Abstract
The nonsense-mediated mRNA decay (NMD) pathway selectively eliminates aberrant transcripts containing premature translation termination codons (PTCs) and regulates the levels of a number of physiological mRNAs. NMD modulates the clinical outcome of a variety of human diseases, including cancer and many genetic disorders, and may represent an important target for therapeutic intervention. Here we have developed a novel multicolored, bioluminescence-based reporter system that can specifically and effectively assay NMD in live human cells. Using this reporter system, we conducted a robust high-throughput small-molecule screen in human cells and, unpredictably, identified a group of cardiac glycosides including ouabain and digoxin as potent inhibitors of NMD. Cardiac glycoside-mediated effects on NMD are dependent on binding and inhibiting the Na+/K+-ATPase on the plasma membrane and subsequent elevation of intracellular calcium levels. Induction of calcium release from endoplasmic reticulum also leads to inhibition of NMD. Thus, this study reveals intracellular calcium as a key regulator of NMD and has important implications for exploiting NMD in the treatment of disease.
The NMD pathway selectively degrades mRNAs harboring PTCs and, in so doing, guards cells against insults from potentially deleterious truncated proteins. In addition to eliminating faulty mRNA transcripts, NMD regulates the levels of many physiological mRNAs possessing features that are recognized by the NMD machinery1,2. By modulating the activity of NMD, cells can enact gene expression programs crucial for normal development or for responding to environmental cues such as hypoxia and amino acid deprivation3,4. Furthermore, approximately one-third of human genetic diseases are the manifestation of PTC mutations5, and whole genome sequencing has recently uncovered numerous somatic nonsense mutations in tumor samples6. Thus, NMD has become an attractive target for the treatment of many human diseases. For example, inhibiting NMD may alleviate the symptoms of certain genetic diseases caused by PTCs if the truncated protein products are functional or partially functional hypomorphs7,8. NMD inhibition also represents a promising cancer therapeutic strategy7. Cancer cells likely have an elevated dependency on NMD for survival due to the production of many nonsense mRNAs as a result of their intrinsic genomic instability. Thus, inhibiting NMD may lead to preferential killing of cancer cells. Moreover, inhibiting NMD may also result in production of new antigens on tumor cells that could induce an anticancer immune response9.
RESULTS
Development of a novel dual-color, bioluminescence-based NMD reporter system
To investigate the NMD pathway and to begin to develop NMD-targeting therapeutics, we constructed a multicolored, bioluminescence-based reporter for assaying NMD in mammalian cells, as illustrated in Fig. 1a and Supplementary Fig. 1. This reporter comprises a single expression vector containing two separate transcription units, each with a luciferase inserted into a TCRβ minigene at the same position within the second exon. The first transcription unit consists of a PTC-containing TCRβ minigene fused to click beetle red luciferase (CBR-TCR(PTC)). The second unit contains a wild-type TCRβ minigene fused to click beetle green 99 luciferase (CBG99, hereafter referred to as CBG for simplicity) (CBG-TCR(WT)). Expression of both fusion reporter genes are controlled by separate CMV promoters, splice sites, and polyadenylation signals of identical sequences. A sequence encoding an HA-tag was included in the first exon of the fusion reporter genes, which provides an independent method to detect the translated fusion protein products through Western blotting. PTCs in the well characterized TCRβ minigene are known to elicit robust NMD (but not 100% efficient as is the case for other reporter genes examined)10,11. The CBR-TCR(PTC) and CBG-TCR(WT) transcription units share > 99% sequence identity at the DNA, pre-mRNA, and mRNA levels (see the reporter sequence in Supplementary Fig. 2). Using this dual-colored reporter, NMD is quantified by the ratio of CBR activity to CBG activity, with an increase in the CBR/CBG (red/green) ratio representing inhibition of NMD. Here, the CBR luciferase activity serves as an indirect measure of the steady-state levels of the CBR-TCR(PTC) fusion mRNA, which is targeted for degradation by NMD, whereas the CBG luciferase activity reflects the steady-state levels of the CBG-TCR(WT) fusion mRNA, which is unresponsive to NMD. The use of CBG-TCR(WT) as an internal control in the same cell ensures that changes in the CBR/CBG ratio reflect effects specifically attributable to NMD, but not indirect effects that result from variations in reporter DNA delivery or from effects on cell viability or various steps of gene expression such as transcription, splicing, polyadenylation, and translation. The use of the highly sensitive and closely related red-emitting CBR and green-emitting CBG luciferases, combined with a spectral deconvolution algorithm for unmixing CBR and CBG signals, allows rapid and accurate measurement of their respective activities simultaneously in a single reaction with the same D-luciferin substrate12.
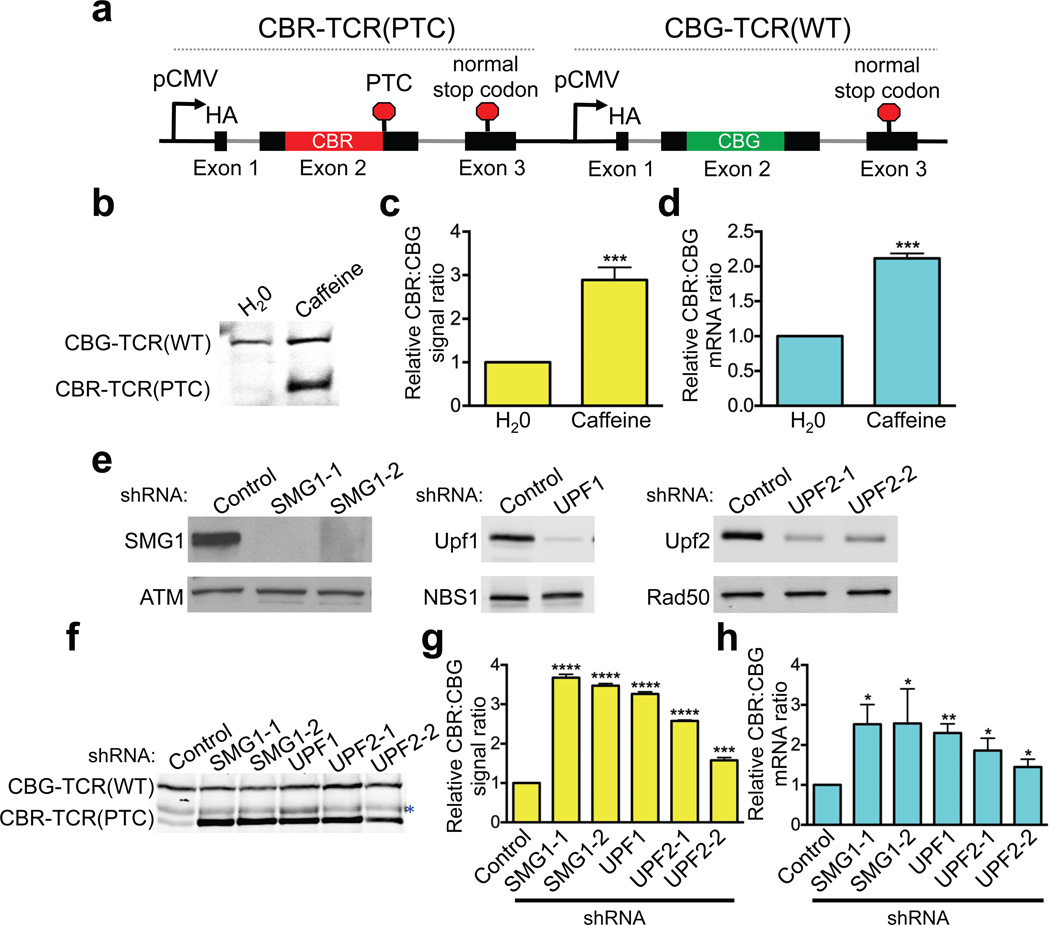
Figure 1. A dual-color bioluminescence-based NMD reporter system.
(a) Schematic diagram of the reporter construct containing two tandem, highly homologous transcription units, CBR-TCR(PTC) and CBG-TCR(WT). See Fig. S1 and text for details. (b–h) Validation of the NMD reporter depicted in a. (b) Western blot analysis of the CBR-TCR(PTC) and CBG-TCR(WT) protein levels in U2OS reporter cells treated with vehicle (H2O) or caffeine. (c) Ratios of CBR/CBG bioluminescence signals in reporter cells treated with vehicle (H2O) or caffeine. The ratio in H2O-treated reporter cells was normalized to 1. Data represent the mean ± SD of four independent experiments. ***P < 0.001 (paired t-test). (d) Ratios of CBR-TCR(PTC)/CBG-TCR(WT) mRNA levels in reporter cells treated with vehicle (H2O) or caffeine. The ratio in H2O-treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. ***P < 0.001 (paired t-test). (e) shRNA-mediated knockdown of the NMD factors SMG1 (2 shRNAs), UPF1 (1 shRNA) or UPF2 (2 shRNAs) in the dual-colored U2OS reporter cells. (f) Western blot analysis of CBR- TCR(PTC) and CBG-TCR(WT) protein levels in reporter cells after control-knockdown or SMG1-, UPF1-, or UPF2-knockdown. *, nonspecific band. (g) Ratios of CBR and CBG bioluminescence signals in reporter cells after control-knockdown or SMG1-, UPF1- or UPF2-knockdown. The ratio in control- knockdown reporter cells was normalized to 1. Data represent the mean ± SD of three biological replicates. ***P < 0.001; ****P < 0.0001 (t-test). (h) Ratios of CBR-TCR(PTC) and CBG-TCR(WT) mRNA levels in reporter cells after control-knockdown or SMG1-, UPF1- or UPF2-knockdown. The ratio in control-knockdown reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. **P < 0.01; *P < 0.05 (paired t-test).
To validate the NMD reporter system, we generated a human U2OS cell line stably expressing the reporter. Western blot results indicate that while the CBG-TCR(WT) fusion protein was efficiently expressed, the basal level of the CBR-TCR(PTC) truncated protein was barely detectable (Fig. 1b). This result is consistent with the prediction that CBR-TCR(PTC) mRNA, but not CBG-TCR(WT) mRNA, is targeted for NMD. Treatment with caffeine, a potent (but not specific) inhibitor of the kinase activity of the NMD factor SMG1, restored the CBR-TCR(PTC) protein levels. Bioluminescence imaging reveals that the CBR/CBG ratio increased by ~ 3-fold following caffeine treatment (Fig. 1c). Importantly, quantitative RT-PCR (RT-qPCR) analysis performed with primers specific to CBR or CBG luciferases in the reporter revealed a similar increase in the ratio of CBR-TCR(PTC)/CBG-TCR(WT) reporter mRNAs (Fig. 1d and Supplementary Table 1b). Furthermore, shRNA-mediated knockdown of NMD factors SMG1, UPF1 and UPF2 also inhibited NMD of the reporter, as measured by Western blot, bioluminescence assay and RT-qPCR (Fig. 1e–h). Taken together, these data demonstrate that this mechanism-based NMD reporter system faithfully recapitulates the characteristics of NMD in human cells and is expected to be specific, rapid and sensitive.
A high-throughput small-molecule compound screen identified cardiac glycosides as potent inhibitors of NMD
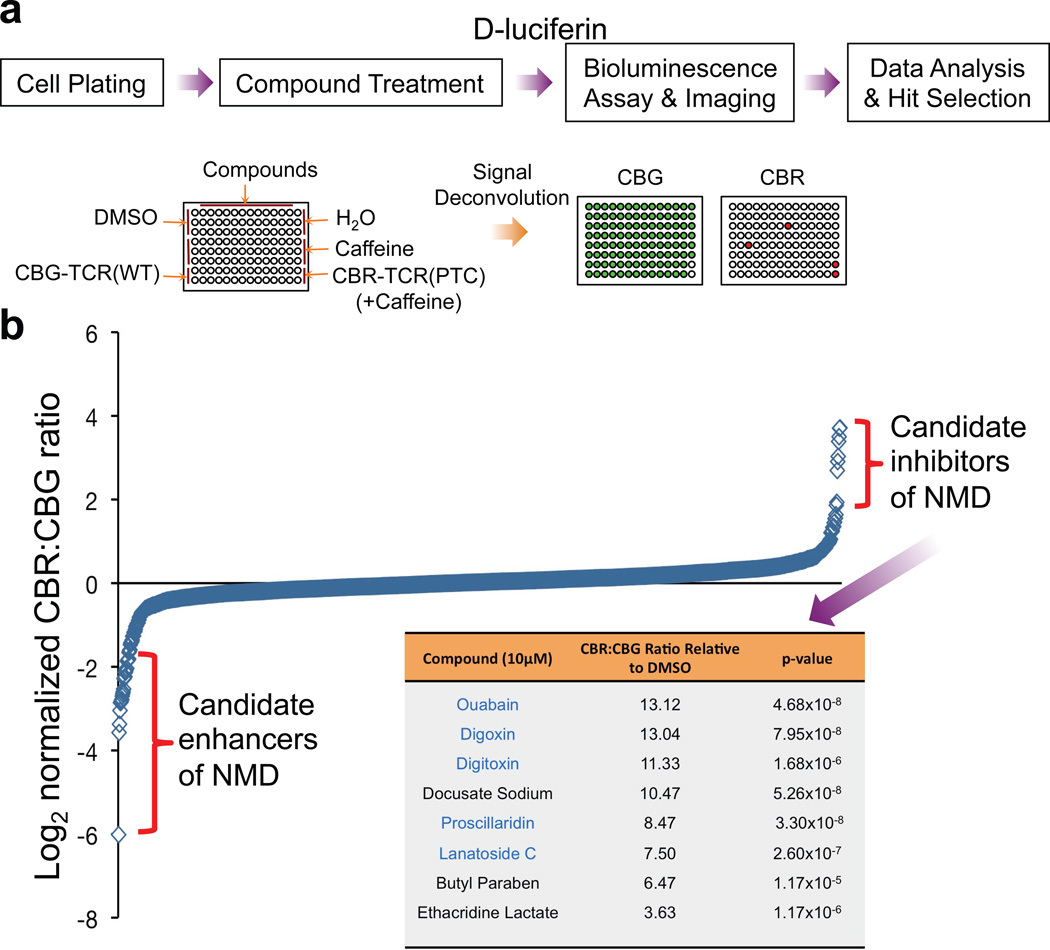
Using the reporter system described above, we next performed a high-throughput screen to identify drug candidates that can alter NMD activity in human cells. The Pharmakon library, which contains a diverse array of 1,600 clinically-evaluated compounds, was used as the source of small molecules in the hope of repurposing existing drug candidates to fast-track the drug development process. Human U2OS cells containing the dual-colored NMD reporter were seeded on 96-well plates and individual drugs were added to each well at a concentration of 10 µM, along with appropriate controls. Twenty-four hours after drug treatment, D-luciferin was added to the medium followed by bioluminescence imaging and spectral unmixing to obtain CBR and CBG signals (Fig. 2a). A Z’ factor of 0.77 was calculated for the reporter assay, demonstrating the robustness of the strategy (see Methods). As shown in Fig. 2b and Supplementary Table 2, the majority of the compounds in the library exhibited little or no effect on NMD of the reporter. However, 8 candidate inhibitors and 14 candidate enhancers of NMD were identified by quartile analysis after employing stringent criteria for hit selection (Fig. 2b and Supplementary Table 3)13. Focusing on NMD inhibitors in this study, the effects of seven of the inhibitor hits were confirmed in follow-up analysis, and each validated compound inhibited NMD in a dose-dependent manner (data not shown). Strikingly, the top five verified hits, including digitoxin, digoxin, lanatoside C, proscillaridin and ouabain, are all cardiac glycosides (CGs), which inhibit Na+/K+-ATPase in cells (Fig. 2b)14.
Figure 2. A high-throughput screen using the NMD reporter identified existing drugs that modulate NMD.
(a) Procedure for a high-throughput screen of the Pharmakon 1600 drug library.
(b) Primary screening data. Data are shown as the log2 of the normalized CBR/CBG ratio for each compound. Each ratio was normalized to the DMSO controls on the same plate. Data represent the average of three biological replicates. Compounds were ordered from left to right with increasing relative CBR/CBG ratios. Eight candidate NMD inhibitors of stringent statistical significance are shown in the table. See Supplementary Table 2 for primary screen data of all compounds in the library.
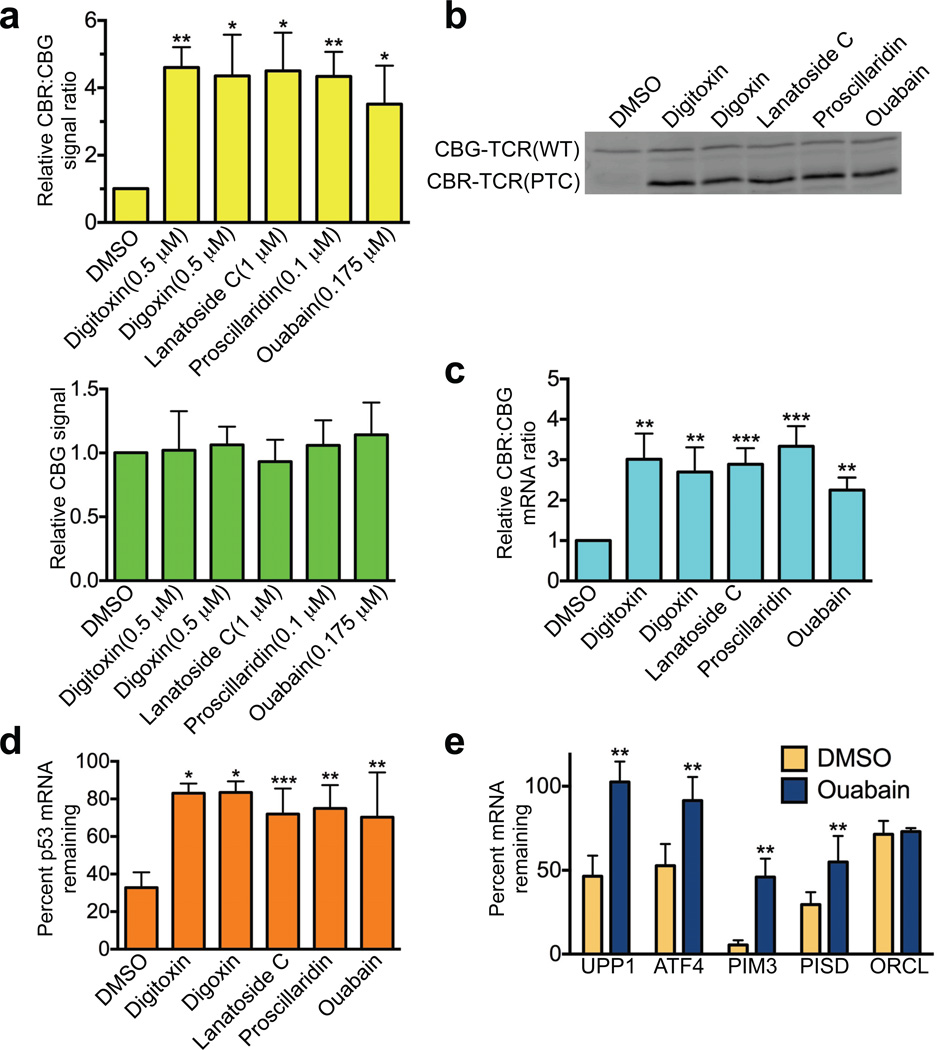
At the initial screening concentration (10 µM), all CGs increased CBR signal compared to DMSO treatment, but these drugs also decreased CBG signal, potentially due to non-specific toxic effects on cell viability (Supplementary Table 2). However, we identified a lower concentration for each drug at which the CBR activity was still dramatically increased while the CBG activity remained largely unaffected (Fig. 3a). At these modest concentrations, these drugs did not significantly impact general translation in cells and had only a mild inhibitory effect on cell viability (Supplementary Fig. 3a, b). Because CBR and CBG proteins are relatively stable, the inhibitory effects of CGs on the signal generated by the NMD reporter was time-dependent, increasing over the 24 h treatment period; CG-mediated blockade of NMD was fully concentration-dependent, showing reporter-generated EC50 values of ~70 nM and ~200 nM for ouabain and digoxin, respectively (Supplementary Fig. 4a, b). Importantly, our bioluminescence imaging results were corroborated by Western blot and reporter-specific RT-qPCR analyses of the CBR-TCR(PTC) and CBG-TCR(WT) protein and mRNA levels, respectively (Fig. 3b, c).
Figure 3. Cardiac glycosides are potent inhibitors of NMD.
(a) CBG bioluminescence signal (lower panel) and ratios of CBR/CBG bioluminescence signals (upper panel) in U2OS reporter cells treated with DMSO or cardiac glycosides for 24 h at the indicated concentrations. The CBG signal and the CBR/CBG ratio in DMSO-treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. *P < 0.05; **P < 0.01 (paired t-test).
(b) Western blot analysis of CBR-TCR(PTC) and CBG-TCR(WT) protein levels in U2OS reporter cells treated with DMSO or cardiac glycosides for 24 h at the concentrations indicated in a.
(c) Ratios of CBR-TCR(PTC)/CBG-TCR(WT) mRNA levels in U2OS reporter cells treated with DMSO or cardiac glycosides for 24 h at the concentrations indicated in a. The ratio in DMSO-treated reporter cells was normalized to 1. Data represent the mean ± SD of four independent experiments. **P < 0.01; ***P < 0.001(paired t-test).
(d) Effects of cardiac glycosides on the stability of the PTC-containing p53 mRNA in Calu-6 cells. Cells were treated with DMSO or cardiac glycosides at the concentrations indicated in a for 16 h before the addition of the transcriptional inhibitor actinomycin D (5 µg ml−1) to block new RNA synthesis. Total RNA was collected immediately before or 6 h after the addition of actinomycin D, and p53 mRNA levels were analyzed by RT-qPCR and normalized to GAPDH mRNA levels. Data represent the percent mRNA remaining 6 h after transcriptional ablation (mean ± SD) from at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001(paired t-test).
(e) Effects of ouabain on the stability of wild-type endogenous NMD target transcripts (UPP1, ATF-4, Pim3 and Pisd) in Calu-6 cells. ORCL mRNA, which is not a NMD target, was used as a control. Cells were treated with DMSO or ouabain, and then actinomycin D; samples were collected and analyzed as described in d. Data represent the percent mRNA remaining 6 h after transcriptional ablation (mean ± SD) from four independent experiments. **P < 0.01 (paired t-test).
To further validate the effects of CGs on NMD using an independent NMD model system, we used the human Calu-6 cell line, which expresses a PTC-containing nonsense mRNA of endogenous p5315,16. Calu-6 cells were first treated with CGs for 16 h. Subsequently, the transcription inhibitor actinomycin D was added to block new mRNA synthesis and the levels of p53 mRNA were determined by RT-qPCR at 0 h and 6 h after actinomycin D treatment. All 5 CGs significantly increased the stability of the mutant p53 transcript, compared with DMSO (Fig. 3d). In further support of the idea that CGs inhibit NMD, we observed increased stability of wild-type endogenous mRNA targets of NMD, including UPP1, ATF4, PIM3 and PISD, in Calu6 cells after treatment with ouabain. In comparison, the stability of endogenous ORCL transcripts, which are not targeted by NMD, was unaffected under the same condition (Fig. 3e)17–20. It has been previously shown that the mRNA levels of many of the NMD factors such as SMG1, UPF1, UPF2, UPF3B, SMG5, SMG6 and SMG7 are controlled by NMD via an autoregulatory loop, as they are themselves targets of NMD20,21. Consequently, these transcripts are upregulated upon ablation of NMD activity21,22. If CGs inhibit NMD, one would expect an increase in these NMD factor transcript levels after CG treatment. Indeed, we found that ouabain treatment led to upregulation of all of these autoregulated NMD factors except SMG6 (whose upregulation was also less obvious upon NMD inhibition after UPF1 knockdown in previous studies21,22) (Supplementary Fig. 4c). Taken together, these data strongly suggest that CGs have a previously unrecognized ability to inhibit NMD in human cells.
Na+/K+-ATPase is a robust regulator of NMD
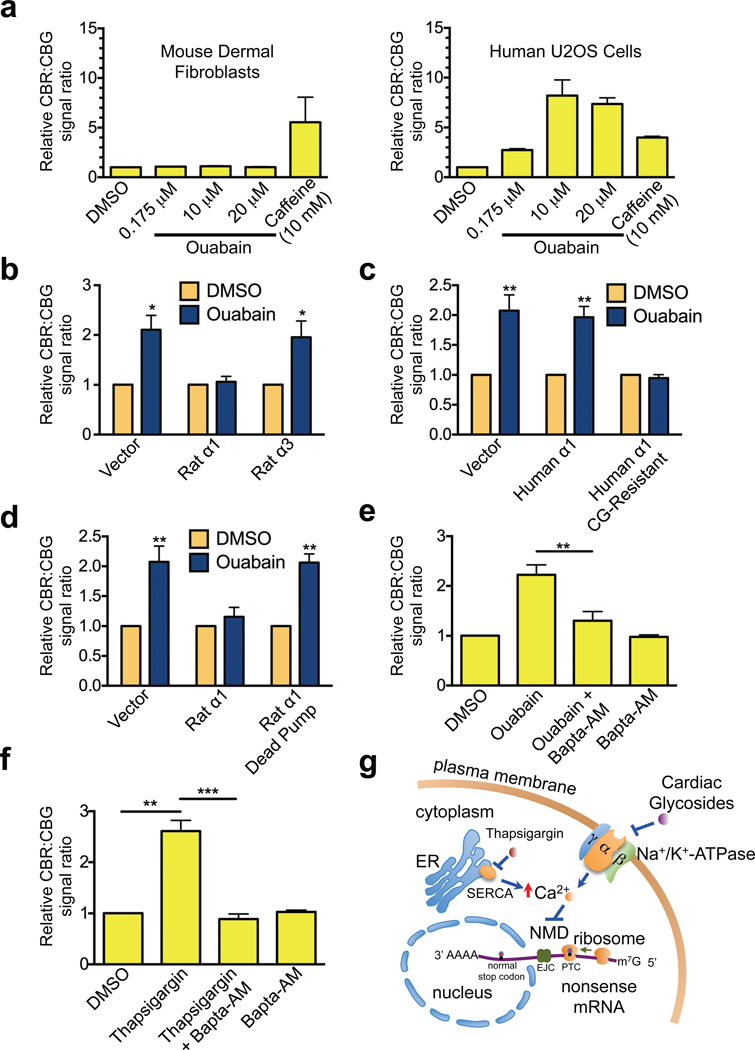
Since the Na+/K+-ATPase (composed of a catalytic α subunit and a structural β subunit, each with various isoforms, and a regulatory γ subunit) is the only known pharmacological target of the identified CGs, our results suggest that CGs inhibit NMD via the sodium-potassium pump23. Consistent with this idea, our reporter assay indicates that mouse skin fibroblasts, which express a naturally CG-resistant α1 subunit of Na+/K+-ATPase, exhibited more than 100 times greater resistance to ouabain, compared to human cells in which all 4 isoforms of the α subunit have a much higher affinity for CGs (Fig. 4a)24. In contrast, NMD of the reporter exhibited a similar level of sensitivity to caffeine in both human and mouse cells (Fig. 4a). Moreover, overexpression in human cells of the rat α1 subunit of Na+/K+-ATPase, which also has a low affinity for CGs, abrogated the inhibitory effects of ouabain on NMD. In contrast, expression of a similar level of rat α3 subunit of Na+/K+-ATPase, which is sensitive to CGs24,25, did not cause resistance of NMD to ouabain (Fig. 4b and Supplementary Fig. 5a). Furthermore, expression of a CG-resistant mutant version of the human α1 subunit also prevented the CG-mediated inhibition of NMD while the wild-type protein failed to do so (Fig. 4c and Supplementary Fig. 5b). To further demonstrate that CGs inhibit NMD through their binding and inhibition of Na+/K+-ATPase, we generated a catalytically inactive mutant of the rat α1 subunit (D376E) that has diminished binding and hydrolysis of ATP25. Unlike the WT rat α1 subunit, expression of this mutant in human cells could not cause resistance of NMD to CGs, indicating that blockade of the sodium/potassium pump enzymatic activity is necessary for efficient CG-induced inhibition of NMD (Fig. 4d and Supplementary Fig. 5c). Taken together, these data strongly suggest that Na+/K+-ATPase is a robust regulator of NMD in mammalian cells and that CGs inhibit NMD through binding and inhibiting this sodium-potassium pump. Given the essential role of Na+/K+-ATPase for cell viability, the observation that CGs strongly inhibited NMD at concentrations where no obvious decrease in CBG activity or cell viability was observed suggests that partial inhibition of the sodium-potassium pump is sufficient to abolish NMD (Fig. 3 and Supplementary Fig. 3b).
Figure 4. Cardiac glycosides block NMD through inhibition of Na+/K+-ATPase and elevation of intracellular calcium.
(a) Ratios of CBR/CBG bioluminescence signals in mouse dermal fibroblasts (left panel) or human U2OS cells (right panel) expressing the NMD reporter after 24 h treatment with DMSO, caffeine, or various concentrations of ouabain. The ratio in DMSO-treated reporter cells was normalized to 1. Data represent the mean ± SD of three biological replicates.
(b) Ratios of CBR/CBG bioluminescence signals in human U2OS reporter cells expressing either empty vector, rat α1 subunit or rat α3 subunit of Na+/K+-ATPase after 24 h treatment with DMSO or ouabain. The ratio in DMSO-treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. *P < 0.05 (paired t-test).
(c) Ratios of CBR/CBG bioluminescence signals in human U2OS reporter cells expressing either empty vector, human α1 subunit, or CG-resistant mutant human α1 subunit of Na+/K+-ATPase after 24 h treatment with DMSO or ouabain. The ratio in DMSO- treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. **P < 0.01 (paired t-test).
(d) Ratios of CBR/CBG bioluminescence signals in human U2OS reporter cells expressing either empty vector, rat α1 subunit, or catalytically-inactive rat α1 subunit of Na+/K+-ATPase after 24 h treatment with DMSO or ouabain. The ratio in DMSO- treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. **P < 0.01 (paired t-test).
(e) Ratios of CBR/CBG bioluminescence signals in human U2OS reporter cells following 24 h treatment with DMSO, ouabain, ouabain and Bapta-AM, or Bapta-AM. Ouabain, 0.175 µM; Bapta-AM, 25 µM. The ratio in DMSO-treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. **P < 0.01 (paired t-test).
(f) Ratios of CBR/CBG bioluminescence signals in human U2OS reporter cells following 4 h treatment with DMSO, thapsigargin, thapsigargin and Bapta-AM, or Bapta-AM. Cells were pre-treated with either DMSO or Bapta-AM for 1 h before addition of thapsigargin. Thasipgargin, 0.2 µM; Bapta-AM, 50 µM. The ratio in DMSO- treated reporter cells was normalized to 1. Data represent the mean ± SD of three independent experiments. **P < 0.01; ***P < 0.001(paired t-test).
(g) A model for the regulation of NMD by cardiac glycosides, Na+/K+-ATPase and intracellular calcium.
Intracellular calcium is a key regulator of NMD
Because a major effect of Na+/K+-ATPase inhibition is the elevation of intracellular calcium levels14, we hypothesized that inhibition of NMD by the cardiac glycosides is due to increased intracellular calcium concentration. To test this, we co-treated U2OS reporter cells with ouabain and Bapta-AM, a cell-penetrating intracellular calcium chelator, to buffer cytosolic free calcium transients. Bapta-AM reversed the inhibitory effects of ouabain on NMD, although Bapta-AM treatment alone did not affect NMD (Fig. 4e). This suggests that calcium indeed mediates the inhibitory effects of cardiac glycosides on NMD. To further establish the regulation of NMD by calcium, we treated reporter cells with multiple drugs that raise intracellular calcium levels by independent mechanisms. We found that thapsigargin, which induces the release of calcium stores from ER into the cytoplasm, abrogated NMD in the reporter cells, and that this effect was also ablated by Bapta-AM (Fig. 4f). Furthermore, the calcium ionophore A23187, which also increases intracellular calcium concentration, exhibited similar inhibitory effects on NMD (data not shown). Taken together, these results strongly suggest that calcium is a crucial regulator of NMD in human cells.
DISCUSSION
In this report, we developed a highly effective NMD bioluminescent reporter system, and using this reporter we identified CGs as potent inhibitors of NMD. Our studies also led to the discovery of a novel calcium-mediated NMD regulatory pathway (Fig. 4g). These findings have important implications in both basic and translational research. Our results suggest a new therapeutic strategy for NMD inhibition by manipulating intracellular calcium levels. CGs or derivatives with low toxicity might also be effective in treating certain genetic diseases such as cystic fibrosis and Duchenne muscular dystrophy, wherein truncated protein products encoded by the corresponding nonsense mRNAs are fully or partially functional7,8. It should be noted that the concentrations of CGs we used in this study to achieve more complete NMD inhibition without causing significant cellular toxicity (e.g., 500 nM for digoxin and 175 nM for ouabain) are much higher than standard clinical doses used for treatment of cardiac failure26. Thus, acute use of these drugs at our current experimental working concentrations cannot directly translate to the clinic due to in vivo toxic effects. However, the benefits of partial NMD inhibition with chronic treatment at clinically-relevant doses may potentially be efficacious, but this will require further clinical pharmacology studies. Other drugs or derivatives of CGs that can efficiently increase intracellular calcium with low toxicity profiles may find use as clinical agents for NMD inhibition.
Interestingly, our results may also suggest a possible explanation for the anticancer properties previously observed for CGs in both epidemiological and laboratory studies14. A recent study indicates that the same group of CGs we identified herein are potent inducers of an anticancer immune response at the concentrations similar to that used in this study27. Mechanistically, inhibition of NMD may result in the synthesis of novel, tumor-specific antigens that could induce antitumor immunity9. Given the antitumor immunity observed in mice after NMD disruption9 and the inhibitory effects of CGs on NMD revealed by this study, it is possible that the antitumor immunity induced by these drugs, in part, results from their chronic effects on NMD. However, the functions of NMD in cancer cells are likely to be complex, and further in-depth studies are needed to determine the potential of NMD inhibition as a cancer therapeutic strategy.
As a key secondary messenger in the cell, calcium is involved in a wide range of signaling pathways. Fluctuation of intracellular calcium concentration dictates many physiological processes, such as development, proliferation, synaptic transmission and immune activation. Our identification of the novel link between intracellular calcium and NMD suggests that NMD is more dynamically regulated in cells than previously thought. Furthermore, given the role of NMD in regulating the levels of many physiological mRNAs, our study also suggests a novel mechanism for the calcium-mediated gene expression programs3,28–34. It is not clear at present how intracellular calcium regulates NMD in cells. Calcium may directly bind and regulate the activities and functions of NMD factors. Alternatively, it may indirectly regulate NMD through calcium-dependent signaling molecules such as kinases and phosphatases. Interestingly, Johansson and Jacobson have previously shown in yeast cells that elevated levels of another divalent cation, magnesium, inhibit NMD by promoting translation read-through at stop codons35. Unlike magnesium, intracellular calcium apparently does not promote translation read-through, as treatment with cardiac glycosides or thapsigargin resulted in the production of a truncated protein terminated at the PTC in the CBR-TCR(PTC) reporter (Fig. 3b and data not shown). Elucidating the precise mechanism of this calcium-mediated NMD regulation will not only broaden our understanding of how NMD activity is fine-tuned, but also provide crucial insights into the cellular processes that are dependent on this novel regulation.
ONLINE METHODS
NMD reporter construction
The dual-color NMD reporter was generated by PCR cloning. The TCRβ mini gene sequence was derived from pβ510 (a gift from Dr. Oliver Mühlemann, University of Bern)10. The CBR and CBG99 ORF sequences were derived from pCBR-Basic and pCBG99-Basic vectors (Promega). The CMV promoter and SV40 polyadenylation signal sequences were derived from pmCherry-N1, a derivative of pEGFP-N1 vector (Promega) in which the EGFP ORF is replaced with mCherry ORF. These elements were first assembled in pmCherry-N1 deleted of the mCherry ORF to generate pN1-(CBR-TCR(PTC)) and pN1-(CBG-TCR(WT)). The CBR-TCR(PTC) transcription unit between the CMV promoter and SV40 polyadenylation signal in pN1-(CBR-TCR(PTC)) was then inserted into pBluescript SK(−) at Sac I (5’) and Spe I (3’) to generate the single-color reporter pBS-(CBR-TCR(PTC)). Similarly, the CBG-TCR(WT) transcription unit between the CMV promoter and SV40 polyadenylation signal in pN1-(CBG-TCR(WT)) was inserted into pBluescript SK(−) at Spe I (5’) and Kpn I (3’) to generate the single-color reporter pBS-(CBG-TCR(WT)). To generate the dual- color reporter pBS-(CBR-TCR(PTC)-CBG-TCR(WT)), the Spe I (5’)-KpnI (3’) fragment containing the CBG-TCR(WT) transcription unit from pBS-(CBG-TCR(WT)) was inserted into pBS-(CBR- TCR(PTC)) at Spe I (5’) and Kpn I (3’) sites. Detailed cloning strategy and PCR primer sequences are available upon request. An annotated, complete sequence of the dual-color NMD reporter is shown in Supplementary Fig. 1.
Cell culture, adenovirus and lentivirus production and infection, expression of α subunits of Na+/K+-ATPase, and chemical inhibitors
Primary fibroblasts from skin explants of newborn mice were kindly provided by Dr. Robert Mecham (Washington University School of Medicine). The cells were maintained in Dulbecco’s modified Eagle’s media (DMEM) supplemented with 100 units ml−1 penicillin, 100 µg ml−1 streptomycin, 0.1 mM nonessential amino acid, and 10% fetal bovine serum (FBS) in a 5% CO2 incubator at 37 °C. Human U2OS cells and HEK293T cells were cultured in DMEM with 10% FBS at 37 °C with 5% CO2.
To generate U2OS cells stably expressing the dual-colored or single-colored NMD reporters, pBS-(CBR-TCR(PTC))-(CBG-TCR(WT)), pBS-(CBR-TCR(PTC)) or pBS-(CBG-TCR(WT)) were co-transfected with a pMXs-puro vector that encodes a puromycin-resistance gene into U2OS cells using the TransIT-LT1 transfection reagent (Mirus). Single colonies were selected with puromycin (1.5 µg ml−1) and expression of the reporters was verified by Western blot, bioluminescence imaging and RT-qPCR.
An adenovirus construct encoding our NMD reporter was generated by inserting the reporter into the pAdenoX-PRLS-ZsGreen1 vector using the In-Fusion HD cloning kit (Clontech), according to the manufacturer’s protocol. Linearized adenoviral vector with exposed inverted terminal repeats was transfected into HEK293 cells for viral production, followed by viral amplification in the same cell line. Target cells were infected with adenoviruses for 48 h before bioluminescence imaging.
To knockdown NMD factors in NMD reporter cells, lentiviruses expressing a non-targeting control shRNA or shRNAs targeting SMG1, UPF1 or UPF2 were generated in HEK293T cells as previously described36. Briefly, HEK293T cells were co-transfected with an shRNA-encoding lentiviral vector and packaging plasmids (pCMV-dR8.2 and pCMV-VSVG) using TransIT-LT1 transfection reagent (Mirus). Virus-containing supernatant was collected 48 and 72 h after transfection. The U2OS cells stably expressing the dual-colored NMD reporter were then infected with the viruses. The lentiviral vectors expressing a control nontargeting shRNA (5’- CAACAAGAUGAAGAGCACCAA-3’), shSMG1-1 (5’-GCCGAGAUGUUGAUCCGAAUA-3’), shSMG1-2 (5’-GCACUGUAACUACGGCUACAA-3’), shUPF1 (5’- GCAUCUUAUUCUGGGUAAUAA-3’), shUPF2-1 (5’-GCGUUAUGUUUGGUGGAAGAA-3’) and shUPF2-2 (5’-GCGAGAUACGUCACAAUGGUA-3’) were purchased from Sigma.
The CG-resistant human α1 mutant containing two mutations (Q118R and N129D) and the catalytic inactive rat α1 subunit mutant containing the D376E mutation were generated by site-directed mutagenesis using the following primers: 5’-TATAGCATCCGAGCTGCTACAGAAGAGGAACCTCAAAACGATGATCTGTACCTGG-3’/5’-CCAGGTACAGATCATCGTTTTGAGGTTCCTCTTCTGTAGCAGCTCGGATGCTATA-3’, and 5’-CATCTGCTCCGAGAAGACTGGAACTC-3’/5’-GAGTTCCAGTCTTCTCGGAGCAGATG-3’, respectively. Because we observed inhibitory effects of a FLAG tag on the function of α subunits (data not shown), we used untagged forms for expression in human cells. All α subunits were expressed in U2OS cells by transfection using the TransIT-LT1 transfection reagent. Twenty fours hours following transfection, cells were re-plated in 96-well plates before drug treatment and bioluminescence imaging (see below).
Cardiac glycosides, including ouabain, digoxin, digitoxin, proscillaridin and lanatoside C, were purchased from MicroSource. Ouabain (O3125) and digoxin (D6003) were also purchased from Sigma. Bapta-AM (A1076), thapsingargin (T9033), A23187 (C7522), actinomycin D (A1410) and caffeine (C0750) were purchased from Sigma. Caffeine was dissolved in H2O, and all other inhibitors were dissolved in DMSO.
High-throughput screening
Human U2OS cells stably expressing our dual-colored NMD reporter were plated in black-walled 96-well dishes with 100 µl media using a Biomek FX liquid handler. Colorless DMEM with 10% FBS was used throughout the screening process. Eighteen hours following cell plating, original media were removed and replaced with media (130 µl) supplemented with individual drugs in the Pharmakon Library (PHARMAKON 1600, MicroSource) at a concentration of 10 µM. The liquid handling robot diluted the drugs from a stock concentration of 2 mM in dimethyl sulfoxide (DMSO) to the final 10 µM concentration. The final concentration of DMSO was 0.5%. Three wells treated with DMSO (0.5%, negative vehicle control for drugs in the Library) or caffeine (10 mM in H2O, positive control) were also included on each plate. For the purpose of spectral unmixing of the CBR and CBG signals in the dual-colored reporter, control U2OS reporter cells stably expressing either the CBG-TCR(WT) alone or the CBR-TCR(PTC) alone (treated with 10 mM caffeine to increase signal) were included on the same plate to provide pure CBR and CBG reference signals. Cells were incubated in the presence of compound for 24 h followed by bioluminescence imaging (see below).
Bioluminescence imaging and spectral deconvolution
Cells were incubated with 150 µg ml−1 D-luciferin for 10 min at 37 °C and bioluminescence signals were measured sequentially using a charge-coupled device (CCD) camera-based bioluminescence imaging system (IVIS 100; Caliper) with appropriate open, red, or green filters and exposure settings (exposure time: 10 s; binning: 8; field of view: 15; f/stop: 1). Regions-of-interest (ROIs) were drawn over images of wells and bioluminescence signals were quantified using Living Image (Caliper) and Igor (Wavemetrics) analysis software packages as described previously37. Spectral unmixing was performed using an ImageJ plugin with an algorithm we developed previously12. The CBR/CBG ratio was calculated for each well and normalized by the CBR/CBG ratio in DMSO-treated wells. Data were presented as the log2 of the fold-change in the CBR/CBG ratio. To document the quality of our NMD reporter assay, we calculated the Z’ factor for a 96-well screening plate based on the CBR/CBG ratios in caffeine-treated (control positive sample) and H2O-treated (control negative sample) cells: Z’ factor = 1 − (3× (standard deviationcaffeine + standard deviationH2O) / (averagecaffeine − averageH2O)) = 1 − (3× (0.1123 + 0.0508) / (3.2356 − 1.0850)) = 0.77
Screen hit selection and statistic analysis
A battery of statistical tests was employed to identify statistically significant screen hits. First, all compounds whose CBR/CBG ratio was less than 2 standard deviations above or below the CBR/CBG ratio for DMSO control wells on the same plate were excluded from further analysis. Compounds passing this initial criterion were then subjected to quartile analysis. The first (Q1), third (Q3), and median (Q2) quartile values were calculated using the log2 of the ratio fold-change for compounds in the data set. Potential NMD inhibitors were selected using an upper boundary for hit selection calculated as Q3 + c(ICQ), where ICQ = Q3 − Q1, and c = 1.7239. This c value corresponds to a high stringency targeted error rate of α = 0.002712. Where multiple independent experiments are presented, p-values were calculated using a paired Student’s t-test. Where biological replicates are presented (including the screen data), p-values were calculated using a unpaired Student’s t-test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
RT-qPCR and Western blotting
To measure levels of NMD reporter mRNAs in U2OS cells, or p53 and other endogenous mRNAs in Calu-6 cells, total RNA was isolated using a NucleoSpin RNA II kit (Clonetch) and cDNA was synthesized using a poly-dT primer and the SuperScript II cDNA synthesis kit (Invitrogen), according to the instructions of the manufacturers. All reactions were performed in at least duplicate using a three step PCR protocol (melting temperature: 95 °C; annealing temperature: 55 °C; extension temperature: 72 °C; cycle number: 40) on an ABI VII7 real-time PCR system with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific). The mRNA levels of the housekeeping gene GAPDH were used for normalization. Primers for qPCR are listed in Supplementary Table 4. Western blot to detect CBR-TCR(PTC) and CBG-TCR(WT) protein products in reporter cells using an HA-antibody (Convance, MMS-101R, 1:1,000) was performed as described previously using a Li-Cor Odyssey system36. Anti-SMG1 antibody raised in rabbits against Xenopus protein cross-reacts with human SMG1, as described previously38. Anti-UPF1 (NBP1-05967, 1:500) and anti-UPF2 (NB2-20813, 1:1,000) antibodies were purchased from Novus Biologicals. Anti-PCNA antibodies were home-made against full-length Xenopus PCNA protein, which also recognize human PCNA39. A pan-Na+/K+-ATPase antibody that recognizes all isoforms of the α subunit of Na+/K+-ATPase in human and rat cells (Cell Signaling Technology, 3010, 1:1,000) was used in Supplementary Fig. 5.
Cell viability analysis
The AlamarBlue assay was used to measure the overall cytotoxicity of cardiac glycoside treatment40. Human U2OS NMD reporter cells were treated with DMSO, ouabain or digoxin in 96-well plates. Two hours prior to the each time point shown in Supplementary Fig. 3B, Alamar Blue was added to each well at a final concentration of 40 µM. Cells were incubated at 37 °C in the presence of AlamarBlue for 2 h and then fluorescence measured (excitation 544 nm; emission 590 nm).
Analysis of general translation
Human U2OS NMD reporter cells were treated with DMSO or ouabain in methionine-free media (CellGro, 17-204-CI) supplemented with 2 mM L-glutamine, 10% dialyzed FBS (Sigma, F0392), and 35S-labeled methionine (10 µCi ml−1) for 24 h prior to protein collection. Protein samples from the same number of cells were run on two separate SDS-PAGE gels, with one for 35S autoradiography analysis (newly synthesized proteins) and one for Coomassie blue staining (total protein levels). Images were quantified using ImageJ and autoradiography intensity was normalized to the total protein level (Coomassie blue intensity).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Peng Wu and Xiaoqing Chen for their contributions in the early stage of this project. We are grateful to Dr. Oliver Mühlemann for providing the pβ510 reporter construct, which we used to obtain the TCRβ mini reporter sequence for our reporter construction, and Dr. Robert Mecham for providing primary mouse skin fibroblasts. This study was supported by a Molecular Imaging Center grant from the US National Institutes of Health to Washington University and The University of Texas M.D. Anderson Cancer Center (P50 CA94056, DP-W), by a Washington University Molecular Imaging Center Pilot Research Project Grant (ZY), and by an Interdisciplinary Research Initiative grant from the Children’s Discovery Institute of Washington University (MC-II-2012-215, ZY and DP-W). The HTS core is supported in part by the Siteman Cancer Center (Cancer Center Support Grant P30 CA91842 from the US National Cancer Institute).
Footnotes
AUTHOR CONTRIBUTIONS
Z.Y. and D.P-W. conceived the project and supervised the studies. A.N. and Z.Y. designed and performed the experiments. E.J. contributed to the bioluminescence imaging and the analysis of screen and imaging data. J.M. assisted with the high-throughput chemical screen. P.N. assisted with the analysis of the screen results. R.W.M. provided expression constructs for rat Na+/K+-ATPase α subunits and technical advice. A.N., Z.Y. and D.P-W. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruno IG, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 6.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 8.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudikote JP, Wilkinson MF. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 2002;21:125–134. doi: 10.1093/emboj/21.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal Chem. 2006;78:1520–1527. doi: 10.1021/ac051999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang XD, et al. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics. 2006;7:299–309. doi: 10.2217/14622416.7.3.299. [DOI] [PubMed] [Google Scholar]
- 14.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 15.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 18.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, Li Y, Li P. MARVELD1 Inhibits Nonsense-Mediated RNA Decay by Repressing Serine Phosphorylation of UPF1. PLoS One. 2013;8:e68291. doi: 10.1371/journal.pone.0068291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dostanic-Larson I, Van Huysse JW, Lorenz JN, Lingrel JB. The highly conserved cardiac glycoside binding site of Na,K-ATPase plays a role in blood pressure regulation. Proc Natl Acad Sci U S A. 2005;102:15845–15850. doi: 10.1073/pnas.0507358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingrel JB. The physiological significance of the cardiotonic steroid/ouabain-binding site of the Na,K-ATPase. Annu Rev Physiol. 2010;72:395–412. doi: 10.1146/annurev-physiol-021909-135725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye J, Chen S, Maniatis T. Cardiac glycosides are potent inhibitors of interferon-beta gene expression. Nat Chem Biol. 2011;7:25–33. doi: 10.1038/nchembio.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith TW, Haber E. Digoxin intoxication: the relationship of clinical presentation to serum digoxin concentration. J Clin Invest. 1970;49:2377–2386. doi: 10.1172/JCI106457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menger L, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med. 2012;4:143ra199. doi: 10.1126/scitranslmed.3003807. [DOI] [PubMed] [Google Scholar]
- 28.Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 30.Long AA, et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci. 2010;123:3303–3315. doi: 10.1242/jcs.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIlwain DR, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 34.Eom T, et al. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson MJ, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 2010;24:1491–1495. doi: 10.1101/gad.1930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- 38.You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Paudyal SC, Chin RI, You Z. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013;41:9325–9338. doi: 10.1093/nar/gkt672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nociari MM, Shalev A, Benias P, Russo C. A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J Immunol Methods. 1998;213:157–167. doi: 10.1016/s0022-1759(98)00028-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.