Hologic, Boxborough, MA) and used for subsequent testing. Frozen tumor tissue specimens were collected immediately after surgical removal of the uterus.
Methylation profiling using bead arrays
Methylation profiling was performed with the Illumina Golden Gate platform using the Standard Cancer Methylation Panel including 1,505 sites from 807 genes (19). Assays were performed in two batches (148 carcinomas from PECS and 23 benign endometrial samples from BRTE and 69 carcinomas from the EH study and 40 benign endometrial tissues from BRTE). Crude lysates were generated from paraffin tissue cores using proteinase K digestion and treated with bisulfite using the Zymo Methylation Gold kit according to the manufacturer’s instructions. Human placenta samples were included as controls across plates. Bisulfite (250 ng)-modified DNA was used for the Illumina bead array methylation assay. Methylation detection for 1505 CpG sites was performed, as described previously (20), according to the manufacturer’s instructions. Image processing and intensity data extraction were performed with the BeadStudio software. Each methylation data point is represented by fluorescent signals from the methylated (M) and unmethylated (U) alleles. Methylation beta values were calculated from raw Cy3 and Cy5 values as described previously (19). Normalization of beta values was performed using the background normalization method. The average signal intensity in the absence of hybridization to a specific target was determined by averaging the signals from built-in negative controls. This average was subtracted from each probe signal such that the expected signal for unmethylated targets is equal to zero. All analyses were performed with raw and normalized beta values, yielding similar results. Analyses with normalized values are presented in the manuscript.
Pyrosequencing of candidate genes
Pyrosequencing was performed on Tao brush samples and frozen tissues samples from hysterectomy specimens. Primers were designed using Pyrosequencing Assay Design Software (Qiagen). Pyrosequencing assays were developed for ADCYAP1, ASCL2, CDH13, HS3ST2, HTR1B, MME, and NPY covering between 2 and 10 CpG sites overlapping with the probes included in the Illumina Golden Gate array. Pyrosequencing assays for SOX1 failed to perform adequately. Amplification was carried out on 20 ng of bisulfate treated DNA using TaqGold DNA polymerase (Applied Biosystems) under the following conditions: 10 min at 95°C, followed by 50 cycles of 35 sec at 95°C, 35 sec at 57,5°C, and 1 min at 72°C. PCR products were checked by gel-electrophoresis to confirm product size. Pyrosequencing reactions were performed on Biotage PyroMark MD System (Qiagen) according to manufacturer’s protocols. Briefly, the incorporated biotinylated primer is immobilized on streptavidin-coated beads used to purify and render the denatured, single stranded and biotinylated PCR product. The single-stranded product was then annealed to 0.3 μM of the sequencing primer complementary to the single-stranded template, and placed at 85 °C for 2 min, cooled to room temperature for 5 min and the pyrosequencing reaction is performed by the sequential addition of single nucleotides in a predefined order. Raw data were analyzed using the provided Pyro Q-CpG 1.0.9 analysis software (Qiagen).
Data analysis
Data collection and analysis were divided into three steps.
Step 1
We compared the methylation levels in tissues from 148 carcinomas from PECS and 23 benign tissues from BRTE. Methylation levels of individual probes between carcinomas and benign tissues were compared by a t-test, and results were summarized by the resulting p-values and false discovery rates (21). After evaluating all genes, we identified a group of eight candidate markers (ADCYAP1, ASCL2, CDH13, HS3ST2, HTR1B, MME, NPY, and SOX1), meeting the following criteria a) p-values <10−7 b) differences in average methylation levels ≥ 0.4 and c) mean methylation values in subjects without carcinoma <0.2. Using the methylation levels from these 8 genes, we created a combined score reflective of a woman’s risk of endometrial carcinoma after averaging across all probes within a gene. Specifically, we fit the logistic model: , where Xij is the methylation level from gene j in subject i and Yij is an indicator of her carcinoma status. For individuals measured in step 2, we used these estimated coefficients, {β̂1,…, β̂J}, to calculate a disease score , which is reflective of the predicted disease risk. Model fitting was performed in R. Furthermore, we constructed a model based on all 807 genes included on the array using a LASSO penalized logistic regression where the penalty-term was chosen by cross-validation (optL1 function). The resulting coefficients from that model, { }, were also used to calculate a disease score in step 2.
Step 2
We compared the methylation levels in tissues from 69 carcinomas from EH and 40 benign tissues from BRTE. Using these samples, we created two receiver-operating characteristic (ROC) curves to determine how well the scores S and S*, based on the models built in step 1, could discriminate carcinomas from benign tissues. In the first curve, sensitivity and specificity is defined as the proportion of carcinomas from benign tissues with a score S (based on eight candidate genes) exceeding a threshold. The second curve repeats the process for S* (based on all genes included on the array). We calculated AUCs and bootstrap-based confidence intervals. For analyses of the eight individual markers, we dichotomized methylation levels into high vs. medium/low tertiles based on the controls and calculated odds ratios comparing the odds that a sample is derived from carcinoma given that it is in the high methylation group with the same odds given that it is in the lower methylation group. We calculated crude and age-adjusted odds ratios for the discovery set (from step 1), the replication set, and both sets combined.
Step 3
We compared the methylation levels among seven candidate markers for which pyrosequencing assays were successfully developed (ADCYAP1, ASCL2, CDH13, HS3ST2, HTR1B, MME, and NPY) in endometrial brush samples from 38 subjects with endometrial carcinoma and 37 subjects without endometrial carcinoma. We quantified the association between each gene and risk of endometrial carcinoma by both adjusted and unadjusted OR and 95% CIs. Furthermore, for each gene individually, we calculated ROC curves and corresponding AUCs using all individuals with non-missing values. To obtain an ROC curve based on all genes, we imputed missing values as previously described (22). For each gene, we ranked all non-missing methylation values. Let rij be the rank of individual i’s methylation level at gene j. For each woman, we calculated her average rank, , where is the number of genes with non-missing methylation levels for subject i. If a woman was missing a value for a given gene, j′, she was assigned a value so such that rij′ = Ri. Then we created 1000 pairs of “test” and “training” datasets. Using the training dataset, we built a model by stepwise logistic regression with the number of included coefficients determined by cross-validation. The methylation levels for the test set, consisting of one carcinoma and benign tissue, were then input into this model to obtain estimates of their disease risk. The sensitivity and specificity were the proportion of women with carcinoma and women with benign disease with an estimated probability exceeding a given threshold.
Methylation marker discovery analyses were performed in BRB array tools 4.2.1 and in R 2.13.2 (23). We performed unsupervised hierarchical clustering of the 25% most variable probes using Euclidean distance, complete linkage, and centering of the probes in BRB array tools. All other statistical analyses were performed in Stata11 (Statacorp, College Station, TX).
Results
Comparison of methylation profiles between endometrial carcinoma and benign tissues
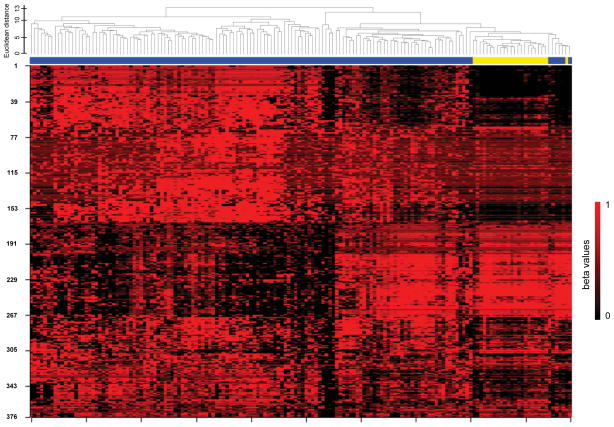
Unsupervised hierarchical clustering of methylation profiling data for 148 endometrial carcinomas from the Polish Endometrial Cancer Study (PECS) and 23 benign endometrial tissues from the Benign Reproductive Tissue Evaluation Study (BRTE) demonstrated near-perfect separation between carcinomas and normal tissues (Figure 1). We observed two distinct clusters of endometrial carcinomas with the smaller cluster showing more similarity to the normal endometrial tissues. We evaluated methylation of MLH1 because it is an established alteration in endometrial carcinoma (16). We observed a bimodal distribution of MLH1 methylation values in carcinomas, with 25% of tumors showing high methylation levels (>0.8), while the remaining 75% of carcinomas and all benign tissues showed very low methylation levels (<0.1). In class-comparison analysis, 114 CpG sites showed methylation differences with p-values of 10−7 or less (the top 50 sites representing 37 genes are shown in Supplementary table 2). Using the same DNA methylation platform, we re-assessed cancer associations for eight genes (ADCYAP1, ASCL2, CDH13, HS3ST2, HTR1B, MME, NPY, and SOX1) from the first analysis (Table 1, Supplementary Figure 1). All eight genes replicated in the independent sample set consisting of 69 endometrial carcinomas from an independent study, the Endometrial Hyperplasia Study (EH), and 40 additional benign tissues from BRTE. We calculated crude and age-adjusted odds ratios for individual probes representing the 8 candidate genes for the discovery set and the replication set (Supplementary Table 3). In analyses in which results for carcinomas in PECS and EH combined (n=214) were compared to all normal tissues analyzed from BRTE (n=63), unadjusted odds ratios ranged from 4.52 (95%-CI: 2.47–8.24) for MME to 31.18 (95%-CI: 14.61–66.53) for HS3ST2 and SOX1. Age-adjustment attenuated associations for most genes, but all remained statistically significant. In age-adjusted analyses, odds ratios ranged from 3.44 (95%-CI: 1.33–8.91) for ASCL2 to 18.61 (95%-CI: 5.50–62.97) for HTR1B (Table 2). We evaluated the discrimination between endometrial carcinoma and normal endometrial tissues achieved by methylation markers using cross-validation receiver-operating characteristic (ROC) curves analyses (Figure 2A). When methylation values from all 807 genes evaluated in the discovery phase were used in the model, we achieved an AUC of 0.91. When we restricted the analysis to the eight genes selected for further replication in clinical specimens, a similar AUC of 0.93 was achieved in the replication set. Using data from The Cancer Genome Atlas (TCGA), we evaluated methylation of the eight candidate genes in 444 endometrial carcinomas and 41 normal tissues. Stratified by histologic subtype, all eight genes showed highly significant differences with high AUCs between endometrioid carcinomas and normal tissue. All candidate markers showed lower methylation values in serous cancers compared to endometrioid cancers. Two candidates (HTR1B and SOX1) showed excellent discrimination between serous cancers and normal tissues with AUCs of 0.96. Three other candidates (ADCYAP1, HS3ST2, and NPY) showed decent discrimination with AUCs between 0.63 and 0.77, while AUCs for ASCL2, CDH13, and MME were not significantly different from 0.5 (Table 3).
Figure 1.
Unsupervised hierarchical clustering of 148 endometrial carcinomas and 23 benign endometrial tissues
Unsupervised hierarchical clustering was based on the 25% most variable probes within the discovery set. Methylation probes are in rows, endometrial cancers (blue) and normal endometrial tissues (yellow) are in columns. The dendrogram shows the Euclidean distance between individual samples.
Table 1.
Methylation probes from eight genes with highest mean methylation differences between endometrial carcinoma and benign endometrium and low methylation levels in benign endometrium
| Probe-ID | Mean beta values in benign endometrium | Mean beta values in endometrial carcinomas | p-value | False discovery rate | Gene |
|---|---|---|---|---|---|
| ADCYAP1_P398_F | 0.09 | 0.61 | < 1e-07 | < 1e-07 | ADCYAP1 |
| ADCYAP1_P455_R | 0.07 | 0.54 | < 1e-07 | < 1e-07 | ADCYAP1 |
| ASCL2_E76_R | 0.12 | 0.78 | < 1e-07 | < 1e-07 | ASCL2 |
| CDH13_P88_F | 0.33 | 0.75 | < 1e-07 | < 1e-07 | CDH13 |
| CDH13_E102_F | 0.14 | 0.54 | 1.80e-06 | 1.89e-05 | CDH13 |
| HS3ST2_E145_R | 0.19 | 0.78 | < 1e-07 | < 1e-07 | HS3ST2 |
| HS3ST2_P171_F | 0.17 | 0.72 | < 1e-07 | < 1e-07 | HS3ST2 |
| HTR1B_E232_R | 0.25 | 0.68 | < 1e-07 | < 1e-07 | HTR1B |
| HTR1B_P107_F | 0.10 | 0.75 | < 1e-07 | < 1e-07 | HTR1B |
| HTR1B_P222_F | 0.12 | 0.67 | < 1e-07 | < 1e-07 | HTR1B |
| MME_P388_F | 0.06 | 0.61 | < 1e-07 | < 1e-07 | MME |
| NPY_P295_F | 0.17 | 0.68 | < 1e-07 | < 1e-07 | NPY |
| SOX1_P294_F | 0.20 | 0.76 | < 1e-07 | < 1e-07 | SOX1 |
Table 2.
Crude and age adjusted odds ratios by probe for 8 candidate genes in carcinomas (n=214) and normal tissues (n=63)
| Crude | Age-adjusted | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |||
| ADCYAP1_P398_F | 20.07 | 10.01 | 40.23 | 12.88 | 4.04 | 41.06 |
| ADCYAP1_P455_R | 25.29 | 12.24 | 52.26 | 14.46 | 4.48 | 46.62 |
| ASCL2_E76_R | 6.49 | 3.51 | 11.98 | 3.44 | 1.33 | 8.91 |
| CDH13_E102_F | 15.13 | 7.77 | 29.46 | 10.27 | 3.31 | 31.86 |
| CDH13_P88_F | 15.13 | 7.77 | 29.46 | 10.27 | 3.31 | 31.86 |
| HS3ST2_E145_R | 31.18 | 14.61 | 66.53 | 11.40 | 3.73 | 34.90 |
| HS3ST2_P171_F | 25.29 | 12.24 | 52.26 | 11.29 | 3.69 | 34.56 |
| HTR1B_E232_R | 22.41 | 11.02 | 45.54 | 12.30 | 3.74 | 40.49 |
| HTR1B_P107_F | 14.51 | 7.48 | 28.15 | 18.61 | 5.50 | 62.97 |
| HTR1B_P222_F | 11.99 | 6.28 | 22.90 | 11.53 | 3.81 | 34.95 |
| MME_P388_F | 4.52 | 2.47 | 8.24 | 7.81 | 2.75 | 22.13 |
| NPY_E31_R | 10.13 | 5.36 | 19.12 | 4.68 | 1.77 | 12.36 |
| NPY_P295_F | 13.40 | 6.96 | 25.82 | 6.97 | 2.50 | 19.46 |
| NPY_P91_F | 7.57 | 4.07 | 14.07 | 4.27 | 1.63 | 11.17 |
| SOX1_P1018_R | 11.18 | 5.88 | 21.25 | 10.08 | 3.45 | 29.45 |
| SOX1_P294_F | 31.18 | 14.61 | 66.53 | 17.76 | 5.09 | 61.97 |
Analysis is based on normalized methylation data. Odds ratios are calculated using univariate and age-adjusted logistic regression; dichotomization of methylation levels is based on distribution in controls, comparing high vs. medium/low tertiles. 95% CI= 95% confidence interval.
Figure 2.
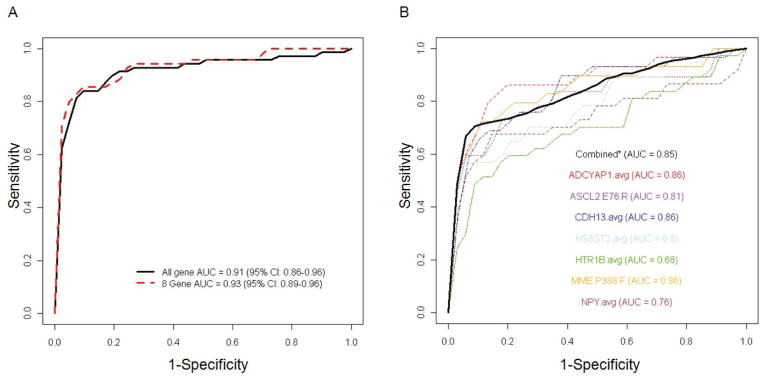
Receiver Operator Characteristic curve analysis to discriminate endometrial carcinoma from benign endometrial tissue using methylation markers
Receiver-operating characteristics (ROC) curves with areas under the curve (AUC) are shown for discrimination between endometrial carcinomas and normal endometrial tissues using methylation markers. (A) Independent replication of all genes (black line) and the top 8 candidate genes (red dashed line) from the discovery effort (including 148 carcinomas from PECS and 25 normal tissues from BRTE) in the replication study (including 69 carcinomas from EH and 40 normal tissue from BRTE). (B) Independent replication of seven candidate genes in prospectively collected endometrial brushings from women with and without endometrial carcinoma. Colored lines show ROC curves for individual genes and solid black line shows the ROC curve for all seven genes combined.
Table 3.
Methylation of eight genes in 444 endometrial carcinomas and 43 normal endometrial tissues from TCGA
| Endometrioid Carcinomas | Serous Carcinomas | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Normal tissues (n=43) | Cases (n=358) | p-value | AUC | Cases (n=86) | p-value | AUC |
| ADCYAP1 | 0.07 | 0.60 | <0.0001 | 0.97 (0.95–0.99) | 0.28 | <0.0001 | 0.77 (0.68–0.86) |
| ASCL2 | 0.12 | 0.55 | 0.0001 | 0.79 (0.75–0.83) | 0.08 | 0.28 | 0.44 (0.34–0.54) |
| CDH13 | 0.16 | 0.54 | <0.0001 | 0.95 (0.92–0.97) | 0.18 | 0.83 | 0.51 (0.41–0.61) |
| HS3ST2 | 0.12 | 0.55 | <0.0001 | 0.95 (0.93–0.97) | 0.24 | 0.0005 | 0.69 (0.60–0.78) |
| HTR1B | 0.12 | 0.56 | <0.0001 | 0.99 (0.98–1.00) | 0.27 | 0.0001 | 0.96 (0.94–0.99) |
| MME | 0.20 | 0.48 | <0.0001 | 0.85 (0.80–0.89) | 0.22 | 0.19 | 0.57 (0.47–0.68) |
| NPY | 0.10 | 0.57 | <0.0001 | 0.92 (0.89–0.94) | 0.20 | 0.02 | 0.63 (0.53–0.72) |
| SOX1 | 0.14 | 0.60 | <0.0001 | 0.99 (0.98–1.00) | 0.45 | <0.0001 | 0.96 (0.93–0.99) |
For each gene, average beta values of all CpG sites included on the TCGA high density methylation platform within 200bp of the sites evaluated in the discovery effort were evaluated (14). Mann-Whitney U p-values and area under the curve (AUC) estimates are shown for each carcinoma subgroup compared to normal tissues.
Evaluation of methylation markers in prospectively collected specimens
Pyrosequencing was performed to measure methylation of seven genes (ADCYAP1, ASCL2, CDH13, HS3ST2, HTR1B, MME, and NPY) in Tao brush samples and matched frozen tissues from women with endometrial carcinoma and from women with benign endometrium. Among women with carcinoma, for most genes, median methylation levels in frozen carcinoma tissues and Tao brush specimens were similar (Table 4). All genes had significantly higher methylation levels in Tao brush samples from endometrial carcinoma compared to Tao brush samples from benign endometrium (Table 4, Supplementary Table 4). Median methylation differences between endometrial carcinomas and benign endometrium ranged from 5.9 for HTR1B (p=0.008) to 23.9 (p<0.000001) for ADCYAP1 and odds ratios ranged from 3.6 (95%-CI: 1.35–9.47) for HTR1B to 10.9 (95%-CI: 3.23–36.91) for ADCYAP1. We stratified the analysis of methylation levels in Tao brush specimens by cancer stage and histologic type (Supplemental Table 5). Stratification resulted in very low numbers for some of the subgroups which limited the interpretation of the results. Carcinomas of all stages showed higher methylation levels compared to benign endometrium, but HTRB1 and NPY were not significantly elevated at higher stages. In analyses stratified by histologic type, methylation in Tao brush samples from both endometrioid and non-endometrioid carcinomas were higher compared to benign endometrium. In endometrioid carcinomas, all genes had significantly higher methylation levels compared to samples from benign endometrium. In non-endometroid carcinomas, ASCL2, CDH13, HS3ST2, and MME had significantly higher methylation levels compared to benign endometrium.
Table 4.
Performance of methylation markers in endometrial brush samples obtained from women with endometrial carcinoma and from women with benign uteri
| Median methylation levels | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer tissue | Brush cancer | Brush benign | Difference brush | p-value | OR | 95% CI | Area under the curve | 95% CI | |||
| ADCYAP1 | 19.4 | 35.0 | 11.1 | 23.9 | <0.00001 | 10.92 | 3.23 | 36.91 | 0.86 | 0.76 | 0.96 |
| ASCL2 | 19.9 | 19.2 | 8.1 | 11.1 | 0.00001 | 7.64 | 2.64 | 22.07 | 0.81 | 0.70 | 0.92 |
| CDH13 | 40.5 | 42.3 | 19.7 | 22.6 | <0.00001 | 9.33 | 2.85 | 30.60 | 0.86 | 0.76 | 0.96 |
| HS3ST2 | 25.5 | 25.3 | 10.7 | 14.6 | 0.00002 | 5.98 | 2.11 | 16.93 | 0.80 | 0.69 | 0.90 |
| HTR1B | 26.7 | 20.1 | 14.3 | 5.9 | 0.00827 | 3.57 | 1.35 | 9.47 | 0.68 | 0.55 | 0.81 |
| MME | 14.7 | 21.1 | 9.9 | 11.2 | <0.00001 | 10.56 | 3.04 | 36.67 | 0.86 | 0.76 | 0.96 |
| NPY | 34.1 | 23.6 | 12.6 | 11.0 | 0.00018 | 5.67 | 2.04 | 15.74 | 0.76 | 0.64 | 0.88 |
Mann-Whitney U p-values are shown for median differences in % methylation levels (based on pyrosequencing) in cancer and benign endometrial brush samples. Odds ratios are calculated using univariate logistic regression; dichotomization of methylation levels is based on distribution in controls, comparing high vs. medium/low tertiles. 95% CI= 95% confidence interval.
Performance of a DNA methylation marker panel to discriminate women with endometrial carcinoma from women without carcinoma
We calculated AUC values for all seven genes individually and for the combination of all seven markers. Five markers had an individual AUC of 0.80 or higher (ADCYAP1, ASCL2, CDH13, HS3ST2, and MME) and three markers (ADCYAP1, CDH13, and MME) had an AUC of 0.86 (Table 4). Using a cross-validation approach, we calculated an unbiased AUC estimate of 0.85 (Figure 2B). Thus, at a sensitivity of >90%, a specificity >50% can be achieved with individual or combinations of methylation markers.
Discussion
We conducted a biomarker discovery and validation study demonstrating that analysis of DNA methylation markers can be used for diagnosing endometrial carcinoma. By requiring that candidate DNA methylation markers demonstrate low values in benign tissues, large differences between carcinomas and benign tissues and highly statistically significant differences by disease status, we were able to identify an eight-marker panel with substantial discrimination. The sensitivity and specificity of top candidate methylation markers in our analysis were robust across three samples sets, whether performed in fixed or frozen tumor tissues, and were also reproducible in data from TCGA (14) and in novel endometrial brush samples prospectively collected from women with endometrial carcinoma and a comparable group without carcinoma. These data provide proof-of-principle that it may be possible to develop diagnostic molecular testing as an adjunct to the classification of endometrial biopsies or brushings performed to assess suspicious vaginal bleeding.
Several observations support the validity of our results. First, in addition to discovering new methylation markers for endometrial carcinoma, our agnostic approach identified well- established alterations, including methylation of MLH1 and CDH13 (13;16;24). Second, several of the novel endometrial carcinoma markers that we identified have been identified previously in other cancers (HS3ST2, (25); HTR1B, (26); MME, (27)) or involve important pathways in carcinogenesis such as Wnt signaling (ASCL2, (28)) and the Hedgehog pathway (ADCYAP1, (29)). In addition, NPY, one of our candidate markers, is proposed to function in energy balance and obesity (30;31), which is notable because elevated body mass is strongly and consistently related to increased endometrial carcinoma risk (32). Further studies are required to understand the role of these genes in endometrial carcinogenesis and to elucidate the etiologic heterogeneity of endometrial carcinomas reflected by the two groups of carcinomas with distinct methylation patterns observed in unsupervised hierarchical clustering.
The optimal method for investigating abnormal vaginal bleeding has been debated (6), but ultimately classification of a tissue or cellular sample is required for diagnosis. A recent study demonstrated that almost 25% of endometrial cancer cases had a preceding benign endometrial biopsy or curettage (33), suggesting that endometrial biopsy may have missed a prevalent carcinoma. Furthermore, pathological diagnosis is limited by both over- and under-estimation of disease severity, leading to over-treatment of some women and delayed diagnosis of others (8–10). Given that the incidence of endometrial carcinoma and its precursors may rise in future years, likely reflecting dramatic increases in the prevalence of obesity (34), a strong endometrial carcinoma risk factor, improved methods for triaging women according to risk may having growing utility.
An approach similar to ours has been previously applied to vaginal tampons, which offers a novel approach to facilitate early clinical triage of women presenting with abnormal vaginal bleeding (35). In the prior report, assessment of a five-marker methylation panel to assess DNA extracted from vaginal tampons collected from women with abnormal vaginal bleeding demonstrated high sensitivity and specificity for the detection of endometrial carcinoma. Two of the five genes included in that panel, RASSF1A and CDH13 ranked highly in our profiling analysis, and the latter was included in our seven-gene replication analysis. Thus, data suggest that the specificity and robustness of DNA methylation markers may be suitable for testing both conventional tissue samples, as well as novel specimen types. Our study included the evaluation of a novel type of endometrial sample obtained by brushing, which has been suggested to provide a more representative sample than biopsy, without the discomfort associated with a biopsy (18;36). With our approach, we were able to detect carcinomas of all stages and we also detected non-endometrioid carcinomas, which are associated with a worse prognosis compared to the endometrioid carcinomas. It will be important to compare the performance of the methylation markers to routine cytological evaluation of Tao brush samples to evaluate whether methylation testing provides additional information; this is currently ongoing. Furthermore, as an extension of the current effort, we are now evaluating the methylation marker candidates in Tampon samples that have the advantage of less invasive sampling procedures.
Strengths of our study include assessment of agnostic profiling for marker discovery, evaluation of endometrial carcinomas collected in two population-based studies, comparisons with epidemiologically annotated benign endometrial tissues, analysis of multiple sample types and independent validation of specific markers using the TCGA resource and a novel brushing method with gene-specific pyrosequencing assays. A potential limitation of our efforts was that there were imbalances in age of the participants across the different studies, however, age-adjusted models led to the same conclusions. The DNA methylation platform included many tumor suppressor genes that are strongly implicated in cancer and we successfully replicated markers in independent replication studies; however, newer methylation profiling platforms have wider CpG site coverage, suggesting that there may be additional candidates worthy of evaluation. The results from our cross-sectional study justify evaluating methylation markers as an adjunct to the diagnosis of endometrial carcinoma (37). In ongoing discovery efforts, we are exploring higher density DNA methylation profiling platforms, addition of DNA mutation analysis and evaluation of a greater number of type II endometrial carcinomas, such as serous carcinomas, which are comparatively uncommon, but portend a worse prognosis. Addition of PTEN and p53 mutational analysis, which are detectable in a high percentage of endometrioid and serous carcinomas, respectively, could further increase the sensitivity of this approach.
In summary, we conducted a rigorous tissue biomarker discovery effort to identify DNA methylation markers associated with endometrial carcinoma, and then replicated the findings in an independent set of samples collected using a novel brushing method. This proof of principle study establishes the promise of adding molecular biomarkers to conventional methods of evaluating women with abnormal vaginal bleeding as an approach for distinguishing women likely to harbor endometrial carcinoma from the vast majority of women who have findings unrelated to neoplastic lesions.
Supplementary Material
Supplemental figure 1: Supervised heatmap of methylation levels of the candidates in the discovery and validation sets.
Samples were clustered with supervision: Case-control pairs are shown for the discovery phase and the validation phase. Probes were clustered hierarchically without supervision using Euclidean distance.
Supplementary Table 1: Key characteristics of included studies
Supplementary Table 2: Top 50 probes with highest mean methylation differences between benign endometrial carcinoma and benign endometrium
Supplementary table 3: Crude and age adjusted odds ratios for all probes representing the 8 candidate genes in the discovery set, replication set, and the combined set
Supplementary table 4: Performance of individual CpG sites measured in endometrial brushings to discriminate women with carcinoma from women with benign endometrium
Supplemental Table 5: Methylation marker levels in endometrial brush samples stratified by endometrial carcinoma stage and histologic type
Novelty statement.
Using a rigorous discovery and replication effort, we identified methylation marker candidates for early detection of endometrial cancer. We successfully translated promising candidates to a study of endometrial brushings. This study establishes the promise of adding methylation biomarkers to conventional methods of evaluating women with abnormal vaginal bleeding to distinguish women with endometrial carcinoma from the majority of women with benign postmenopausal bleeding.
Acknowledgments
This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health.
Footnotes
The authors have declared no conflicts of interest.
References
- 1.American Cancer Society. Cancer Facts and Figures 2012. 2012 [Google Scholar]
- 2.Wright JD, Barrena Medel NI, Sehouli J, Fujiwara K, Herzog TJ. Contemporary management of endometrial cancer. Lancet. 2012;379:1352–60. doi: 10.1016/S0140-6736(12)60442-5. [DOI] [PubMed] [Google Scholar]
- 3.Kosary CL. Cancer of the Corpus Uteri. In: Gloeckler Ries LA, Young JL, Keel GE, Eisner MP, Lin YD, Horner MD, editors. SEER Survival Monograph: Cancer Survival Among Adults 2007. pp. 123–32. [Google Scholar]
- 4.Lacey JV, Jr, Ioffe OB, Ronnett BM, Rush BB, Richesson DA, Chatterjee N, Langholz B, Glass AG, Sherman ME. Endometrial carcinoma risk among women diagnosed with endometrial hyperplasia: the 34-year experience in a large health plan. Br J Cancer. 2008;98:45–53. doi: 10.1038/sj.bjc.6604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacey JV, Jr, Sherman ME, Rush BB, Ronnett BM, Ioffe OB, Duggan MA, Glass AG, Richesson DA, Chatterjee N, Langholz B. Absolute risk of endometrial carcinoma during 20-year follow-up among women with endometrial hyperplasia. J Clin Oncol. 2010;28:788–92. doi: 10.1200/JCO.2009.24.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van HN, Breijer MC, Khan KS, Clark TJ, Burger MP, Mol BW, Timmermans A. Diagnostic evaluation of the endometrium in postmenopausal bleeding: an evidence-based approach. Maturitas. 2011;68:155–64. doi: 10.1016/j.maturitas.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 7.van HN, Breijer MC, Opmeer BC, Mol BW, Timmermans A. Prediction models in women with postmenopausal bleeding: a systematic review. Womens Health (Lond Engl) 2012;8:251–62. doi: 10.2217/whe.12.10. [DOI] [PubMed] [Google Scholar]
- 8.Sherman ME, Ronnett BM, Ioffe OB, Richesson DA, Rush BB, Glass AG, Chatterjee N, Duggan MA, Lacey JV., Jr Reproducibility of biopsy diagnoses of endometrial hyperplasia: evidence supporting a simplified classification. Int J Gynecol Pathol. 2008;27:318–25. doi: 10.1097/PGP.0b013e3181659167. [DOI] [PubMed] [Google Scholar]
- 9.Trimble CL, Kauderer J, Zaino R, Silverberg S, Lim PC, Burke JJ, Alberts D, Curtin J. Concurrent endometrial carcinoma in women with a biopsy diagnosis of atypical endometrial hyperplasia: a Gynecologic Oncology Group study. Cancer. 2006;106:812–9. doi: 10.1002/cncr.21650. [DOI] [PubMed] [Google Scholar]
- 10.Trimble CL, Method M, Leitao M, Lu K, Ioffe O, Hampton M, Higgins R, Zaino R, Mutter GL. Management of endometrial precancers. Obstet Gynecol. 2012;120:1160–75. doi: 10.1097/aog.0b013e31826bb121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bach PB, Schrag D, Brawley OW, Galaznik A, Yakren S, Begg CB. Survival of blacks and whites after a cancer diagnosis. JAMA. 2002;287:2106–13. doi: 10.1001/jama.287.16.2106. [DOI] [PubMed] [Google Scholar]
- 12.Gallicchio L, Harvey LA, Kjerulff KH. Fear of cancer among women undergoing hysterectomy for benign conditions. Psychosom Med. 2005;67:420–4. doi: 10.1097/01.psy.0000160472.69303.56. [DOI] [PubMed] [Google Scholar]
- 13.Jiang SW, Li J, Podratz K, Dowdy S. Application of DNA methylation biomarkers for endometrial cancer management. Expert Rev Mol Diagn. 2008;8:607–16. doi: 10.1586/14737159.8.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih I, Kurman R, Dao F, Levine DA, Giuntoli R, Roden R, Eshleman JR, et al. Evaluation of DNA from the papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5:167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou XC, Dowdy SC, Podratz KC, Jiang SW. Epigenetic considerations for endometrial cancer prevention, diagnosis and treatment. Gynecol Oncol. 2007;107:143–53. doi: 10.1016/j.ygyno.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Brinton LA, Sakoda LC, Lissowska J, Sherman ME, Chatterjee N, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Garcia-Closas M. Reproductive risk factors for endometrial cancer among Polish women. Br J Cancer. 2007;96:1450–6. doi: 10.1038/sj.bjc.6603731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kipp BR, Medeiros F, Campion MB, Distad TJ, Peterson LM, Keeney GL, Halling KC, Clayton AC. Direct uterine sampling with the Tao brush sampler using a liquid-based preparation method for the detection of endometrial cancer and atypical hyperplasia: a feasibility study. Cancer. 2008;114:228–35. doi: 10.1002/cncr.23636. [DOI] [PubMed] [Google Scholar]
- 19.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–63. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 20.Killian JK, Bilke S, Davis S, Walker RL, Killian MS, Jaeger EB, Chen Y, Hipp J, Pittaluga S, Raffeld M, Cornelison R, Smith WI, Jr, et al. Large-scale profiling of archival lymph nodes reveals pervasive remodeling of the follicular lymphoma methylome. Cancer Res. 2009;69:758–64. doi: 10.1158/0008-5472.CAN-08-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 22.Munoz JF, Rueda M. New imputation methods for missing data using quantiles. Journal of Computational and Applied Mathematics. 2009;232:305–17. [Google Scholar]
- 23.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 24.Nieminen TT, Gylling A, Abdel-Rahman WM, Nuorva K, Aarnio M, Renkonen-Sinisalo L, Jarvinen HJ, Mecklin JP, Butzow R, Peltomaki P. Molecular analysis of endometrial tumorigenesis: importance of complex hyperplasia regardless of atypia. Clin Cancer Res. 2009;15:5772–83. doi: 10.1158/1078-0432.CCR-09-0506. [DOI] [PubMed] [Google Scholar]
- 25.Shivapurkar N, Sherman ME, Stastny V, Echebiri C, Rader JS, Nayar R, Bonfiglio TA, Gazdar AF, Wang SS. Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecol Oncol. 2007;107:549–53. doi: 10.1016/j.ygyno.2007.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takai D, Yagi Y, Wakazono K, Ohishi N, Morita Y, Sugimura T, Ushijima T. Silencing of HTR1B and reduced expression of EDN1 in human lung cancers, revealed by methylation-sensitive representational difference analysis. Oncogene. 2001;20:7505–13. doi: 10.1038/sj.onc.1204940. [DOI] [PubMed] [Google Scholar]
- 27.Archer KJ, Mas VR, Maluf DG, Fisher RA. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol Genet Genomics. 2010;283:341–9. doi: 10.1007/s00438-010-0522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Sousa E, Melo Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, Rodermond H, Dekker E, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476–85. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Cohen JR, Resnick DZ, Niewiadomski P, Dong H, Liau LM, Waschek JA. Pituitary adenylyl cyclase activating polypeptide inhibits gli1 gene expression and proliferation in primary medulloblastoma derived tumorsphere cultures. BMC Cancer. 2010;10:676. doi: 10.1186/1471-2407-10-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Bijker MS, Herzog H. The neuropeptide Y system: pathophysiological and therapeutic implications in obesity and cancer. Pharmacol Ther. 2011;131:91–113. doi: 10.1016/j.pharmthera.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Nguyen AD, Lee IC, Yulyaningsih E, Riepler SJ, Stehrer B, Enriquez RF, Lin S, Shi YC, Baldock PA, Sainsbury A, Herzog H. NPY modulates PYY function in the regulation of energy balance and glucose homeostasis. Diabetes Obes Metab. 2012;14:727–36. doi: 10.1111/j.1463-1326.2012.01592.x. [DOI] [PubMed] [Google Scholar]
- 32.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–78. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 33.Torres ML, Weaver AL, Kumar S, Uccella S, Famuyide AO, Cliby WA, Dowdy SC, Gostout BS, Mariani A. Risk factors for developing endometrial cancer after benign endometrial sampling. Obstet Gynecol. 2012;120:998–1004. doi: 10.1097/aog.0b013e31826b9fef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans T, Sany O, Pearmain P, Ganesan R, Blann A, Sundar S. Differential trends in the rising incidence of endometrial cancer by type: data from a UK population-based registry from 1994 to 2006. Br J Cancer. 2011;104:1505–10. doi: 10.1038/bjc.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widschwendter A, Gattringer C, Ivarsson L, Fiegl H, Schneitter A, Ramoni A, Muller HM, Wiedemair A, Jerabek S, Muller-Holzner E, Goebel G, Marth C, et al. Analysis of aberrant DNA methylation and human papillomavirus DNA in cervicovaginal specimens to detect invasive cervical cancer and its precursors. Clin Cancer Res. 2004;10:3396–400. doi: 10.1158/1078-0432.CCR-03-0143. [DOI] [PubMed] [Google Scholar]
- 36.Wu HH, Harshbarger KE, Berner HW, Elsheikh TM. Endometrial brush biopsy (Tao brush). Histologic diagnosis of 200 cases with complementary cytology: an accurate sampling technique for the detection of endometrial abnormalities. Am J Clin Pathol. 2000;114:412–8. doi: 10.1093/ajcp/114.3.412. [DOI] [PubMed] [Google Scholar]
- 37.Wentzensen N, Wacholder S. From differences in means between cases and controls to risk stratification: a business plan for biomarker development. Cancer Discov. 2013;3:148–57. doi: 10.1158/2159-8290.CD-12-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1: Supervised heatmap of methylation levels of the candidates in the discovery and validation sets.
Samples were clustered with supervision: Case-control pairs are shown for the discovery phase and the validation phase. Probes were clustered hierarchically without supervision using Euclidean distance.
Supplementary Table 1: Key characteristics of included studies
Supplementary Table 2: Top 50 probes with highest mean methylation differences between benign endometrial carcinoma and benign endometrium
Supplementary table 3: Crude and age adjusted odds ratios for all probes representing the 8 candidate genes in the discovery set, replication set, and the combined set
Supplementary table 4: Performance of individual CpG sites measured in endometrial brushings to discriminate women with carcinoma from women with benign endometrium
Supplemental Table 5: Methylation marker levels in endometrial brush samples stratified by endometrial carcinoma stage and histologic type