Summary
Plk4 family kinases control centriole assembly. Plk4s target to mother centrioles through an interaction between their cryptic polo box (CPB) and acidic regions in the centriolar receptors SPD-2/Cep192 and/or asterless/Cep152. Here, we report a crystal structure for the CPB of C. elegans ZYG-1, which forms a Z-shaped dimer containing an intermolecular β-sheet with an extended basic surface patch. Biochemical and in vivo analysis revealed that electrostatic interactions dock the ZYG-1 CPB basic patch onto the SPD-2-derived acidic region to promote ZYG-1 targeting and new centriole assembly. Analysis of a new crystal form of the Drosophila Plk4 (DmPlk4) CPB suggests that it also forms a Z-shaped dimer. Comparison of the ZYG-1 and DmPlk4 CPBs revealed structural changes in the ZYG-1 CPB that confer selectivity for binding SPD-2 over asterless-derived acidic regions. Overall, our findings suggest a conserved mechanism for centriolar docking of Plk4 homologs that initiate daughter centriole assembly.
Keywords: centriole, cryptic polo box, crystallography, polo-like kinase, ZYG-1, Plk4
Introduction
Centrioles are small cylindrical organelles with two critical functions - to recruit pericentriolar material to form centrosomes and to serve as basal bodies during ciliogenesis (Gönczy, 2012; Brito et al., 2012; Azimzadeh and Marshall, 2010; Loncarek and Khodjakov, 2009). In dividing cells, duplicated centrosomes promote bipolar spindle assembly by nucleating microtubules on opposite sides of the aligning chromosomes. Centrioles duplicate precisely once per cell cycle to ensure that cells entering mitosis have exactly two centrosomes. However, cells in solid tumors and many cancer cell lines have extra centrosomes, suggesting that dysregulation of centriole duplication may contribute to tumorigenesis (Nigg and Raff, 2009; Anderhub et al., 2012).
Studies in eukaryotic model systems and vertebrate cells have defined a small set of core components required for centriole duplication including: SAS-4/CPAP, SAS-5/Ana2/STIL, SAS-6, SPD-2/Cep192, asterless/Cep152, and Plk4/ZYG-1 (Gönczy, 2012; Brito et al., 2012; Azimzadeh and Marshall, 2010; Loncarek and Khodjakov, 2009; Carvalho-Santos et al., 2011). Two of these, SAS-6 and SAS-4, which are structural components of the cartwheel and outer centriole wall (Gönczy, 2012), are conserved across eukaryotes (Carvalho-Santos et al., 2011). In contrast, Plk4 kinase and its centriolar receptors, asterless/Cep152 and SPD-2/Cep192, are only found in animals and fungi (Carvalho-Santos et al., 2011), likely because they represent a regulatory module that couples centriole assembly to cell division.
In metazoans, Plk4 kinases control daughter centriole assembly. Plk4 homologs have an N-terminal kinase domain, a C-terminal polo box, and a central domain termed the “cryptic polo box” (CPB) (Swallow et al., 2005) that has been shown to dimerize, to be sufficient for centriole localization and to be required for Plk4 to promote centriole assembly (Leung et al., 2002; Habedanck et al., 2005; Slevin et al., 2012). One of the most divergent Plk4 kinases, only recently appreciated to be a bona fide Plk4 homolog, is C. elegans ZYG-1 (Jana et al., 2012). In vertebrates, Plk4 is recruited to mother centrioles through interactions of its CPB with acidic regions in SPD-2/Cep192 and asterless/Cep152 (Kim et al., 2013, Sonnen et al., 2013; Hatch et al., 2010; Cizmecioglu et al., 2010), whereas in other organisms, Plk4 is recruited via interactions with either SPD-2 (C. elegans; Pelletier et al., 2006; Delattre et al., 2006) or asterless (Drosophila, Dzhindzhev et al., 2010). A recent structural study showed that the DmPlk4 CPB consists of two tandem polo-boxes and proposed that it forms a butterfly-like side-by-side dimer (Slevin et al., 2012). However, this structure did not explain how electrostatic interactions could mediate an interaction between the Plk4 CPB and the acidic regions in SPD-2/Cep192 and asterless/Cep152.
Here, we report a crystal structure for the CPB of C. elegans ZYG-1. The ZYG-1 CPB formed a Z-shaped end-to-end dimer containing a 12-stranded inter-molecular β-sheet with a conserved basic surface patch. Parallel in vitro and in vivo analysis demonstrated that electrostatic interactions between the basic patch on the ZYG-1 CPB dimer and the SPD-2 acidic region dock ZYG-1 onto centrioles to promote new centriole assembly. Analysis of a new crystal form of the DmPlk4 CPB and of the dimer in solution using small-angle X-ray scattering suggest that the DmPlk4 CPB also forms a Z-shaped dimer with a basic surface patch. A comparison of the ZYG-1 and DmPlk4 CPBs revealed structural changes in the ZYG-1 CPB dimer that confer selectivity for binding SPD-2 over asterless-derived acidic regions. Overall, our work has elucidated the native dimeric conformation of the CPBs of ZYG-1 and DmPlk4, and suggests that Plk4 homologs dock onto their centriolar receptors via a conserved basic patch on the CPB dimer.
Results
The ZYG-1 cryptic polo box forms a Z-shaped end-to-end dimer
The ZYG-1 CPB (aa 338-564) was expressed in bacteria, purified, and crystallized. The resulting crystals diffracted to 2.3-Å resolution with space group P2 (a = 53.38 Å, b = 60.09 Å, c = 87.52 Å; β = 93.31°). The structure was solved by single-wavelength anormalous dispersion (SAD) using selenomethionine (SeMet)-substituted protein. The final structure contains residues 351-562, with segments 510-515 and 548-550 disordered, and has Rwork and Rfree of 24.4% and 27.6%, respectively (Table 1). The structure revealed that the ZYG-1 CPB contains two tandem polo boxes (PB1 and PB2), each containing a six-stranded β-sheet with an α-helix packed against one side (Figure 1A). The two PBs are organized like an open clamshell with the β-sheet surfaces not covered by helices facing each other. While covered by the long β5-β6 loop in PB1, this β-sheet surface is solvent exposed in PB2 (Figure 1A).
Table 1.
Data collection and refinement statistics
| ZYG-1 CPB | DmPlk4 CPB | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9792 | 0.9394 |
| Space group | P2 | P6422 |
| Cell dimensions | ||
| a, b, c (Å) | 53.38, 60.09, 87.51 | 132.21, 132.21, 134.25 |
| α, β, γ (°) | 90, 93.31, 90 | 90, 90, 120 |
| Resolution (Å) | 46-2.30 (2.44-2.30) * | 39-3.40 (3.60-3.40) * |
| No. observed reflections | 45,979 (7,427) | 9,997 (1,518) |
| No. unique reflections | 15,326 (2,476) | 241 (37) |
| Rmeas | 0.092(0.882) | 0.148(3.52) |
| CC(1/2) | 99.7 (72.1) | 100.0 (74.4) |
| I / σI | 9.7 (1.5) | 25.77 (2.06) |
| Completeness (%) | 97.5 (96.2) | 99.5 (97.4) |
| Redundancy | 3.0 (3.0) | 41.5 (41.3) |
| Refinement | ||
| Resolution (Å) | 25-2.3 | 20-3.4 |
| No. reflections | 45,953 | 9,938 |
| Rwork / Rfree (%) | 24.4/27.6 | 22.6/24.6 |
| No. atoms | ||
| Protein | 3,244 | 1,705 |
| Ligand/ion | 27 | - |
| Water | 226 | - |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.016 |
| Bond angles (°) | 0.876 | 1.051 |
Values in parentheses are for highest-resolution shell.
Figure 1. The ZYG-1 CPB forms a Z-shaped end-to-end dimer with a conserved basic patch.
(A) Ribbon diagram of the ZYG-1 CPB color-ramped from blue to red (N- to C-terminus). The two polo boxes (PB1 and PB2) are indicated and the two disordered loops in PB2 are shown as dashed lines. (B) Crystal-packing suggests two possible dimer interfaces. (C) SAXS of the ZYG-1 CPB in solution. The logarithm of the scattering intensity is plotted as a function of momentum transfer s = 4π sin(θ)/λ, where θ is the scattering angle and λ is the X-ray wavelength. Inset shows the boxed region at higher magnification. (D) Two views of the representative ab initio low resolution shape reconstructed by GASBOR. The most probable model, chosen from a dozen reconstructions by DAMAVER (Volkov & Svergun, 2003), is shown (transparent beads) superimposed with the Z-dimer structure (green ribbons). The overall normalized spatial discrepancy (NSD) was 1.3 and all models were consistent. (E-H) Surface plots of the ZYG-1 CPB Z-dimer highlighting conserved basic (blue) and acidic (magenta) residues (E, G) and electrostatic potential (F, H). Views showing the basic patch (E, F; dashed line in F) and the reverse side (G, H) of the dimer are shown.
The asymmetric unit contains two copies of the ZYG-1 CPB arranged in a compact, U-shaped dimer (U-dimer). A second more extended Z-shaped dimer (Z-dimer) can be assembled based on crystal packing interactions (Figure 1B). The surface area buried by the Z- and U-dimers is nearly identical (838 versus 819 Å2, PISA server; Krissinel and Henrick, 2007). As attempts to disrupt dimer formation by mutating residues on either interface were unsuccessful due to protein insolubility/instability, we decided to use to small-angle X-ray scattering (SAXS) to examine the shape of CPB dimers in solution (Svergun and Koch, 2002). Synchrotron SAXS data was collected and the scattering pattern processed and extrapolated to infinite dilution (Figure 1C, Experimental). This yielded a molecular mass of 55 ± 6 kDa, confirming that the CPB is dimeric in solution (monomer = 26.2 kDa). The radius of gyration Rg = 32 ± 1 Å and maximum particle size Dmax = 110 ± 10 Å closely corresponded to the values computed from the extended Z-dimer (Rg = 31.7 Å, Dmax = 115 Å) rather than the compact U-dimer (Rg = 24.5 Å, Dmax = 76 Å). In addition, whereas the experimental data superimposed well with the scattering pattern calculated from the Z-dimer (discrepancy χ = 1.25), it deviated substantially from that calculated from the U-dimer (χ = 3.60; Figure 1C). The Z-dimer structure also superimposed well with the low resolution ab initio shape determined from the experimental data (Figure 1D). We conclude that the ZYG-1 CPB dimer in solution has a quaternary structure similar to that of the Z-dimer.
A conserved basic patch runs across the ZYG-1 CPB dimeric interface
The majority of the acidic and some of the basic residues conserved across nematode species (Figure S1A) are buried in the interior, implying a role in protein folding; notably, a few of these are buried under the β5-β6 loop in PB1 and within the PB1-PB2 interface. However, a subset of conserved positive residues - including K454, R463, R470, R471, R489, K491 and K500 - form an extensive basic patch on the CPB dimer surface (Figure 1E, F). In contrast, very few conserved hydrophilic residues are present on the reverse side of the dimer (Figure 1G, H). Nine of thirteen temperature sensitive (ts) zyg-1 mutations (O’Connell et al., 2001; Kemp et al., 2007; O’Rourke et al., 2011) are in the CPB (Figure S1A, B). In contrast to the conserved basic residues, which cluster near the dimer midline, the majority of ts CPB mutations (8/9) are in non-conserved residues in PB1 near the dimer periphery (Figure S1C, D). We hypothesize that these mutations locally perturb PB1 structure at elevated temperatures, compromising CPB function. Thus, an analysis of charged residues revealed a conserved basic patch that extends approximately 65 Å across one surface of the 12-stranded inter-molecular β-sheet that constitutes the midline of the CPB dimer.
The ZYG-1 CPB interacts with the CeSPD-2 N-terminus through a series of electrostatic interactions
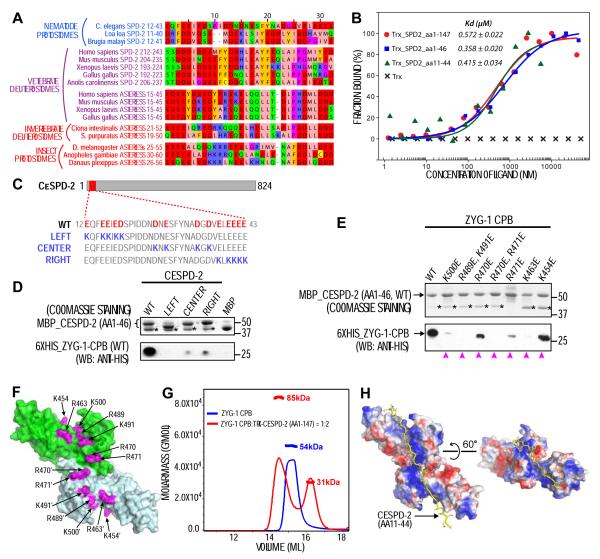
In vertebrates, Plk4 is recruited to mother centrioles through interactions of its CPB with acidic regions in SPD-2/Cep192 and asterless/Cep152 (Kim et al., 2013; Sonnen et al., 2013; Hatch et al., 2010; Cizmecioglu et al., 2010). In Drosophila, SPD-2 lacks an acidic region and Plk4 targeting depends on asterless (Dzhindzhev et al., 2010). C. elegans lacks an asterless homolog and recruits ZYG-1 via CeSPD-2 (Pelletier et al., 2006; Delattre et al., 2006), which contains an N-terminal acidic region similar to those in other SPD-2 and asterless homologs (Figures 2A, S2A,B). Microscale thermophoresis (MST; Seidel et al., 2013) using fluorescently labeled ZYG-1 CPB as reporter revealed that thioredoxin fusions with three CeSPD-2 N-terminal fragments (aa 1-147, 1-46 and 11-44) bound with nearly identical affinities (Kd ~ 0.5 μM; Figure 2B), indicating that residues 11-44 of CeSPD-2 are sufficient for interaction.
Figure 2. The ZYG-1 CPB interacts with the CeSPD-2 N-terminus through a series of electrostatic interactions.
(A) Acidic regions in SPD-2 and asterless homologs were manually selected and aligned using Muscle WS in Jalview. Color code: negatively charged (red), positively charged (blue), aromatic (gold), hydrophilic (green), conformationally special (pink) and cysteine (yellow). (B) MST binding curves of thioredoxin (Trx) fusions with regions of the CeSPD-2 N-terminus to the ZYG-1 CPB. (C) Schematic showing the CeSPD-2 AR (red box) and residue changes in charge reversal mutants. (D, E) In vitro pull-down experiments. Resin-immobilized WT or mutant MBP-CeSPD-2 AR (aa 1-46) (coomassie gels; top panels) were used to pull down WT or mutant 6×His-ZYG-1 CPB (Western blots; bottom panels). Asterisks mark MBP released due to degradation. Equivalent solubility of all ZYG-1 CPB mutants in (E) was independently confirmed. Magenta arrowheads mark CPB mutants that reduced the interaction. (F) Location of positively charged residues on the ZYG-1 CPB dimer that contribute to the interaction with the CeSPD-2 AR. (G) Static light scattering results for the ZYG-1 CPB alone (blue) or mixed in a 1:2 ratio with Trx fusion with the CeSPD-2 N-terminus. (H) In silico docking result using the ZYG-1 CPB dimer (electrostatic surface plot) as receptor and the CeSPD-2 N-terminus (aa 11-44; yellow stick model) as ligand.
To find out whether the ZYG-1 CPB interacts with all or part of the CeSPD-2 acidic region (CeSPD-2 AR), we generated maltose binding protein (MBP)-tagged WT CeSPD-2 (aa 1-46) and three mutant versions (LEFT, CENTER and RIGHT; Figure 2C) that charge reversed negatively charged residues in the center or in the left or right flanking sides of the CeSPD-2 AR. All three mutants pulled down significantly less ZYG-1 CPB than the WT AR, suggesting that negatively charged residues across the entire CeSPD-2 AR interact with the ZYG-1 CPB (Figures 2D, S3A). Analysis of seven charge reversal CPB mutants similarly revealed that all mutations compromised binding to the CeSPD-2 AR; only K454E - located at the very distal tips of the basic patch - retained a substantial ability to be pulled down (Figure 2E,F). Static light scattering (SLS) demonstrated that ZYG-1 CPB dimers (54 kDa) each bind one Trx-CeSPD-2 AR monomer (31 kDa) to form an 85 kDa complex (Figure 2G). Far-UV circular dichroism and nuclear magnetic resonance confirmed that the CeSPD-2 AR is intrinsically disordered with a classical random-coil structure (Figure S2C-E). Docking trials using the ZYG-1 CPB dimer structure as receptor and the CeSPD-2 AR as ligand (ClusPro 2.0, electrostatic-favored mode; Kozakov et al., 2010) docked the CeSPD-2 AR neatly on the conserved basic patch across the dimeric interface (Figure 2H). Overall, our data strongly suggest that the CeSPD-2 AR interacts with the ZYG-1 CPB through an extensive series of electrostatic contacts that run along the length of the inter-molecular β-sheet on the CBP dimer.
A distributed array of negatively charged residues in the CeSPD-2 N-terminus recruits ZYG-1 to mother centrioles to initiate centriole duplication in vivo
Next, we investigated the role of charges on both sides of the proposed interface in ZYG-1 centriolar targeting and daughter centriole assembly in vivo. To analyze the CeSPD-2 AR, we generated mCherry-tagged single copy spd-2 transgenes encoding WT SPD-2 and the same charge reversal mutants that we tested in vitro (Figure 2C). Transgenes were engineered to be resistant to RNA interference (RR: RNAi-resistant) targeting endogenous spd-2 by altering 622 bp of nucleotide sequence without affecting protein coding (Figures 3A, S3B). Western blotting confirmed that the WT and mutant proteins were comparably expressed and that RNAi targeting the re-encoded region selectively depleted endogenous CeSPD-2 (Figure 3B). Quantitative confocal microscopy revealed that WT mCherry::CeSPD-2 and the LEFT, CENTER and RIGHT charge reversal mutants all accumulated at centrosomes to the same extent when endogenous SPD-2 was depleted (Figure 3C).
Figure 3. A distributed array of negatively charged residues in the CeSPD-2 N-terminus recruits ZYG-1 to mother centrioles to initiate centriole duplication in vivo.
(A) Single copy mCherry::spd-2RR transgene. (B) Western blot of mCherry::spd-2RR worm lysates, with (+) or without (−) endogenous SPD-2 depletion, probed for SPD-2 (top) and α-tubulin (bottom). Asterisk marks a background band. (C) (left) Confocal images of one-cell stage embryos expressing mCherry::SPD-2 fusions. (right) Graph of mean centrosomal mCherry::SPD-2 fluorescence for the indicated strains (n = number of centrosomes; error bars = SEM). (D, E) Deconvolved immunofluorescence images of first (left) and second division (right) embryos lacking (D) or expressing the indicated spd-2 transgenes (E). Embryos were depleted of the indicated protein (D) or endogenous SPD-2 (E) by RNAi and were stained for microtubules (green) and DNA (red). n = number of embryos imaged. (F) Plots showing percent embryonic viability (left) and frequency of second division monopolar spindles (right). Error bars = SD of percent lethality per worm; N = number of worms, n = total number of embryos (left) or second division cells (right). (G) Deconvolved immunofluorescence images of embryos stained for DNA and SAS-4 (red and green; left panels) and ZYG-1 (right panels). Embryos expressing the indicated spd-2 transgenes were depleted of endogenous SPD-2 by RNAi. (H) Graph plotting mean centrosomal ZYG-1 fluorescence. Values are percentage of the mean control value. n = number of centrosomes. Error bars = SEM. Scale bars, 10 m.
CeSPD-2 is recruited to the two sperm centrioles following fertilization and is required for them to recruit pericentriolar material (PCM) to form centrosomes, as well as for them to initiate daughter centriole formation. Centrioles in CeSPD-2 depleted embryos recruit only a small shell of PCM and the resulting centrosomes are insufficient to support spindle assembly; thus, chromosome segregation and the first embryonic cytokinesis fail (Figure 3D; Kemp et al., 2004; Pelletier et al., 2004). In contrast, in embryos depleted of ZYG-1, which is required for centriole duplication but not PCM assembly, the sperm centrioles recruit PCM and a normal bipolar spindle forms during the first embryonic division. However, since the sperm centrioles fail to duplicate, each daughter cell inherits only a single centriole and monopolar, rather than bipolar, spindles form during the second embryonic division (Figure 3D; O’Connell et al., 2001). Whereas WT mCherry::SPD-2 rescued embryonic viability following endogenous SPD-2 depletion, the LEFT, CENTER and RIGHT mutants did not (Figure 3F). Immunofluorescence revealed that all three mutants exhibited the ZYG-1 depletion phenotype - 100% second division monopolar spindles (Figure 3E, F). Thus, the LEFT, CENTER and RIGHT CeSPD-2 mutants target to mother centrioles and support PCM recruitment but fail to promote centriole duplication. To determine whether duplication failure was due to a defect in ZYG-1 docking, we quantified ZYG-1 levels at mother centrioles in fixed mitotic embryos expressing WT mCherry::CeSPD-2 or the LEFT, CENTER or RIGHT mutants. All three mutants abolished ZYG-1 recruitment (Figure 3G,H); the effect was the same in embryos at a stage coincident with new centriole assembly (Figure S3C). We conclude that an array of negatively charged residues in the CeSPD-2 N-terminus recruits ZYG-1 to mother centrioles; selective alteration of charges in the beginning, middle or end of this region disrupts ZYG-1 docking and centriole duplication.
The ability of ZYG-1 CPB mutants to target to centrioles and support centriole assembly in vivo parallels their ability to bind to the CeSPD-2 N-terminus in vitro
To examine residues in the basic surface patch on the ZYG-1 CPB dimer, we generated transgenes encoding untagged ZYG-1, as both N- and C-terminal ZYG-1 GFP fusions are non-functional (Figure 4A; Lettman et al., 2013). As injection of DNA containing the WT zyg-1 locus is toxic (Lettman et al., 2013), transgenes also included the well-characterized P442L temperature-sensitive point mutation (Kemphues et al., 1988; Kemp et al., 2007). Strains were maintained at the non-permissive temperature (23.5°C) to keep the transgenic proteins inactive. After dsRNA injection to deplete endogenous ZYG-1, strains were shifted to the permissive temperature of 16°C to replace endogenous ZYG-1 with transgene-encoded Control or mutant ZYG-1. We constructed a transgene encoding ZYG-1 lacking the CPB (ΔCPB; deletion of aa 350-560) and mutants that charge reversed basic residues that were centrally positioned (R470E & R471E), more peripherally positioned (R463D & R489D) or at the distal tips (K454E) of the basic patch (Figure 4B). ZYG-1 is exceedingly rare and difficult to detect by immunoblotting, however, an ultrasensitive detection method revealed a band at the predicted size of ZYG-1 that was lost following RNAi-mediated depletion in the absence of a transgene (Figure 4C) and reduced (as expected due to depletion of endogenous ZYG-1) but not lost in the transgenic strains (Figure 4C). Whereas the Control zyg-1 transgene fully rescued the embryonic inviability and monopolar second division phenotype resulting from endogenous ZYG-1 depletion, the ΔCPB and central (R470E & R471E) and peripheral (R463D & R489D) basic patch mutants exhibited 100% monopolar second divisions and 0% embryonic viability, indicating failure of centriole duplication (Figure 4D, E). The mutant charge reversing the distal residue K454 exhibited a more mild phenotype with 52% of second divisions being monopolar and a small number of embryos (4.9%) even surviving to hatching (Figure 4D, E). Quantitative immunofluorescence revealed that whereas the ΔCPB and the central (R470E & R471E) and peripheral (R463D & R489D) charge reversal mutations reduced the amount of ZYG-1 recruited to centrioles to the background level seen following depletion of endogenous ZYG-1 (Figure 4F, G), charge reversing the distal K454 residue reduced, but did not abolish ZYG-1 targeting. We conclude that the ability of the CPB mutants to target to centrioles and support centriole assembly in vivo (Figure 4E, G) parallels the ability of purified CPB with the equivalent mutations to bind to the SPD-2 N-terminus in vitro (Figure 2E). Thus, our in vivo data strongly support the conclusion that the association between the SPD-2 N-terminus and the ZYG-1 CPB is mediated by electrostatic interactions involving most of the conserved charges in both proteins.
Figure 4. Basic patch residues on the ZYG-1 CPB dimer are required to target ZYG-1 to mother centrioles and promote daughter centriole assembly.
(A) Single copy zyg-1RR transgene and predicted ZYG-1 protein carrying the temperature sensitive P442L mutation. (B) Surface plot of the ZYG-1 CPB dimer showing the three tested mutant clusters. (C) Western blot of lysates from worms lacking a transgene (No Transgene) or with the indicated zyg-1RR transgenes with (+) or without (−) endogenous ZYG-1 depletion. The blot was probed for ZYG-1 (top) and α-tubulin (bottom). Asterisks mark variable background bands due to the bacterial food that the worms eat. Serial dilutions of lysate prepared from worms without a transgene (first five lanes) were used to assess endogenous ZYG-1 depletion. Numbers are percentage loaded relative to the 100% no transgene control. (D, E) Plots showing percent embryonic viability (D) and the frequency of second division monopolar spindles (E). Error bars in D = SD of percent lethality per worm; N = number of worms, n = total number of embryos (D) or the number second division cells (E). (F) (left) Deconvolved immunofluorescence images of control and zyg-1(RNAi) embryos stained for DNA and SAS-4 (red and green; left panels) and ZYG-1 (right panels). (right) Graph plotting mean centrosomal ZYG-1 fluorescence. Values are percentage of the mean control value. n = number of centrosomes. Error bars = SEM. (G) (left) Representative deconvolved widefield immunofluorescence images of embryos stained for DNA and SAS-4 (red and green; left panels) and ZYG-1 (right panels). Embryos expressing the indicated zyg-1 transgenes were depleted of endogenous ZYG-1 by RNAi. (right) Graph plotting mean centrosomal ZYG-1 fluorescence. Values are percentage of the mean control value. n = number of centrosomes. Error bars = SEM. Scale bars, 10 m.
The DmPlk4 CPB forms a Z-shaped end-to-end dimer analogous to that formed by the ZYG-1 CPB
Although a crystallographic interface that would generate a Z-shaped dimer analogous to that formed by the ZYG-1 CPB was present in the previously characterized crystal of the CPB from Drosophila Plk4 (DmPlk4), the DmPlk4 CPB was proposed to dimerize through a different interface that generates a side-by-side butterfly-like dimer (Slevin et al., 2012). As we already had crystals for a similar construct (aa 383-601, compared to 382-602 in the published construct), we characterized them to determine if they would provide insight into the DmPlk4 CPB dimerization interface. Interestingly, the small sequence change generated a different crystal form with space group P6422 (a = b = 132.21 Å, c = 134.25 Å; β = 120°), compared to the P21212 for the previous structure. We determined the DmPlk4 CPB structure by molecular replacement using the previous structure, 4G7N.pdb (Slevin et al., 2012). The structural model was refined to 3.4-Å resolution with final Rwork and Rfree of 22.6% and 24.6%, respectively (Table 1). The structure contains residues 383-596 of DmPlk4 with a short disordered loop (aa 549-552) between β11 and α2 (Figure 5A). The Drosophila and C. elegans CPB monomers were topologically identical (compare Figures 5A and 1A) and could be neatly superimposed with a r.m.s.d. of 2.1 Å for backbone atoms in the 164 aligned residues.
Figure 5. The DmPlk4 CPB forms a Z-shaped end-to-end dimer analogous to that formed by the ZYG-1 CPB.
(A) Ribbon diagram of the DmPlk4 CPB structure, color-ramped from blue to red (N- to C-terminus). The disordered loop (aa 549-552) in PB2 is shown as a dashed line. (B, C) Top (B) and side (C) views of a single hexagonal unit in the DmPlk4 CPB crystal, which contains 12 CPBs forming 6 pairs (yellow, blue, pink, green, red, and cyan) arranged in a spiral along the vertical screw axis (c-axis). (D) Side view of the previously reported butterfly-like side-by-side dimer (Slevin et al., 2012). (E) The inter-molecular contact mediated by the β5-β6 loops (arrows) of adjacent PB1 domains in the hexagonal unit. (F) The Z-dimer formed by the DmPlk4 CPB. (G, H) Superposition of the Z-dimer (G) and butterfly-dimer (H) onto the ab-initio shell reconstructed from SAXS data. The ab inito model is the most representative model selected by DAMAVER out of 40 DAMMIN runs. The mean NSD was 0.542. (I) SAXS analysis of the DmPlk4 CPB in solution. Inset shows the boxed region at a higher magnification.
In contrast to the numerous intermolecular contacts in the published crystal, our crystal form had only three, none of which resembled the proposed butterfly dimer interface (Figures 5D, S4C, D; Slevin et al., 2012). In our crystal, twelve DmPlk4 CPBs form a hexagonal spiral (Figure 5B, C). The smallest contact (interface area: 359 Å2) is located between neighboring spirals with contributions from two vector-derived residues (Figure S4A, B). The other two contacts are within the hexagonal spiral (Figure 5B, C). One is primarily mediated by the loop connecting β5 and β6 in PB1 and has an interface area of 607 Å2 (Figure 5E, inset). The last contact, mediated by the C-terminal β12 strand and the N-terminus of α2 in PB2, has a surface area of 794 Å2 and generates a Z-shaped end-to-end dimer (Figure 5F). The fact that the only intermolecular interface in common between our and the published crystal forms generates the Z-shaped dimer, suggests that DmPlk4, like ZYG-1 uses this dimerization interface. In support of this conclusion, experimental SAXS data for the DmPlk4 dimer in solution superimposed well with the scattering pattern calculated from the Z-dimer (χ = 0.935), but not with that calculated from the butterfly dimer (χ = 1.96). The low resolution ab initio shapes determined from the experimental data also provided an extended particle envelope consistent with the Z-dimer but not the butterfly-dimer (Figure 5G-I).
Thus, analysis of a new crystal form for the DmPlk4 CPB, combined with the SAXS analysis of the dimer in solution, suggests that the DmPlk4 CPB forms a Z-shaped end-to-end dimer analogous to the one formed by the ZYG-1 CPB.
Interaction with the acidic region in Drosophila asterless requires residues in the center and around the sides of the DmPlk4 CPB dimer
Mapping conserved hydrophilic residues revealed an extensive basic patch on the side of the DmPlk4 CPB dimer containing the solvent-exposed surface of the inter-molecular β-sheet, whereas no conserved charge clusters were present on the reverse side of the dimer (Figures 6A,B and S5A-C). SLS further demonstrated that each DmPlk4 CPB dimer binds to one acidic region from Drosophila asterless (aa 21-60, DmAsl AR; Figure S5D). To map residues important for binding, we generated 10 charge reversal mutants in the DmPlk4 CPB. Five of the ten mutations, corresponding to 6 residues in the basic patch (magenta in Figure 6D), significantly reduced the ability of resin-immobilized 6×His-DmPlk4 CPB to pull down MBP-DmAsl AR (Figure 6D). In addition to residues in the dimer center (K510, R523, R595 and R594), the more distal residues K498 and R490 on the dimer sides were also critical for DmAsl AR binding; the R490E mutation reduced binding to the same extent as the central R594E/R595E combination. These results suggest that the DmAsl AR makes electrostatic contacts both in the center and around the sides of the DmPlk4 CPB dimer, in a fashion similar to the interaction between the CeSPD-2 AR and the ZYG-1 CPB (Figure 6C, D).
Figure 6. The SPD-2/Cep192 acidic region interacts with a more compact region of the MmPlk4 CPB than the asterless/Cep152 acidic region.
(A) Surface plot (front view) of the DmPlk4 CPB Z-dimer with the two CPBs shown in green and pale cyan. Completely and highly (80-95% identity) conserved basic (dark and light blue) and acidic (red and pink) residues are shown. Residues in the basic surface patch (dashed line) are labeled. (B) In silico docking result using the DmPlk4 CPB Z-dimer (electrostatic surface plot) as receptor and the DmAsl N-terminus (aa 21-60; yellow stick model) as ligand. (C) Location of positively charged residues on the ZYG-1 CPB dimer that contribute to the interaction with the CeSPD-2 AR, repeated from Figure 2F for comparison. An end view is shown on right. (D-F) In vitro pull down experiments. (D, left) Resin-immobilized WT or mutant 6×His-DmPlk4 CPB (Coomassie gel; top panel) was used to pull down MBP-DmAsl AR (aa 21-60; Western blot; bottom panel). (E-F, left) Resin-immobilized GST-MmAsl/Cep152 AR (E) or GST-MmSPD-2/Cep192 AR (F) (Coomassie gels; top panels) were used to pull down WT and mutant versions of the MBP-MmPlk4 CPB (Western blots; bottom panels). (D-F, right) Location of positively charged residues on the DmPlk4 CPB dimer (D) or the modeled MmPlk4 CPB dimer (E,F; see Figure S5H) whose mutation reduced (magenta) or did not affect (pale yellow) interaction with the tested ARs. Arrows indicate a mutant outside the conserved patch that did not significantly affect the binary interactions. Empty triangles mark mutants that significantly enhanced interactions.
The SPD-2/Cep192 acidic region interacts with a more compact region of the MmPlk4 CPB than the asterless/Cep152 acidic region
The CPB of mammalian Plk4 interacts with acidic regions in both asterless/Cep152 and SPD-2/Cep192 (Dzhindzhev et al., 2010; Hatch et al., 2010; Cizmecioglu et al., 2010; Sonnen et al., 2013; Kim et al., 2013). We therefore sought to determine whether the asterless/Cep152 and SPD-2/Cep192 ARs interact with the same or different sets of CPB residues. Our attempts to crystallize the M. musculus Plk4 (MmPlk4) CPB were not successful. Nevertheless, homology-based structural modeling is generally reliable when primary sequences are more than 30% identical (Xiang, 2006). Since MmPlk4 and DmPlk4 share 38% primary sequence identity, we employed the (PS)2-v2 protein structure prediction server (Chen et al., 2009), using the DmPlk4 CPB as a template, to generate a high confidence model for the MmPlk4 CPB (aa 557-770; Figure S5E). Assembly of a homologous dimer based on the DmPlk4 CPB dimer revealed an extensive basic patch similar to that on the DmPlk4 CPB (Figure S5F-H).
To map residues that interact with the MmAsl/Cep152 and MmSPD-2/Cep192 ARs, we generated charge reversal mutations for all of the positively charged basic patch residues in the MmPlk4 CPB, along with a control mutant (K752E/R753E) that charge reversed two basic residues on the reverse side of the β-sheet and a mutant (W687A) that removed a bulky aromatic side-chain. Five of the eight mutations in the conserved basic patch dramatically reduced the ability of MBP-MmPlk4 CPB to be pulled down by resin-immobilized MmAsl AR, whereas only three significantly reduced binding to the MmSPD-2 AR (Figure 6E, F). The control K752E/R753E mutation and the W687A mutant (Figure 6E, F) did not affect binding to either acidic region. Interestingly, three of the charge reversal mutations, R564E/K566E, K660E, and K568E/K569E appeared to enhance CPB binding to the MmAsl or MmSPD-2 ARs (Figure 6E, F; empty triangles). The MmPlk4 CBP residues required for binding to the MmAsl AR, like the DmPlk4 CPB residues required for binding to the DmAsl AR, formed an elongated cleft that ran across the surface of the dimer and included residues on the dimer sides (K660, R652; Figure 6D, E). In contrast, the MmSPD-2 AR interacted with a subset of the residues required for interaction with the MmAsl AR that formed a more limited cluster in the dimer center (Figure 6F). Our results suggest that the MmSPD-2 AR interacts with a more compact region of the Plk4 CPB than the MmAsl AR.
Structural changes in the ZYG-1 CPB dimer confer the ability to selectively bind SPD-2-derived acidic regions
The different binding footprints of the MmSPD-2 and MmAsl ARs, prompted us to investigate whether the ZYG-1 or DmPlk4 CPBs are specifically adapted to bind SPD-2 versus asterless-derived peptides since the CPBs in these species no longer need to bind both acidic regions. We began by using microscale thermophoresis to measure the binding affinities of the ZYG-1 and DmPlk4 CPBs for the acidic regions of CeSPD-2, DmAsl, MmSPD-2, and MmAsl (Figures 7A, S6A). The results demonstrated that the ZYG-1 and DmPlk4 CPBs have a similar binding affinity for their natural ligands, with Kds of 0.42 and 0.88 μM for the CeSPD-2 and DmAsl ARs, respectively. The DmPlk4 CPB bound to the MmSPD-2 AR (Kd = 0.53 μM) and MmAsl AR (Kd = 0.72 μM) with similar affinity to the DmAsl AR (Kd = 0.88 μM), suggesting that the DmPlk4 CPB retains the ability to bind both asterless and SPD-2-derived acidic regions. In contrast, the ZYG-1 CPB showed remarkable specificity for SPD-2-derived ARs. The ZYG-1 CPB bound to MmSPD-2 AR with an affinity only ~12× lower than its affinity for its native ligand (Kd = 5.17 μM); in contrast, its affinities for the DmAsl and MmAsl ARs were substantially lower (Kd = 197.0 μM and too weak to measure, respectively). Cumulatively, these results suggest that the DmPlk4 CPB has remained similar to the MmPlk4 CPB, whereas the ZYG-1 CPB has evolved specificity for SPD-2-derived ARs. The reduced affinity of the DmPlk4 CPB for the CeSPD-2 AR (Kd = 26.0 μM) compared to its affinity for MmSPD-2 AR (Kd = 0.53 μM), further suggests that the CeSPD-2 acidic region has co-evolved with the ZYG-1 CPB.
Figure 7. Structural changes in the ZYG-1 CPB dimer confer the ability to selectively bind SPD-2-derived acidic regions.
(A) Kds measured by MST for different CPB-AR pairs. (B) Superposition of the DmPlk4 and ZYG-1 CPB dimers aligning one of the two CPBs. (left) A side view demonstrates that the ZYG-1 CPB dimer bends 35° away from the long axis relative to the DmPlk4 dimer. (right) A view of the plane containing the intermolecular β-sheet shows that the second ZYG-1 CPB is rotated counterclockwise towards the long axis by 15°. (C,D) (left) Electrostatic surface plots showing side and top views of the DmPlk4 (C) and ZYG-1 (D) CPB dimers. The DmPlk4 dimer has a linear long axis and the blunt angle at the dimeric junction is 105°. The ZYG-1 CPB dimer is curved, with a 35° distortion of the long axis and a blunt angle of 120°. (right) The distributions of basic residues involved in the DmAsl AR-DmPlk4 and CeSPD-2 AR-ZYG-1 interactions are shown for comparison. (E) Schematics illustrating docking modes for asterless and SPD-2-derived acidic regions onto the ZYG-1 and Plk4 CPB dimers. The two CPBs in each dimer are colored green and pale cyan. Conserved basic clusters are shown as blue patches. Acidic regions are shown as curved lines with the rough positions of the acidic (red) and basic (blue) segments highlighted. (F) Sequence comparison of the acidic regions in asterless and SPD-2 homologs. Acidic (red) and basic (blue) residues are highlighted.
To understand how the ZYG-1 CPB selectively binds SPD-2-derived ARs, we compared the ZYG-1 and DmPlk4 CPBs. Superimposing one monomer in each of the dimeric structures shows that the other monomers cannot be placed on top of each other (Figure 7B). A side-view showed that, whereas the two CPB molecules are linearly arranged along the long axis of the DmPlk4 dimer, the second CPB molecule bends 35° away from the long axis of the ZYG-1 dimer (Figure 7B-D). The view down onto the β-sheet plane further revealed that the second CPB molecule in the ZYG-1 dimer is rotated 15° counter-clockwise relative to the second CPB molecule in the DmPlk4 dimer (Figure 7B-D). This change in blunt angle narrows the central basic cleft, which adopts an elongated “S” shape, and likely explains the selectivity of the ZYG-1 CPB for binding SPD-2 over asterless-derived acidic regions (see discussion).
Discussion
The CPBs from C. elegans ZYG-1 and Drosophila Plk4 form Z-shaped dimers with a central intermolecular β-sheet
Our data suggest that the CPBs from C. elegans ZYG-1 and Drosophila Plk4 both form Z-shaped end-to-end dimers. The only intermolecular interface in common between the crystal form of the DmPlk4 CPB that we characterize here and the previously published crystal form (Slevin et al., 2012) generates the Z-shaped dimer, which is also the conformation adopted by the ZYG-1 CPB. SAXS analysis of the dimers in solution also supported Z-shaped versus other possible dimer conformations for both the ZYG-1 and DmPlk4 CPBs. The Z-dimer is further supported by our in vitro and in vivo analyses showing that the 12-stranded intermolecular β-sheet in the Z-dimer is decorated with an extended cluster of basic residues that are functionally important for CPB binding to the centriolar receptors SPD-2/Cep192 and Asterless/Cep152 in vitro and for ZYG-1 targeting and daughter centriole assembly in vivo.
Electrostatic interactions generate an interface that docks CPBs onto their centriolar receptors
Our work suggests that centriolar Plk4 docking is mediated by electrostatic interactions between basic residues on the CPB dimer and acidic residues in N-terminal unstructured regions of centriolar receptors that cumulatively result in a strong binding affinity (Kd ~ 0.5 μM). In vitro pull down assays demonstrated that, in many cases, reversal of even a single charge in the CPB basic patch significantly reduced the CPB-AR interaction. For the ZYG-1 CPB, these reductions in CeSPD-2 AR binding translated into failure to recruit ZYG-1 to centrioles and failure of centriole duplication. Similar to the distributed array of basic charges on the CPB surface, acidic charges in all regions of the CeSPD-2 AR were required for CPB binding, centriolar ZYG-1 targeting, and centriole duplication. Cumulatively, these results suggest that tight CPB-AR binding is achieved by cooperative action of individual opposite charge attractions.
Regulation of CPB docking at centrioles
For the ZYG-1-CeSPD-2 interaction, we know that ZYG-1 is present at vanishingly low levels whereas CeSPD-2, which is also a component of the PCM, does not appear to be. If CeSPD-2 in the cytoplasm and on centrioles interacted equivalently with the ZYG-1 CPB, one might expect that cytoplasmic CeSPD-2 would titrate ZYG-1 away from centrioles. In addition, whereas CeSPD-2 targets to the PCM and centrioles, ZYG-1 appears to target more specifically to centrioles. These results suggest that availability of the CeSPD-2 AR might be regulated so that only CeSPD-2 docked onto centrioles, and not CeSPD-2 in the cytoplasm or PCM, has the ability to interact with ZYG-1. CeSPD-2 AR availability could be negatively regulated by an intramolecular interaction with residues in the central and/or C-terminal regions of CeSPD-2; indeed, the CeSPD-2 sequence in these regions contains several conserved basic clusters and is overall positively charged. During centriole duplication, binding of CeSPD-2 to the mother centriole might relieve this intramolecular inhibition, releasing the AR to interact with the ZYG-1 CPB.
Structural changes in the ZYG-1 CPB and CeSPD-2 AR have led to specificity for SPD-2 derived peptides and a more asterless-like binding mode
In contrast to SPD-2-derived ARs, asterless-derived ARs have conserved basic, as well as acidic residues, near their centers (Figures 7F, 2A, S2A, S2B). While ARs containing basic residues can be accommodated by the broad basic cleft in the DmPlk4 and MmPlk4 CPBs, they are likely excluded by the narrower cleft in the ZYG-1 CPB. This idea is supported by docking trials, which show that while the CeSPD-2 and MmSPD-2 ARs make numerous contacts when docked onto the ZYG-1 CPB dimer, far fewer contacts are made in the best docking solutions for asterless-derived ARs (Figure S6B-G).
Pull down experiments revealed that basic residues on the sides of the DmPlk4/MmPlk4 CPB dimers are critical for binding to the DmAsl and MmAsl ARs (Figures 6D, E, 7E). This suggests that binding of asterless-derived ARs to the CPB dimer has a pincer-like quality that may involve the patches of acidic residues on the two sides of the DmAsl and MmAsl ARs (Figure 7F), an idea consistent with the results of docking trials (Figure S6B). In contrast, only the basic residues in the center of the CPB dimer are required for binding to the MmSPD-2 AR (Figure 6F), which lacks distal patches of acidic residues (Figure 7F). Interestingly, the CeSPD-2 AR has distal acidic patches on both sides (Figure 7E, F) that are functionally important in vitro and in vivo (Figures 2C, 2D, 3) and basic residues on the sides of the ZYG-1 CPB dimer are also important for SPD-2 AR binding, ZYG-1 targeting, and daughter centriole assembly (Figures 2E, 2F, 4). Cumulatively, these results suggest that co-evolution of the ZYG-1 CPB and the SPD-2 acidic region have conferred a more asterless-like mode of binding in which distal acidic patches contact residues on the sides of the CPB dimer (Figure S6E).
An interesting question is why vertebrates have CBP-binding acidic regions in both asterless/Cep152 and SPD-2/Cep192, whereas other species use one or the other, and whether these two complexes function redundantly and/or collaboratively in Plk4 targeting (Kim et al., 2013; Sonnen et al., 2013). The fact that the basic residues on the sides of the MmPlk4 CPB are essential for asterless/Cep152 but not SPD-2/Cep192 AR binding suggests that characterization of side residue CPB mutants might provide a means to analyze the relative roles of Plk4 binding to SPD-2/Cep192 versus asterless/Cep152 in vivo.
Experimental Procedures
Protein expression and purification
Proteins were expressed in E. coli BL21(DE3) after induction with 0.5 mM IPTG for 20 h at 16°C; vectors are described in the Supplemental Experimental Procedures. ZYG-1 and DmPlk4 CPBs were purified by nickel affinity chromatography and exchanged into 50 mM HEPES-NaOH (pH 7.5), 250 mM NaCl after proteolytic removal of the 6×His tag. For crystallization, lysine methylated proteins were concentrated to ~5 mg/ml after purification on a Superdex-200 16/60 column (GE Healthcare) pre-equilibrated with buffer containing 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5% (v/v) glycerol, and 2 mM dithiothreitol (DTT). SeMet-substituted ZYG-1 CPB was expressed following published protocols (Doublie, 1997), and purified as above, except 10 mM DTT was added to the gel filtration buffer. Ligands for SAXS and MST binding assays were isolated by nickel affinity chromatography. After removal of the Trx tag, proteins were further purified on a Superdex-200 16/60 column equilibrated as described above and concentrated to 5-10 mg/ml. For details see Supplemental Experimental Procedures.
Crystallization and data collection
SeMet-substituted ZYG-1 CPB was crystallized at 4°C by the hanging drop method against a reservoir solution containing 100 mM tri-sodium citrate (pH 5.6), 15% (w/v) PEG 4,000 and 200 mM (NH4)2SO4. Crystals were soaked in reservoir solution containing increasing amounts of glycerol with a final concentration of 20% (v/v), loop mounted, and flash frozen in liquid nitrogen. Diffraction data were collected at the beamline ID29 at the European Synchrotron Radiation Facility (ESRF). DmPlk4 CPB was crystallized at 4°C using the sitting drop method against a reservoir solution containing 100 mM HEPES-NaOH (pH 7.0), 2.5 M NaCl and 2 mM MgCl2. Crystals were soaked in reservoir solution containing increasing amounts of ethylene glycol with a final concentration of 20% (v/v), loop mounted, and flash frozen in liquid nitrogen. Diffraction data were collected at the beamline ID14-4 of the ESRF.
Structure determination
Diffraction data were integrated and scaled using XDS (Kabsch, 2010). The ZYG-1 CPB structure was determined by the SAD methods. Selenium sites were located and experimental maps calculated using Phenix (Terwilliger et al., 2009). ARP/wARP (Langer et al., 2008), COOT (Emsley and Cowtan, 2004), and Phenix (Terwilliger et al., 2009) were used to build the initial model, check and manually build missing loops/gaps, and refine the structure, respectively. The DmPlk4 CPB structure was determined by molecular replacement using the previously determined DmPlk4 CPB structure (4G7N.pdb; Slevin et al., 2012). COOT and Phenix were used to check and manually rebuild and refine the structure, respectively.
Static light scattering (SLS)
SLS was carried out on a Mini-DAWN Treos light scattering instrument coupled with 1260 Infinity HPLC system (Agilent Technologies) and Superdex-200 10/300 GL gel filtration column (GE Healthcare) equilibrated in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2 mM DTT, and 2 mM MgCl2. Data were collected and analyzed by Wyatt Astra (5.3.4.14) software.
Small angle X-ray scattering (SAXS)
SAXS data collection and processing are described in detail in the Supplemental Experimental Procedures. Data were collected at the EMBL beamline X33 (DESY, Hamburg), Germany and the ESRF beamline BM29 (Pernot et al., 2013) in Grenoble, France. Prior to data collection, samples were purified by gel filtration chromatography and checked for monodispersity by SLS. Individual X-ray exposures were monitored for radiation sensitivity. The data was processed and merged using PRIMUS (Konarev et al., 2003), and the pair distribution function was computed using GNOM (Svergun, 1992). Multiple rounds of ab initio shape reconstruction by both DAMMIN (Svergun, 1999) and GASBOR (Svergun et al., 2001) were consistent. Theoretical scattering was calculated from atomic structures using CRYSOL (Svergun et al., 1995).
Microscale thermophoresis (MST)
MST was performed using a Monolith NT.115 (Nanotemper Technologies® GmBH). Labeled CPBs (Monolith NT.115 Labeling Kit RED-NHS) were mixed with binding partners in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5% (v/v) glycerol, 2 mM DTT, 2 mM MgCl2, 0.5 mg/ml BSA, and 0.2% (v/v) Tween-20. Dissociation constants were determined using the NTA analysis software 1.4.27 (Nanotemper Technologies® GmbH) with subsequent nonlinear fitting in SigmaPlot (Systat Software®).
In vitro pull-down assays and Western blots
For additional details see Supplemental Experimental Procedures. Western blots for pull-downs were performed using mouse monoclonal antibodies against 5×His (1:2000; QIAGEN) and MBP (1:10,000; NEB). Goat anti-mouse HRP secondary (NEB) was visualized using SuperSignal® West Pico (Thermo Scientific). Worm lysate blots were performed as described (Lettman et al., 2013) using rabbit antibodies to SPD-2 (aa 2-200; 1 μg/ml) and ZYG-1 (aa 250-371; Lettman et al., 2013). HRP-conjugated secondary (1:10,000; GE Healthcare Life Sciences) was visualized using ECL-Prime (GE Healthcare; SPD-2) or WesternBright Sirius (Advansta; ZYG-1). α-tubulin was detected using DM1α (1:500; Sigma-Aldrich) followed by an alkaline-phosphatase-conjugated anti-mouse secondary (1:3,750; Jackson ImmunoResearch Laboratories, Inc.).
C. elegans strains and RNA-mediated interference
C. elegans strains (listed in Table S1) and dsRNAs are described in the Supplemental Experimental Procedures. L4 hermaphrodites were injected with dsRNA and incubated at 16°C for 40-44 h before dissection and imaging of their embryos. For viability assays, L4 worms were injected with dsRNA and incubated at 16°C for 48 hours. Adults were singled and allowed to lay eggs for 24 hours before removing them from the plate. Hatched larvae and unhatched embryos were counted 24-30 hours later.
Microscopy, immunofluorescence, and image analysis
For details on microscopy and immunofluorescence, see Supplemental Experimental Procedures. Centrosomal ZYG-1 and mCh::SPD-2 fluorescence was quantified from maximum intensity projections of the portion of the z-stack containing the centrosomes using MetaMorph software (Molecular Devices). A 10 (ZYG-1) or 14 (mCh::SPD-2) pixel wide box was drawn around each centrosome, along with a box one pixel larger on each side in both dimensions. The per-pixel background was calculated as [(integrated intensity in the larger box - integrated intensity in the smaller box)/(area of larger box - area of smaller box)]. The centrosomal ZYG-1/mCh::SPD-2 signal was the integrated intensity in the smaller box minus the area of the smaller box multiplied by the per pixel background.
Supplementary Material
Highlights.
Cryptic polo boxes (CPBs) from Plk4 kinases (ZYG-1 and DmPlk4) form Z-shaped dimers
A basic patch on CPB dimers docks onto acidic regions in centriolar receptors
The CPB basic patch is essential for ZYG-1 targeting and centriole assembly in vivo
The ZYG-1 CPB preferentially binds SPD-2 over asterless-derived acidic regions
Acknowledgements
This work was supported by grants from the Austrian Science Fund (P23440-B20 to GD) and from the National Institutes of Health (R01-GM074207 to KO) and by institutional support from the Max F. Perutz Laboratories to GD and from the Ludwig Institute for Cancer Research to KO. We acknowledge the ESRF and EMBL for providing synchrotron radiation facilities, Arshad Desai for critical discussions, Alexander Dammermann for identifying ZYG-1 homologs, and the CSF (www.csf.ac.at) for help with the CD and MST experiments.
Footnotes
Accession code The coordinates for the ZYG-1 and DmPlk4 CPBs have been deposited to the Protein Data Bank (PDB) under accession codes of 4NKB and 4NK7, respectively.
Author Contributions GD and KO conceived and designed the experiments. ES and JL carried out the crystallizations. GD and ES determined the crystal structures. GD performed the in silico dockings. RQ generated the mutations of MmPLK4. ES generated all other mutations and performed the in vitro pull-down, MST, and SLS experiments. VV, MML and KO performed the in vivo experiments. DS and AR analyzed the SAXS data. GD, KO, ES, DS and AR contributed to writing the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderhub SJ, Krämer A, Maier B. Centrosome amplification in tumorigenesis. Cancer Lett. 2012;322:8–17. doi: 10.1016/j.canlet.2012.02.006. doi:10.1016/j.canlet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. Building the centriole. Curr. Biol. 2010;20:R816–25. doi: 10.1016/j.cub.2010.08.010. doi:10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. Deconstructing the centriole: structure and number control. Curr. Opin. Cell Biol. 2012;24:4–13. doi: 10.1016/j.ceb.2012.01.003. doi:10.1016/j.ceb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. doi:10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Hwang J-K, Yang J-M. PS)2-v2: template-based protein structure prediction server. BMC Bioinformatics. 2009;10:366. doi: 10.1186/1471-2105-10-366. doi:10.1186/1471-2105-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. doi:10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Müller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. doi:10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gönczy P. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. doi:10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. doi:10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. doi:10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. doi:10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. doi:10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana SC, Bazan JF, Bettencourt-Dias M, Bettencourt-Dias M. Polo boxes come out of the crypt: a new view of PLK function and evolution. Structure. 2012;20:1801–1804. doi: 10.1016/j.str.2012.10.008. doi:10.1016/j.str.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. doi:10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Song MH, Addepalli MK, Hunter G, O’Connell K. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics. 2007;176:95–113. doi: 10.1534/genetics.107.071803. doi:10.1534/genetics.107.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Kusch M, Wolf N. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics. 1988;120:977–986. doi: 10.1093/genetics/120.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Park JE, Shukla A, Choi S, Murugan RN, Lee JH, Ahn M, Ree K, Bang JK, Kim BY, Loncarek J, Erickson RL, Lee KS. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc Natl Acad Sci USA. 2013;110:E4849–4857. doi: 10.1073/pnas.1319656110. doi: 10.1073/pnas.1319656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, Shen Y, Li K, Zheng J, Vakili P, Paschalidis IC, Vajda S. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins. 2010;78:3124–3130. doi: 10.1002/prot.22835. doi:10.1002/prot.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. doi:10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen S-H, Shiau AK, Zhou H, Desai A, Oegema K. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell. 2013;25:284–298. doi: 10.1016/j.devcel.2013.03.011. doi:10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat. Struct. Biol. 2002;9:719–724. doi: 10.1038/nsb848. doi:10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol. Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. doi:10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. doi:10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR, Osterberg VR, Lyczak R, Madison EE, Nguyen MH, Sandberg NA, Sedghi N, Willis JH, Yochem J, Johnson EA, Bowerman B. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS ONE. 2011;6:e16644. doi: 10.1371/journal.pone.0016644. doi:10.1371/journal.pone.0016644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Müller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. doi:10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlü N, Hannak E, Cowan C, Habermann B, Ruer M, Müller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. doi:10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Pernot P, Round A, Barrett R, De Maria Antolinos A, Gobbo A, Gordon E, Huet J, Kieffer J, Lentini M, Mattenet M, et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J Synchrotron Radiat. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel SAI, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, Katritch I, Melo FA, Ladbury JE, Schreiber G, Watts A, Braun D, Duhr S. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. doi:10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC. The Structure of the Plk4 Cryptic Polo Box Reveals Two Tandem Polo Boxes Required for Centriole Duplication. Structure. 2012 doi: 10.1016/j.str.2012.08.025. doi:10.1016/j.str.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk A-M, Anselm E, Stierhof Y-D, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell. Sci. 2013;126:3223–3233. doi: 10.1242/jcs.129502. doi:10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch M. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;77:2896–2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun DI, Koch M. Advances in structure analysis using small-angle scattering in solution. Curr. Opin. Struct. Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch M. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306–312. doi: 10.1038/sj.onc.1208275. doi:10.1038/sj.onc.1208275. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta. Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. doi:10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z. Advances in homology protein structure modeling. Curr. Protein Pept. Sci. 2006;7:217–227. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.