Abstract
To study the role of late secretion in Candida albicans pathogenesis, we created conditional mutant C. albicans strains in which the t-SNARE-encoding genes SSO2 or SEC9 were placed under the control of a tetracycline-regulated promoter. In repressing conditions, C. albicans tetR-SSO2 and tetR-SEC9 mutant strains were defective in cytokinesis and secretion of aspartyl proteases and lipases. The mutant strains also exhibited a defect in filamentation compared to controls, and thus we followed the fate of the C. albicans Spitzenkörper, an assembly of secretory vesicles thought to act as a vesicle-supply center for the growing hyphae. In the absence of Ca Sso2p, the Spitzenkörper dissipated within 5 hours and thin-section electron microscopy revealed an accumulation of secretory vesicles. Moreover, the hyphal tip developed into a globular yeast-like structure rather than maintaining a typical narrow hyphae. These studies indicate that late secretory t-SNARE proteins in C. albicans are required for fundamental cellular processes and contribute to virulence-related attributes of C. albicans pathogenesis. Moreover, these results provide direct evidence for a key role of SNARE proteins in vesicle-mediated polarized hyphal growth of C. albicans.
Keywords: SNARE, Spitzenkörper, aspartyl protease, lipase, hyphae
INTRODUCTION
In prior studies, we have investigated the role of the pre-vacuolar secretory pathway, regulated by the vacuolar protein sorting genes VPS1, VPS4, and PEP12, in the secretion of virulence-associated proteins and biofilm formation in C. albicans (Lee, et al., 2007, Bernardo, et al., 2008, Lee, et al., 2009, Palanisamy, et al., 2010). These investigations have demonstrated that pre-vacuolar secretion plays a key role in the pathogenesis of C. albicans. In order to expand our understanding of the contribution of C. albicans secretion to pathogenesis, we have now focused our studies on trafficking genes involved in exocytosis.
The final stages of secretion leading to exocytosis depend on the fusion of late secretory vesicles to the plasma membrane, which is regulated by the exocyst complex. This octameric complex mediates tethering of secretory vesicles to the plasma membrane, followed by membrane fusion enabled by assembly and disassembly of a SNARE complex. In the model yeast Saccharomyces cerevisiae, the exocyst is encoded by SEC3, SEC5, SEC6, SEC8, SEC15, EXO70, and EXO84 (TerBush, et al., 1996). Once late secretory vesicles reach the plasma membrane, assembly of the SNARE complex occurs, whereas in yeast exocyst mutants, SNARE assembly fails to occur (Grote, et al., 2000). The SNARE complex is composed of the Snc1/2 v-SNARE proteins on the secretory vesicles and Sso1/2 and Sec9 t-SNARE proteins on the plasma membrane (Gerst, 1999). Snc1/2p and Sso1/2p each contribute one helix and Sec9p contributes two helices to the SNARE complex (Sutton, et al., 1998). In vitro studies have indicated that a hetero-oligomeric complex of Sso1p and Sec9p binds Snc1p rather than the individual t-SNARE proteins (Rossi, et al., 1997). Tethering of the vesicle to the exocyst occurs first and is required for subsequent SNARE assembly, which then permits the “zipping” together of each plasma membrane to allow vesicle fusion and final exocytosis to occur.
Although S. cerevisiae SSO1 and SSO2 were generated evolutionarily by whole genome duplication, this duplication did not occur in C. albicans. In S. cerevisiae, the duplicated genes SSO1 and SSO2 play a key role in secretion. While neither null mutant in S. cerevisiae is inviable, the double null mutant is synthetic lethal. A S. cerevisiae temperature-sensitive mutant lacking functional Sso1/2p accumulates secretory vesicles and is defective in invertase secretion (Aalto, et al., 1993). Although Sso1p and Sso2p have substantial functional overlap, SSO1 is required for sporulation whereas SSO2 is not, thus indicating an important divergence in function (Jantti, et al., 2002).
In S. cerevisiae, SEC9 and SPO20 were also generated by whole genome duplication; however, Sec9p and Spo20p are highly divergent and share only 37% identity. Spo20p is required for sporulation only, whereas Sec9p is required for both secretion during vegetative growth and during sporulation (Kienle, et al., 2009). In contrast, C. albicans has only one SEC9/SPO20 homolog; this homolog is more similar to SEC9 than to SPO20. Strains carrying mutations in S. cerevisiae SEC9 were first isolated in studies of the yeast secretory pathway (Novick, et al., 1980). These temperature-sensitive strains were defective in invertase secretion and accumulated membrane-bound secretory organelles. Functional analyses of Sec9p reveal that the functional domain is found at the C-terminal end of the protein (Brennwald, et al., 1994). The temperature-sensitive sec9-4 strain carries a Gly458Asp mutation that reduces its ability to complex with either Sso1/2p or Snc1/2p, and thus disrupts the formation of the SNARE complex (Brennwald, et al., 1994). Interestingly, functional studies by Rossi and colleagues (Rossi, et al., 1997) provide evidence for an additional function of SEC9 that is independent of SNARE formation. The sec9-7 and sec9-Δ17 alleles result in temperature-sensitive and lethal phenotypes, respectively, but retain the ability to form SNARE complexes in vitro. These mutant alleles act in a dominant-negative manner when expressed under a strong promoter suggesting that SNARE complex formation also occurs in vivo (Rossi, et al., 1997). Thus, while these altered Sec9p are able to bind with Sso1/2p and Snc1/2p to form the SNARE complex, they lack a function of Sec9p that is required for wild-type cell growth. Although this specific role for Sec9p has yet to be explicitly defined, Williams and Novick (2009) have suggested a constitutive role that is intimately linked to the physiological state of the cell.
To study the final stages of secretion in C. albicans, we identified close structural homologs of several S. cerevisiae SNARE complex genes. We present here the results of our studies on the t-SNAREs encoded by C. albicans SSO2 and SEC9. We performed complementation studies in yeast, followed by more detailed analysis of conditional mutant strains in C. albicans in order to define the role of late secretion in key aspects of Candida cell biology and pathogenesis.
MATERIALS AND METHODS
Strains and media
All strains used in this study are listed in Table 1. Strains were grown at 30 °C in YPD (1% yeast extract, 2% peptone, 2% glucose) or complete synthetic medium (CSM: 0.67% yeast nitrogen base without amino acids [YNB], 2% glucose, 0.079% Complete Synthetic Mixture), supplemented with uridine (80 μg ml−1) as required. Cells were expanded in liquid YPD overnight at 30 °C with shaking at 250 rpm prior to use in all phenotypic characterizations/assays. Uracil auxotrophs were selected on 5-fluoro-orotic acid (FOA) agar medium (complete synthetic medium [CSM] supplemented with 0.1 mg ml−1 uridine and 0.7 mg FOA ml−1). Doxycycline (DOX) was added to a final concentration of 20 μg ml−1 where required. Solid media were prepared by adding 2% (w/v) agar. S. cerevisiae sso1Δ sso2-1 (gift from J. Jantti, University of Helsinki, Finland) and sec9-4 (gift from P. Novick, UCSD, San Diego, CA) are temperature-sensitive at 30 and 35 °C, respectively (Novick, et al., 1980, Brennwald, et al., 1994, Jantti, et al., 2002).
Table 1.
Candida albicans strains used in this study
| Strain Name | Parent | Genotype | Source |
|---|---|---|---|
| H603 | sso1-Δ1::HIS3 sso2-1 ura3-1 | (Jantti, et al., 2002) | |
| NY57 | MATa ura3-52 sec9-4 | (Finger & Novick, 2000) | |
| SC5314 | wild-type | ||
| THE1 | CAI8 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 SSO2/SSO2 SEC9/SEC9 |
(Nakayama, et al., 2000) |
| THE1-CIp10 | THE1 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 RP10/RP10::URA3 SSO2/SSO2 SEC9/SEC9 |
(Bernardo, et al., 2008) |
| SSO2 Δ/+ | THE1 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 SSO2/sso2Δ::dpl200-URA3-dpl200 |
This study |
| SEC9 Δ/+ | THE1 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 SEC9/sec9Δ::dpl200-URA3-dpl200 |
This study |
| SSO2 Δ/+ FOA | SSO2 Δ/+ |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 SSO2/sso2Δ::dpl200 |
This study |
| SEC9 Δ/+ FOA | SEC9 Δ/+ |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 SEC9/sec9Δ::dpl200 |
This study |
| tetR-SSO2 | SSO2 Δ/+ FOA |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 tetR-URA3-SSO2/sso2Δ::dpl200 |
This study |
| tetR-SEC9 | SEC9 Δ/+ FOA |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 tetR-URA3-SEC9/sec9Δ::dpl200 |
This study |
| T-M1gfp | THE1-CIp10 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 RP10/RP10::URA3 SSO2/SSO2 SEC9/SEC9 |
This study |
| tS2-M1gfp | tetR-SSO2 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 tetR-URA3-SSO2/sso2Δ::dpl200 MLC1/MLC1-GFP::NAT1 |
This study |
| tS9-M1gfp | tetR-SEC9 |
ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 tetR-URA3-SEC9/sec9Δ::dpl200 MLC1/MLC1-GFP::NAT1 |
This study |
Preparation of plasmid and genomic DNA
Plasmids were expanded in Escherichia coli DH5α competent cells (Invitrogen, Carlsbad, CA) grown in LB medium with ampicillin (100 μg ml−1) at 37 °C. Plasmid DNA was prepared from E. coli strains using the PureYield™ Plasmid Miniprep System (Promega, Madison, WI) following the manufacturer’s instructions. Genomic DNA was extracted from yeast cells using the MasterPure™ Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer’s instructions with the exception of an additional incubation step (1 hr on ice) performed after the addition of the MPC Protein Precipitation Reagent (Bernardo, et al., 2008).
Extraction of mRNA and Reverse-Transcriptase PCR analysis
Overnight cultures were used to prepare cultures at a starting OD600nm of 0.1 in YPD, in the presence or absence of DOX. Following incubation at 30 °C, with shaking at 250 rpm, cells were harvested and total RNA was extracted from 108 cells using the RiboPure™ Yeast RNA kit (Ambion®, Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. The integrity of total RNA was assessed on 1% denaturing agarose gel as described previously (Bernardo, et al., 2008). Next, mRNA was prepared from 1 μg of total RNA using the PolyATtract® mRNA Isolation System (Promega, Madison, WI) and quantified on a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, Delaware). Reverse-transcriptase PCR was performed with Promega’s AccessQuick™ RT-PCR System (Promega, Madison, WI) according to the manufacturer’s instructions using primers sets SSO2-5RT and SSO2-3RT or SEC9-5RT and SEC9-3RT, and 50 ng mRNA as template, to amplify SSO2 and SEC9 transcripts, respectively. The absence of contaminating DNA was tested in parallel reactions in which the reverse transcriptase enzyme was excluded.
Promoter replacement strategy in C. albicans
We first identified C. albicans orthologs of S. cerevisiae SSO1 (SSO2) and SEC9 from a BLASTP search of the Candida Genome Database (Assembly 21) (http://www.candidagenome.org). The search revealed a single ortholog of the S. cerevisiae t-SNAREs Sso1p and Sso2p in C. albicans, orf19.1376, which encodes a 295-aa protein whose sequence shares 50.5% (52.8%) identity and 67.8% (69.1%) similarity with S. cerevisiae Sso1p and Sso2p, respectively. Similarly, a 530-aa Sec9p ortholog (orf19.117) sharing 47% identity and 64% similarity with S. cerevisiae was identified in C. albicans. Next, one allele of each gene of interest was deleted in strain THE1 (Nakayama, et al., 2000), using primers YFG-5DR and YFG-3DR (Table 2), according to the PCR-based gene disruption method described previously (Wilson, et al., 2000) to generate heterozygous null mutant strains, YFG/yfgΔ (YFG/yfgΔ::dpl200- URA3-dpl200) (Table 1). To regenerate ura3Δ auxotrophy, the YFG/yfgΔ strains were plated onto solid FOA medium containing uridine. Next, FOA-resistant strains were screened by PCR for the YFG/yfgΔ::dpl200 genotype (Table 1). The regulatable tetracycline promoter was inserted upstream of the remaining YFG allele according to the PCR-based strategy described previously (Bates, et al., 2006) and utilizing primers TetYFG-5DR and TetYFG-4DR. Transformants carrying the tetR-YFG allele were screened using primers TetYFG-5Det and TetYFG-3Det. All other transformants were screened for the genotype of interest using primers YFG-5Det and YFG-3Det. The correct genotype of these strains was subsequently confirmed by Southern blot analysis following standard protocols (Ausubel, et al., 1987). Table 2 lists primers used in this study.
Table 2.
Primer sequences used in this study
| Primer | Primer sequence (5′-3′) |
|---|---|
| ScSSO2-5BamHI | CCACAGGGATCCAGATTTGGATCTAGGTCCGCTATT |
| ScSSO2-3HindIII | GTTCAGAAGCTTGGGGGCAAAAAGAATCACGGGACT |
| ScSEC9-5BamHI | CATTTGGCTGTGGCGGTTTA |
| ScSEC9-3HindIII | GTTCAGAAGCTTCTTACTGAGAGGTTTTCCTC |
| CaSSO2-5BamHI | CCACAGGGATCCACTAAACTGTAATGTCAATT |
| CaSSO2-3HindIII | GTTCAGAAGCTTAGTCAACAACAATTAGTGTG |
| CaSEC9-5BamHI | CCACAGGGATCCTACTTTCATAAATAGTGATT |
| CaSEC9-3HindIII | GTTCAGAAGCTTATTTATGAAGAGAGTTTTCT |
| SSO2-5DRs | AGTTTGATTTTTCACTTTTCATTTTTTTGGTTTTTACATTCATATCATTCAACTTGACGTTTATTTAACTAGAAACAACCGTTTTCCCAGTCACGACGTT |
| SSO2-3DRs | CCAAAAGACAATACTATTTTCGTGCTTGCTTAGATAACCACAATCAAGTGCATAAAAATAAAACTACGATACTGTGGAATTGTGAGCGGATA |
| SEC9-5DR | ACTAATCTCAAGAACTAACTTACTTCCTTTTATAATACTACAACAACATATATTATACAACTAATCAAAGGTTTTCCCAGTCACGACGTT |
| SEC9-3DR | ATAATTTATTTCCTATTCTTTTTATTAACAAACCTTATACATATATATATTGTTTAAATTGTTTTTTTGT TGTGGAATTGTGAGCGGATA |
| SSO2-5DetS | GTGCCAAATCAGATATCGCC |
| SSO2-3DetS | TCAACACTTTCCATCGCAGC |
| SEC9-5Det | CCTTCTACAAACCATTCAGA |
| SEC9-3Det | CTTTTCAAGCCACCCCCACC |
| tetSSO2-5DRs | CGGGGGCGGAGGGGAAGGCAAATTAAAATTACCTTCCATTGTTTCCTACAAATGAGCTGCGTCGTTCCTGGTAATACGACTCACTATAGGG |
| tetSSO2-3DRs | GGATAATTGTTCAACTCATATGAGTTGTTTTGTTGATAACCATTCTGGGAATTCTGATACGGGTTGCTCATGGTTGCTAGTTTTCTGAGATAAAGCTG |
| tetSEC9-5DR | TCAACAAACTAATCTCAAGAACTAACTTACTTCCTTTTATAATACTACAACAACATA TATTATACAACTAGTAATACGACTCACTATAGGG |
| tetSEC9-3DR | CTCGACTTAATTCTTCACGAATTTCTTGTTCGGTTGGTTCTTTCTTTTGAAACATTTTCTTGATACCCATCTAGTTTTCTGAGATAAAGCTG |
| tetSSO2-5DetS | TGTACGACATGACGGTGGTC |
| tetINS-3Det | CTAGTTTTCTGAGATAAAGCTG |
| tetSEC9-5Det | CATCACGTCCCACGGTATAA |
| tetSEC9-3Det | GATCTTGTAGAAATGCCA |
| SSO2-5SB | CAATGGAGTCAACGGTTGTG |
| SSO2-3SB | CAGGAACGACGCAGCTCATT |
| SEC9-3SB | AGGACTAGGTGATGATCCTC |
| SSO2-5RT | CAACCCGTATCAGAATTCCC |
| SSO2-3RT | TAGTACCGAAGTATGCACCC |
| SEC9-5RT | CGACAAGAGAAGTTTGGTGC |
| SEC9-3RT | CGTTCCAGTTGCGATCCTAA |
| MLC1-GFP-5DR | AAAAGGGGTCAATGTAACTTCTGATGGAAATGTGGATTATGTTGAATTTGTCAAATCAATTTTAGACCAAGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| MLC1-GFP-3DR | TCAAGTACTACATAAAACTTCAAATAAACGGTATCCAATTCGAACAAGACTATACAATAACTATAATTTGCGTTAGTATCGAATCGACAGC |
| MLC1-5Det | GATTCCAAGGTGTCAACTTTC |
| MLC1-3Det | GGCATATATTACTCTCCAAAG |
Complementation studies in S. cerevisiae
High-fidelity PCR was used to amplify each open reading frame of interest from wild-type yeast strains, including upstream and downstream untranslated regions, and the resulting amplicons were subcloned into pCRII-TOPO (Invitrogen, Carlsbad, CA). The primers used included linker sequences to introduce BamHI and HindIII recognition sites at the end of each amplicon, with the exception of ScSEC9-5BamHI whose amplicon had an intrinsic BamHI site in the 5′ untranslated region (Table 2). Sequences −500 to +512, −549 to +429, −800 to +800, and −500 to +500 of S. cerevisiae SSO2 (YPL232W) and SEC9 (YGR009C), and C. albicans SSO2 (orf19.1376) and SEC9 (orf19.117), respectively, were cut and gel purified from each pCRII-TOPO clone with BamHI and HindIII and subsequently cloned into pRS316 (Sikorski & Hieter, 1989) linearized with the same set of restriction enzymes. The resulting plasmids, pRS-ScSSO2 and pRS-CaSSO2, pRS-ScSEC9 and pRS-CaSEC9 were each used to transform the temperature-sensitive mutant strains S. cerevisiae sso1Δ sso2-1 and sec9-4, respectively, using the lithium acetate method (Ausubel, et al., 1987). Cells were plated on complete synthetic medium (CSM) without uracil, supplemented with adenine (20 μg ml−1), and incubated at room temperature. Transformants were screened by colony PCR (with T7 promoter and M13 reverse primers) to confirm the presence of each pRS316 clone. Control strains in which each S. cerevisiae mutant carried the empty vector pRS316 were also generated. The ability of the C. albicans homologs to complement the temperature-sensitivity of the corresponding S. cerevisiae mutant strains were then tested by streaking strains bearing pRS-CaSSO2 (pRS-ScSSO2), pRS-CaSEC9 (pRS-ScSEC9), and pRS316 on CSM lacking uracil, supplemented with adenine, and assessed for growth for 2 days at the restrictive and permissive temperatures.
Analysis of growth
Growth of the conditional mutant and control strains was assessed in both liquid and agar medium. Growth on agar medium was carried out by spotting serial dilutions of cells on CSM with supplemented with uridine, in the presence or absence of doxycycline, where cells were prepared as described previously (Rane, et al., 2013). The plates were then incubated at 30 °C for 48 hours. Growth in liquid media was carried out by measuring OD600nm at fixed intervals, after the overnight strains grown in YPD were washed and transferred to fresh CSM supplemented with uridine (in the presence or absence of DOX) and diluted to a starting OD600nm of 0.1. Optical densities were measured at given hourly intervals and growth curves were generated using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, California). In addition, at each time point, cells were counted using a haemocytometer and a fixed number of cells were plated, in triplicate, onto YPD agar to determine colony forming units. Determination of colony forming units was performed at least twice, independently.
Thin-section electron microscopy
Yeast cells were prepared as follows: standard overnight cultures were used to inoculate cultures in YPD, in the presence or absence of DOX, which were incubated at 30 °C with shaking at 250 rpm for 5 hours. Filamentous cells were prepared following the filamentation assay described below, incubated for 5 hours. Following incubation, cells were collected by centrifugation and washed with 1x Phosphate Buffered Saline (PBS), then resuspended in 1ml of fixative [0.1M phosphate buffered 1% (v/v) glutaraldehyde - 4% (v/v) formaldehyde, pH 7.2] and incubated at room temperature for one hour. Subsequent fixation, dehydration, and staining with uranyl (acetate) and lead (acetate) are described previously (Kushida, 1961, Venable & Coggeshall, 1965, Glauert, 1975, Bozzola & Russell, 1992). Images were acquired using a JEOL 1230 transmission electron microscope (JEOL USA, Inc., Peabody, MA) operating at 80kV.
Assay for secreted aspartyl protease
Liquid assays for secreted aspartyl proteases were performed as follows: Cells from 5-ml overnight YPD cultures were washed and resuspended in 30-ml of BSA medium (0.34% (w/v) YNB without amino acids and without ammonium sulfate, 2% (w/v) glucose, 0.1% (w/v) BSA) and incubated for 24 hours to induce production of Sap2p. Next, spent medium was removed and the cells were resuspended to a final OD600nm of 30, with fresh BSA medium containing only 0.01% (w/v) BSA. The cell suspension was divided equally to which DOX was added to one of the two, and cultures were incubated at 30 °C for 5 hours with shaking at 250 rpm. Cell-free culture supernatant was taken for SDS-PAGE analyses as described previously (Bernardo, et al., 2008).
Assay for lipase secretion
The assay performed for secreted lipases is a modification of the turbidimetric esterase assay developed by von Tigerstrom and Stelmaschuck (von Tigerstrom & Stelmaschuk, 1989). First, secretion of lipase was induced by growing cells from a standard overnight culture (diluted 1:100) in Tween 80 medium (0.54% (w/v) YNB without amino acids, 2.5% (v/v) Tween 80). Following incubation at 37 °C for 24 hours with shaking at 250 rpm, cells were washed twice in 1XPBS, concentrated to a final OD600nm of 10 in Tween 80 medium, and incubated for 5 hours in the presence or absence of doxycycline at 37 °C with shaking (250rpm). Supernatants from the 5-hour cultures were then tested for secreted lipases by performing a kinetic assay as follows: 500μl of cell-free supernatant was added to 5 ml of Tween 20 Substrate (2% (v/v) Tween 20 in 20mM Tris-HCl, pH 8.0 and 120 mM CaCl2). The mixture was then incubated at 37 °C with shaking at 250 rpm, with OD500nm readings taken every 15 minutes for up to 75 minutes. Tween 20 substrate, without lipase, was similarly treated and was used as a control to correct for background absorbance of the substrate at each timepoint. The kinetic profiles of the secreted lipases from each condition were visualized and compared by plotting the OD500nm readings over time. The lipase activity is directly proportional to the amount of enzyme in the reaction (von Tigerstrom & Stelmaschuk, 1989).
Filamentation assays
Filamentation was assayed at 37 °C in liquid RPMI-1640 supplemented with L-glutamine (Cellgro, Manassas, VA) and buffered with 165 mM MOPS (Sigma, St. Louis, MO) to pH 7.0 (referred to as “buffered RPMI-1640” throughout the manuscript). Briefly, liquid medium was inoculated with cells from overnight cultures to achieve a starting density of 5×106 cells ml−1, followed by incubation at 37 °C with shaking at 200 rpm. Cells were visualized by DIC microscopy hourly, up to 6 hours, with a Zeiss EC Plan-NeoFluar 63x/1.25 Oil objective (Carl Zeiss AG, Germany). Calcofluor white staining was performed by adding an equal volume of Calcofluor White Stain (Sigma, St. Louis, MO) directly to a sample of cell culture. Images were then acquired under fluorescence with a DAPI filter set (λEx 365 nm, λEm 445 nm).
Visualization of the C. albicans Spitzenkörper
Strains T-M1gfp, tS2-M1gfp, and tS9-M1gfp (Table 1) carry an allele of C. albicans MLC1 (orf19.2416.1) that bear an in-frame insertion of the green fluorescent protein at the C-terminus. Briefly, primers MLC1-GFP-5DR and MLC1-GFP-3DR were used to amplify the GFP-NAT1 cassette from plasmid pGFP-NAT1 (Milne, et al., 2011) for transformation of strains THE1-CIp10, tetR-SSO2, and tetR-SEC9. Transformation was carried out as described by Milne and colleagues (Milne, et al., 2011) and transformants were selected for on Difco™ Sabouraud-Dextrose agar (BD, Franklin Lakes, NJ) containing 200 μg ml−1 nourseothricin (Gold Biotechnology, St. Louis, MO). Primers flanking the MLC1 ORF (Table 2) were used to screen for isolates carrying the MLC1-GFP allele. For visualization of the Spitzenkörper, hyphal formation was induced in strains carrying the MLC1-GFP allele (Table 1) and visualized immediately with a Zeiss Axio Imager (Carl Zeiss AG, Germany) after 2, 4, and 5 hours of hyphal growth. DIC and GFP fluorescence images were acquired with the AxioVision 4.7 software (Carl Zeiss AG, Germany) using a Zeiss EC Plan-NeoFluar 63x/1.25 Oil objective (Carl Zeiss AG, Germany) and GFP filter set (λEx 470 nm, λEm 525 nm).
Statistical analysis
Statistical significance was assessed with an Analysis of Variance followed by Tukey’s multiple comparison of means using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, California). In general, differences between comparable controls were considered statistically significant with a resulting p-value less than 0.05.
RESULTS
C. albicans SSO2 and SEC9 complement the corresponding temperature-sensitive S. cerevisiae mutant strains
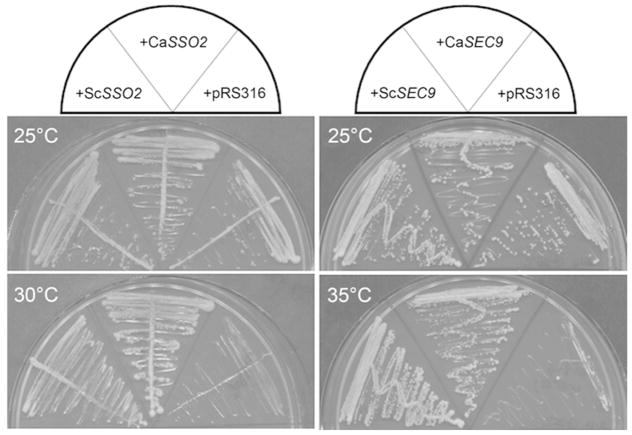
To determine if C. albicans SSO2 and SEC9 are functionally conserved with respect to S. cerevisiae SSO2 and SEC9, C. albicans SSO2 and SEC9 were cloned into the low-copy yeast shuttle vector pRS316 and transformed into the temperature- sensitive S. cerevisiae strains sso1Δ sso2-1 and sec9-4, respectively. The ability of the C. albicans SSO2 and SEC9 homolog to complement the temperature-sensitive phenotype of the corresponding mutant strain was assayed on plates incubated at the corresponding restrictive temperatures (30 and 35 °C, respectively). Strains bearing plasmids carrying S. cerevisiae SSO2 and SEC9 grew at both the permissive and restrictive temperatures (Fig. 1). Similarly, strains bearing plasmids with C. albicans SSO2 and SEC9 grew at both permissive and restrictive temperatures. As expected, strains bearing the empty vector, pRS316, only grew at the permissive temperature. Thus, C. albicans SSO2 and SEC9 are functional homologs of S. cerevisiae SSO2 and SEC9, respectively.
Figure 1. Complementation of S. cerevisiae sso2 and sec9 temperature-sensitive mutants.
The S. cerevisiae and C. albicans SSO2 and SEC9 genes were cloned into the low-copy yeast vector, pRS316, and used to transform temperature-sensitive mutant strains sso1Δ sso2-1 (left column) and sec9-4 (right column), respectively. Transformants carrying the plasmid of interest were then streaked onto agar growth medium and incubated at permissive (top) and restrictive (bottom) temperatures. Control strains, which carried empty vector pRS316, were also included. Temperatures at which the plates were incubated are indicated. Diagrams at the top of each column indicate the position of each strain as they appear on the test plates.
SSO2 and SEC9 are likely essential for viability in C. albicans
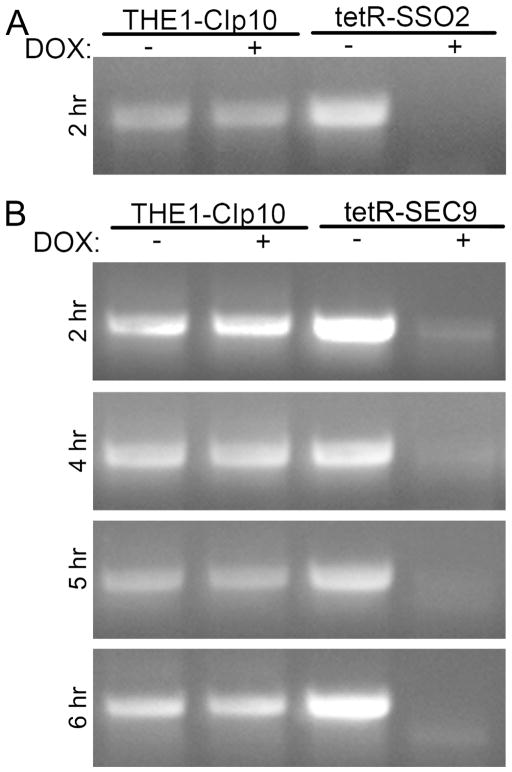
In S. cerevisiae, SEC9 is an essential gene (Brennwald, et al., 1994). Neither SSO1 nor SSO2 are essential but the double null mutant is synthetic lethal (Aalto, et al., 1993). We hypothesized that both genes of interest would be essential in C. albicans, and therefore generated the tetracycline-regulated conditional mutant strains tetR-SSO2 and tetR-SEC9 in order to study gene function. This system has been previously shown to tightly regulate gene expression, such that gene transcription is turned off completely when doxycycline (DOX) is present (Nakayama, et al., 2000, Bernardo, et al., 2008, Rane, et al., 2013). The correct genotypes of all strains were identified using PCR and confirmed with Southern blot analyses (data not shown). We also confirmed that transcription of each of these genes is completely repressed by reverse-transcriptase PCR. No SSO2 mRNA transcript was detectable after 2 hours of growth in the presence of DOX. However, low levels of SEC9 mRNA were still detectable at 2 and 4 hours of growth under repressing conditions (Fig. 2). After 5 hours of growth under repressing conditions, the SEC9 transcript was not detected (Fig. 2).
Figure 2. Reverse-transcriptase PCR analysis of SSO2 and SEC9 transcripts.
mRNA prepared from strains THE1-CIp10, tetR-SSO2, and tetR-SEC9, grown under derepressing and repressing conditions were used as template for reverse-transcriptase PCR to amplify a 875- and 950-bp amplicon from the (A) SSO2 and (B) SEC9 mRNA transcript, respectively. RT-PCR was performed on mRNA extracted from yeast cells grown for 2, 4, 5, and 6 hours, as indicated.
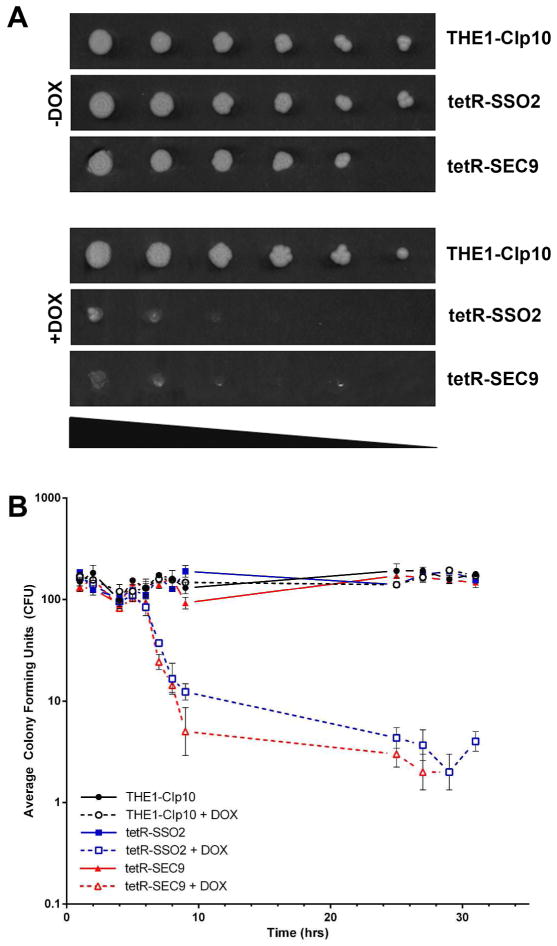
We next examined the in vitro growth of the tetR-SSO2 and tetR-SEC9 strains in the presence or absence of DOX. Under standard growth conditions, either in liquid culture (data not shown) or agar plates (Fig. 3A), suppression of either the SSO2 or SEC9 gene leads to impaired growth compared to the wild-type and control strains. We next sought to determine if this was a result of cell death or reduced fitness. Analysis of growth by enumerating colony forming units from liquid cultures indicated that cell death occurred after 6 hours of incubation under repressing conditions (Fig. 3B). These results suggest that SSO2 and SEC9 are essential for viability in C. albicans. Thus, all subsequent gene function analyses were conducted within a 6-hour period preceding loss of cell viability.
Figure 3. In vitro growth and viability of C. albicans tetR-SSO2 and tet-SEC9 mutants.
(A) Growth was assessed on CSM agar medium, in the presence and absence of doxycycline, by spotting serial dilutions of each strain. Cells were replicated to each agar plate in five-fold serial dilutions ranging from 108 to 3.2×104 cells ml−1. Images are representative of results following 48 hr incubation at 30 °C. (B) Colony forming units were determined at the indicated time points by plating a fixed number of cells from liquid cultures on YPD agar medium. Data was collected in triplicate and the average colony forming units were plot in log-scale (y-axis) against time (x-axis). Error bars indicate standard deviation values.
C. albicans SSO2 and SEC9 expression is required for secretion and proper cytokinesis
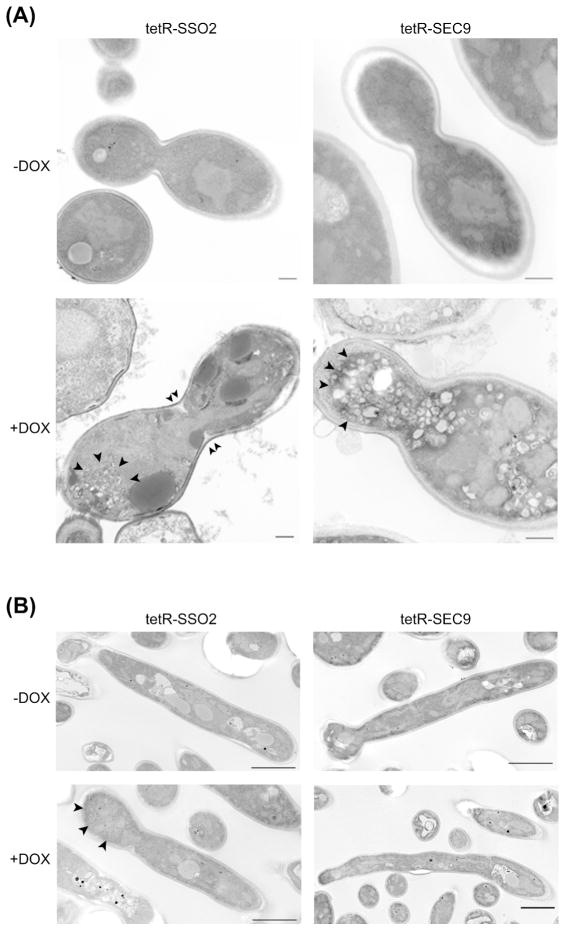
The temperature sensitive S. cerevisiae mutants sso1Δ sso2-1 and sso2Δ sso1-1 accumulate secretory vesicles at a partially formed septum, with marked defects in bud neck closure and cytokinesis at restrictive temperatures (Jantti, et al., 2002). We observed strikingly similar phenotypes in the C. albicans tetR-SSO2 strain. Accumulation of secretory vesicles and the presence of budding cells with abnormally widened bud necks occurred in the tetR-SSO2 strain grown under repressing conditions, but not in derepressing conditions (Fig. 4A). S. cerevisiae sec9 mutants accumulate toroidal or cup-shaped structures known as Berkeley bodies (Novick, et al., 1980, Novick, et al., 1981, Finger & Novick, 1997). The C. albicans tetR-SEC9 strain grown in the presence of DOX also accumulated late secretory vesicles and had widened bud necks compared to cells grown in the absence of DOX (Fig. 4A). Thus, analogous to S. cerevisiae SSO1/2 and SEC9, C. albicans SSO2 and SEC9 are required for cytokinesis and late secretion.
Figure 4. Cell morphology and cytokinesis defect in the absence of SSO2 and SEC9.
Overnight cultures of strains grown in YPD were shifted to fresh media, with or without DOX. Cells were fixed and processed for ultrastructural analysis using thin-section electron microscopy. Note that images for hyphae are representative cross-sections; full length hyphae were not visualized in these TEMs due to curvature of the hyphae in and out of the z-axis. (A) Following incubation in YPD, tetR-SSO2 and tetR-SEC9 cells grown in the presence of DOX accumulate secretory vesicles (arrows) and produce wide-necked buds during cell division. Partial inward growth at the bud neck is also evident in tetR-SSO2 in the presence of DOX (double arrows). Bars represent a length of 500nm. tetR-SSO2 micrographs were imaged under a magnification of 20kX; tetR-SEC9 micrographs were imaged under a magnification of 30kX. (B) Following incubation in buffered RPMI-1640 to induce filamentation, tetR-SSO2 hyphal cells also accumulated secretory vesicles when grown in the presence of DOX (arrows), in contrast to tetR-SEC9 cells which appear like the wild-type controls (de-repressed state). Bars indicate 2 μm. All micrographs were imaged under a magnification of 12kX, except the tetR-SEC9 with DOX micrograph which was imaged under 6kX magnification.
Loss-of-function of either SSO2 or SEC9 results in defects in secretion of degradative enzymes
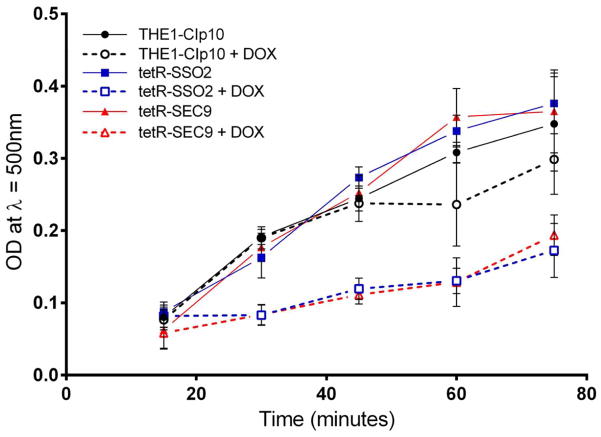
We next examined the contribution of SSO2 and SEC9 to the secretion of secreted aspartyl proteases and lipases, degradative enzymes secreted by C. albicans that play an important role in C. albicans pathogenesis (Ghannoum, 2000, Naglik, et al., 2003, Schaller, et al., 2005). Under derepressing conditions, the tetR-SSO2 strain degraded extracellular bovine serum albumin (BSA) to the same degree as the wild-type THE1-CIp10 strain (Fig. 5). However, under repressing conditions, BSA degradation was significantly reduced compared to the control strains. The tetR-SEC9 strain exhibited a similar phenotype (Fig. 5). When assessed for the ability to secrete lipases, both the tetR-SSO2 and tetR-SEC9 strains secreted lipases at comparable levels to the wild-type strain THE1-CIp10 under derepressing conditions, as indicated by the kinetic profile of secreted lipases (Fig. 6). However, when gene transcription was turned off with the addition of DOX, the tetR-SSO2 and tetR-SEC9 strains secreted significantly less lipase compared to the wild-type controls (Fig. 6). These results provide direct evidence that the t-SNAREs Sso2p and Sec9p play a substantial role in the secretion of virulence-associated extracellular enzymes such as Saps and lipases in C. albicans. In both cases, however, it was noted that the defect in secretion was only partial. Compared to the triple deletion mutant sapΔ (1–3), there was less intact BSA in the supernatant of either the tetR-SSO2 or tetR-SEC9 strains grown under repressing conditions (Fig. 5). Similarly, 50% of lipase activity was still detected under repressing conditions (Fig. 6).
Figure 5. Secretion of degradative proteases.
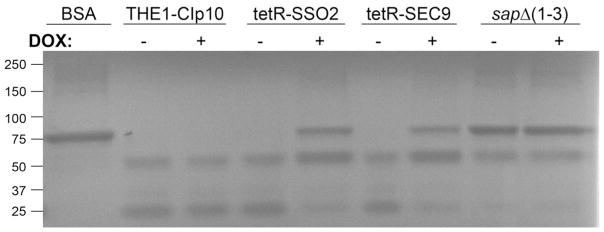
Cells from overnight cultures were transferred to medium to induce secretion of secreted aspartyl proteases encoded by SAP1, SAP2, and SAP3 (Saps 1–3). Degradation of BSA, which served as the sole nitrogen source, was followed by Coomasie staining of supernatants collected from cultures after 5 hours incubation, in the presence (+) and absence (−) of DOX. Intact BSA present in the culture medium was loaded as a reference (BSA). The triple deletion mutant sapΔ (1–3) was also included as a control. Bands of intact BSA are indicative of reduced amounts of Saps secreted by the strain. Band sizes indicate reference protein markers from Precision Plus Protein ™ All-Blue Standard (Bio-Rad, Hercules, CA).
Figure 6. Secretion of degradative lipases.
Cells from overnight cultures were transferred to medium to induce secretion of lipases. The resulting supernatants were used in a turbidimetric kinetic assay for esterases described by von Tigerstrom and Stelmaschuck (von Tigerstrom & Stelmaschuk, 1989). Optical densities (λ=500nm) measured precipitates of calcium salts of fatty acid which results from the hydrolysis of the substrate, Tween 20. The rate of increase in optical density is proportional to the concentration of lipase present in the supernatant. Data presented is the average of three independent experiments and error bars indicate standard deviation values.
Loss-of-function of either SSO2 or SEC9 results in defects in hyphal extension
Next, we assessed the role of C. albicans SSO2 and SEC9 in filamentation, using a liquid assay at specified time points to 6 hours. Initial hyphal growth was similar to the wild-type strain, THE1-CIp10, in the first several hours of growth (data not shown). However, after the fourth hour, the tetR-SSO2 strain grown in the presence of DOX exhibited a clear defect in filamentation compared to itself grown in the absence of DOX, and to the wild-type control strain, with and without DOX (Fig. 7). Of interest, a globular yeast-like structure formed at the hyphal tip, which stained with calcofluor white indicating the presence of the cell wall component chitin that suggests a return to isotropic growth (Fig. 8). This globular structure was not observed in the tetR-SEC9 strain grown under repressing conditions, whose rate of hyphal growth was also reduced (Fig. 7). We further assessed if the defect in hyphal growth was due to cell death by enumerating colony forming units from each culture. In this assay we included the conditional mutant strain, tetR-VPS1, as an additional control strain. C. albicans VPS1 is a non-essential gene that is required for wild-type filamentation (Bernardo, et al., 2008). Colony forming units were similar between all strains under both repressing and de-repressing conditions (data not shown), indicating that the defect in filamentation is not a result of cell death. Interestingly, thin-section electron microscopy of cells grown in hyphal inducing medium in the presence of DOX revealed accumulation of secretory vesicles in the tetR-SSO2 strain, but not in the tetR-SEC9 strain (Fig. 4B).
Figure 7. Filamentation of tetR-SSO2 and tetR-SEC9 in liquid medium.
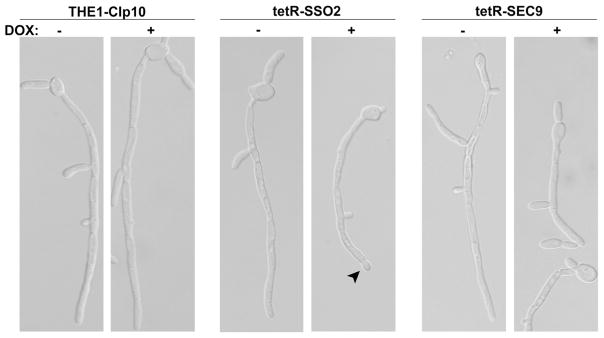
Cells at a concentration of 5×106 cells ml−1 were grown in buffered RPMI-1640 at 37 °C with shaking at 200rpm. Filamentation was followed at hourly intervals by DIC microscopy. Representative DIC images using a 63X oil objective, taken at T=5 hr, are shown. Formation of the bulbous tip in the tetR-SSO2 strain grown in the presence of DOX is typical (arrow). Scale bar shown represents 5 μm.
Figure 8. Staining the cell wall of the tetR-SSO2 hyphae.
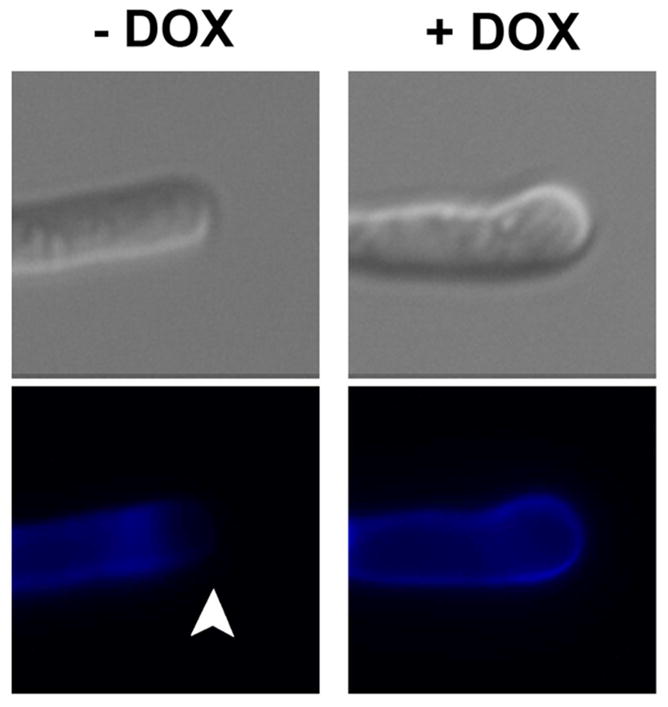
The tetR-SSO2 strain was grown in buffered RPMI-1640, in the absence (−DOX) and presence (+DOX) of doxycycline for 5 hours. Following incubation, the cells were stained with calcofluor white (0.5 μg μl−1) and subsequently imaged under fluorescence with a DAPI filter. Calcofluor white predominantly stains chitin in the fungal cell wall, a component that confers rigidity to the cell. The white arrow highlights the absence of chitin from the growing hyphal tip of the tetR-SSO2 strain grown under de-repressing conditions.
To further analyze whether the t-SNARE protein Sso2p is involved in the initial formation of the hyphal germ tube, tetR-SSO2 cells were grown for 5 hours under repressing conditions that favored vegetative/yeast growth to allow complete repression of the SSO2 transcript and turn-over of Sso2p. These cells were then switched to CSM medium containing 20% (v/v) fetal calf serum to induce hyphal formation within 40 minutes of incubation (Jones & Sudbery, 2010). Germ tube emergence was observed in all control strains and conditions, and in the tetR-SSO2 strain grown under repressing conditions (data not shown) suggesting that SSO2 plays a role in hyphal extension but not initial germ tube formation.
The C. albicans Spitzenkörper dissipates in the tetR-SSO2 mutant
Filamentous fungi accumulate vesicles at the tip of the extending hyphae, within a dynamic structure known as the Spitzenkörper (Virag & Harris, 2006). The Spitzenkörper is thought to act as the vesicle-supply center for membrane components and enzymes required for the growing hyphae. Additionally, the exocyst complex and polarisome have been implicated in polarized growth of C. albicans hyphae, where vesicles from the Spitzenkörper dock with the exocyst complex (Sudbery, 2011). To further define the defect in hyphal extension with the loss-of-function of either SSO2 or SEC9, we sought to directly examine the role of these t-SNARE proteins in polarized growth of the hyphal structure in C. albicans by examining Spitzenkörper dynamics during hyphal formation. To visualize the C. albicans Spitzenkörper, green-fluorescent protein was used to tag the myosin light chain protein, Mlc1p, which has been previously shown to co-localize with the Spitzenkörper (Crampin, et al., 2005). Under de-repressing conditions, the C. albicans Spitzenkörper was similarly observed in all background strains, THE1-CIp10, tetR-SSO2, and tetR-SEC9, for up to 5 hours of incubation in filamentation-inducing medium (Fig. 9). Under repressing conditions, the Spitzenkörper dissipated at the 5-hour timepoint in the tetR-SSO2 strain (Fig. 9). In contrast, the Spitzenkörper was still intact in the tetR-SEC9 strain grown under repressing conditions. These results suggest that SSO2 is required to maintain the integrity of the C. albicans Spitzenkörper.
Figure 9. Effect of loss-of-function of SSO2 or SEC9 on the C. albicans Spitzenkörper.
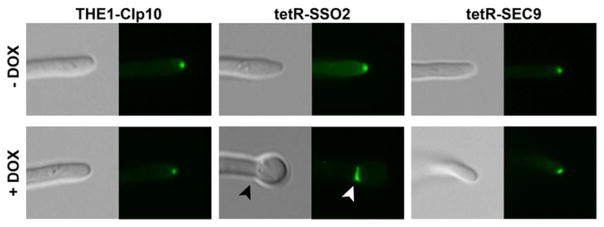
C. albicans MLC1-GFP was used to visualize the C. albicans Spitzenkörper in background strains THE1-CIp10, tetR-SSO2, and tetR-SEC9. Filamentation was induced in buffered RPMI-1640, in the presence (+) or absence (−) of DOX, and the fate of the Spitzenkörper was assessed over 5 hours of incubation. Fluorescent images at the tip of the growing hyphae at the 5-hr timepoint are shown, with the corresponding DIC image. C. albicans Mlc1p-GFP localized to septin rings in the tetR-SSO2 cell grown under repressing conditions (arrow heads); the same structure stains with calcofluor white (data not shown).
DISCUSSION
Complementation and functional studies indicate that C. albicans SSO2 is required for secretion and cell division, and is functionally similar to S. cerevisiae SSO1/SSO2 in its trafficking role. In the repressed state, the tetR-SSO2 stain was markedly defective in cytokinesis, with widened bud necks very similar to that seen in the S. cerevisiae sso1Δ sso2-1 temperature-sensitive mutant. Mlc1p has been shown to localize to septin rings in the budding yeast S. cerevisiae (Boyne, et al., 2000). Under repressing conditions, we have observed similar Mlc1p-GFP localization and calcofluor white staining in the yeast form of tetR-SSO2 (data not shown) despite incomplete formation of a septum. We also demonstrated that C. albicans SSO2 is likely an essential gene, which we had anticipated given the absence of the gene duplication that occurred in S. cerevisiae. Analogously, we have demonstrated functional homology of C. albicans and S. cerevisiae SEC9. In the absence of functional C. albicans SEC9, the strain produces cells with wide bud necks and accumulates secretory vesicles, which is similar to the morphological defects found in the S. cerevisiae sec9-3 and sec9-4 strains (Novick, et al., 1980, Novick, et al., 1981, Finger & Novick, 1997). As in S. cerevisiae, C. albicans SEC9 is also likely essential for viability. Importantly, analogous to the secretory defects described in these S. cerevisiae mutants, we demonstrated that the C. albicans t-SNAREs Sso2p and Sec9p were required for secretion of the virulence-associated degradative enzymes secreted aspartyl proteases and lipases.
We performed time-course experiments to determine when gene transcription is repressed after the addition of doxycycline. Interestingly, while this promoter replacement system is known to tightly regulate transcription, results of reverse-transcriptase PCR revealed that mRNA transcript of SEC9 is still present up to 5 hours post-exposure to DOX. This translates to the presence of functional Sec9p, albeit likely at lower levels compared to wild-type. Despite the presence of Sec9p at low levels, the tetR-SEC9 mutant strain exhibited similar phenotypes to the tetR-SSO2 mutant strain in terms of secretion of degradative enzymes and hyphal elongation, indicating that these cellular processes are likely sensitive to the levels of Sec9p. The dynamics of hyphal induction and the corresponding presence of the Spitzenkörper in the SSO2 mutant, including the partial defects in secretion observed, could be indicative of either prolonged Sso2p protein turnover or the presence of a compensatory protein. However, such a compensatory protein would be transient, possibly recycled from the plasma membrane by endocytic mechanisms, and thus would be unable to sustain cellular requirements for growth, secretion, and hyphal elongation.
In filamentous fungi, hyphal growth is associated with polarized growth and secretion. There are currently three models that are proposed to describe hyphal growth: the vesicle-supply-center model, the amoeboid model, and the steady-state model (Steinberg, 2007). In the vesicle-supply-center model, a constant supply of Golgi-derived proteins, packaged in vesicles, arrive at the hyphal tip transported along actin cables by microtubule-based motors. These vesicles accumulate as a body known as the Spitzenkörper (Virag & Harris, 2006) before docking at the plasma membrane with components of the exocyst complex and finally delivering its cargo by SNARE-mediated membrane fusion events (Sudbery, 2011). Recycling of membrane proteins, such as components of the exocyst complex, via endocytic mechanisms is another important aspect of this dynamic process (Read & Hickey, 2001, Steinberg, 2007, Jones & Sudbery, 2010, Sudbery, 2011). Previous studies have provided evidence for the role of SNAREs in hyphal development in filamentous fungi such as Neurospora crassa, Aspergillus nidulans, and Trichoderma reesei. Specifically, Sso1p homologues have been shown to localize to the plasma membrane at the apical tip of the actively growing hyphae (Taheri-Talesh, et al., 2008), form complexes with the vesicle SNARE Snc1p at this site (Valkonen, et al., 2007), and furthermore, that low level expression of either of the N. crassa homologues results in altered tip growth rate and morphology (Gupta, et al., 2003). In these studies, we have demonstrated that both SSO2 and SEC9 are required to maintain hyphal growth in the dimorphic fungus C. albicans. Germ tube emergence still occurs, but the rate of hyphal extension decreases after 4 hours of growth. This is exemplified by the effect of SSO2 repression, where the disassembly of the Spitzenkörper corresponded to cells exhibiting accumulated secretory vesicles and abnormal hyphal growth. Following the interruption of hyphal growth, a presumed septin ring forms that precludes septation, based on the localization of Mlc1p (Boyne, et al., 2000, Crampin, et al., 2005), and the hyphal tip of the tetR-SSO2 mutant strain becomes globular, reminiscent of the return to isotropic growth of the C. albicans sec3Δ mutant strain where polarized growth is lost following initial germ tube evagination (Li, et al., 2007). In contrast to a study performed in the filamentous fungus Aspergillus nidulans, where the secretory defect resulting in the loss of function of rabC results in a more prominent (i.e., larger) Spitzenkörper (Pantazopoulou & Penalva, 2011), we saw dissipation of the C. albicans Spitzenkörper in the tetR-SSO2 mutant strain. One possibility for this difference could be directly related to the rate of growth of hyphal tips, as it is known that the Spitzenkörper is not visible in slow growing fungi (reviewed by Riquelme, 2013). We observed a marked decrease in hyphal growth of both tetR-SSO2 and tetR-SEC9 strains between 4 and 5 hours of incubation. Alternatively, the presence of a functional SNARE assembly at the plasma membrane is essential for maintaining polarized growth. If this was the case, we would predict that mutations affecting the early stages of exocytosis will result in accumulation of secretory vesicles in the Spitzenkörper, but mutations affecting the late stages of exocytosis (such as in the SNARE and exocyst complex) will result in loss of polarized growth and dissipation of the Spitzenkörper and subsequent accumulation of secretory vesicles within the cytoplasm. In all, these results are in agreement with previous findings that the Spitzenkörper is present throughout polarized growth; disruption of polarized growth results in the loss of the Spitzenkörper and inhibits filamentous growth (Crampin, et al., 2005).
It is not clear why there are differences in the stability of the Spitzenkörper, given that Sso2p and Sec9p form a part of the same complex involved in mediating fusion of secretory vesicles to the plasma membrane. However, we presented evidence through RT-PCR analyses of each transcript that these genes are differentially regulated. This difference in gene regulation may reflect subtle differences in gene function and/or cellular requirements during yeast and filamentous growth. From a pathogenesis standpoint, these data indicate that both SSO2 and SEC9 contribute to two key components of C. albicans virulence: filamentation and secretion of extracellular degradative proteins. Both the tetR-SSO2 and tetR-SEC9 strain are able to form filaments in repressing conditions, however hyphal extension is not maintained following loss of gene expression. In the tetR-SSO2 strain there is also dramatic accumulation of late secretory vesicles in filaments, which demonstrates a clear relationship between secretion and filamentation. Our results are consistent with the vesicle-supply-center model of filamentous growth and provide direct evidence for the role of t-SNAREs in polarized growth of the hyphal structure in C. albicans.
Acknowledgments
We thank Hironobu Nakayama (Suzuka University of Medical Science, Japan) for providing strain THE1 and plasmid p99CAU1; Aaron P. Mitchell (Carnegie Mellon University) for providing plasmid pDDB57; Steven Bates (University of Exeter) for the pNAT1-GFP plasmid; Jussi Jantti (University of Helsinki) and Peter Novick (University of California at San Diego) for S. cerevisiae strains sso1Δ sso2-1 and sec9-4, respectively; and Barbara Hunter (University of Texas Health Science Center at San Antonio) for assistance with transmission electron microscopy.
Sequence data for C. albicans was obtained from the Candida Genome Database website at http://www.candidagenome.org. Sequencing of C. albicans was accomplished with the support of the NIDCR, NIH, and the Burroughs Wellcome Fund.
This work was supported by funding from the Department of Veterans’ Affairs (Merit Award 5I01BX001130-03 to SAL), Biomedical Research Institute of New Mexico (SAL), and the UNM IDIP T32 institutional training grant NIH 5 T32 AI007538-13 (SMB).
Footnotes
The authors declare that they have no competing interests.
Contributor Information
Stella M. Bernardo, Email: SBernardo@salud.unm.edu.
Hallie S. Rane, Email: HallieRane@unm.edu.
Alba Chavez-Dozal, Email: AlbaChavez@unm.edu.
Samuel A. Lee, Email: SamALee@salud.unm.edu.
References
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. Wiley; New York: 1987. [Google Scholar]
- Bates S, Hughes HB, Munro CA, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Bernardo SM, Khalique Z, Kot J, Jones JK, Lee SA. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet Biol. 2008;45:861–877. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113:4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- Bozzola JJ, Russell LD. Electron Microscopy. Jones & Bartlett Publishers; Boston, MA: 1992. [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Finger FP, Novick P. Sec3p is involved in secretion and morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:647–662. doi: 10.1091/mbc.8.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Novick P. Synthetic interactions of the post-Golgi sec mutations of Saccharomyces cerevisiae. Genetics. 2000;156:943–951. doi: 10.1093/genetics/156.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell Mol Life Sci. 1999;55:707–734. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauert AM. Fixation, Dehydration and Embedding of Biological Specimens. North-Holland Publishing Co; New York, NY: 1975. [Google Scholar]
- Grote E, Carr CM, Novick PJ. Ordering the final events in yeast exocytosis. J Cell Biol. 2000;151:439–452. doi: 10.1083/jcb.151.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Free SJ, Levina NN, Keranen S, Heath IB. Two divergent plasma membrane syntaxin-like SNAREs, nsyn1 and nsyn2, contribute to hyphal tip growth and other developmental processes in Neurospora crassa. Fungal Genet Biol. 2003;40:271–286. doi: 10.1016/s1087-1845(03)00109-9. [DOI] [PubMed] [Google Scholar]
- Jantti J, Aalto MK, Oyen M, Sundqvist L, Keranen S, Ronne H. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J Cell Sci. 2002;115:409–420. doi: 10.1242/jcs.115.2.409. [DOI] [PubMed] [Google Scholar]
- Jones LA, Sudbery PE. Spitzenkörper, exocyst, and polarisome components in Candida albicans hyphae show different patterns of localization and have distinct dynamic properties. Eukaryot Cell. 2010;9:1455–1465. doi: 10.1128/EC.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle N, Kloepper TH, Fasshauer D. Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol. 2009;9:19. doi: 10.1186/1471-2148-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida H. Propylene Oxide as a Dehydrating Agent for Embedding with Epoxy Resins. Journal of Electron Microscopy. 1961;10:203– 204. [Google Scholar]
- Lee SA, Jones J, Khalique Z, Kot J, Alba M, Bernardo S, Seghal A, Wong B. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. FEMS Yeast Res. 2007;7:973–985. doi: 10.1111/j.1567-1364.2007.00253.x. [DOI] [PubMed] [Google Scholar]
- Lee SA, Jones J, Hardison S, Kot J, Khalique Z, Bernardo SM, Lazzell A, Monteagudo C, Lopez-Ribot J. Candida albicans VPS4 is required for secretion of aspartyl proteases and in vivo virulence. Mycopathologia. 2009;167:55–63. doi: 10.1007/s11046-008-9155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Lee RT, Wang YM, Zheng XD, Wang Y. Candida albicans hyphal morphogenesis occurs in Sec3p-independent and Sec3p-dependent phases separated by septin ring formation. J Cell Sci. 2007;120:1898–1907. doi: 10.1242/jcs.002931. [DOI] [PubMed] [Google Scholar]
- Milne SW, Cheetham J, Lloyd D, Aves S, Bates S. Cassettes for PCR-mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast. 2011;28:833–841. doi: 10.1002/yea.1910. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect Immun. 2000;68:6712–6719. doi: 10.1128/iai.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Palanisamy SK, Ramirez MA, Lorenz M, Lee SA. Candida albicans PEP12 is required for biofilm integrity and in vivo virulence. Eukaryot Cell. 2010;9:266–277. doi: 10.1128/EC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou A, Penalva MA. Characterization of Aspergillus nidulans RabC/Rab6. Traffic. 2011;12:386–406. doi: 10.1111/j.1600-0854.2011.01164.x. [DOI] [PubMed] [Google Scholar]
- Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, Lee SA. Candida albicans VMA3 Is Necessary for V-ATPase Assembly and Function and Contributes to Secretion and Filamentation. Eukaryot Cell. 2013;12:1369–1382. doi: 10.1128/EC.00118-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read ND, Hickey PC. The Vesicle Trafficking Network and Tip Growth in Fungal Hyphae. In: Geitmann A, Cresti M, Heath IB, editors. Cell Biology of Plant and Fungal Tip Growth. IOS Press; Netherlands: 2001. pp. 137–148. [Google Scholar]
- Riquelme M. Tip growth in filamentous fungi: a road trip to the apex. Annu Rev Microbiol. 2013;67:587–609. doi: 10.1146/annurev-micro-092412-155652. [DOI] [PubMed] [Google Scholar]
- Rossi G, Salminen A, Rice LM, Brunger AT, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot Cell. 2007;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Taheri-Talesh N, Horio T, Araujo-Bazan L, Dou X, Espeso EA, Penalva MA, Osmani SA, Oakley BR. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Valkonen M, Kalkman ER, Saloheimo M, Penttila M, Read ND, Duncan RR. Spatially segregated SNARE protein interactions in living fungal cells. J Biol Chem. 2007;282:22775–22785. doi: 10.1074/jbc.M700916200. [DOI] [PubMed] [Google Scholar]
- Venable JH, Coggeshall R. A Simplified Lead Citrate Stain for Use in Electron Microscopy. J Cell Biol. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A, Harris SD. The Spitzenkörper: a molecular perspective. Mycol Res. 2006;110:4–13. doi: 10.1016/j.mycres.2005.09.005. [DOI] [PubMed] [Google Scholar]
- von Tigerstrom RG, Stelmaschuk S. The use of Tween 20 in a sensitive turbidimetric assay of lipolytic enzymes. Can J Microbiol. 1989;35:511–514. doi: 10.1139/m89-079. [DOI] [PubMed] [Google Scholar]
- Williams DC, Novick PJ. Analysis of SEC9 suppression reveals a relationship of SNARE function to cell physiology. PLoS One. 2009;4:e5449. doi: 10.1371/journal.pone.0005449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]