Abstract
Azole resistance in Candida glabrata, a pathogenic yeast, has prompted studies of compounds that have therapeutic potential by reversing azole resistance. Milbemycin A4 oxime blocked azole efflux and enhanced azole susceptibility four-fold in 28 clinical isolates of C. glabrata. Specificity of the milbemycin A4 oxime effect depended on the drug transporter and the substrate being effluxed. The major effect of milbemycin A4 oxime was inhibition of azole and rhodamine 6G efflux by the ATP-Binding Cassette (ABC) transporters CgCDR1 and PDH1. Milbemycin A4 oxime effect did not extend to oligomycin, transported by the ABC transporter YOR1 or to benomyl, transported by the Major Facilitator Superfamily transporter, CgFLR1. Milbemycin A4 oxime did not suppress transcription of CgCDR1 but increased CgCDR1 expression 126-fold. Selectivity of the effect is compatible with the concept that milbemycin A4 oxime may interact directly with a one or more drug-binding sites of the major azole transporters.
Introduction
Azoles are a major class of drug for treatment of superficial and deep mycoses. Drug efflux is an important mechanism of azole resistance in several species and can limit therapeutic effect (Cannon et al., 2009). Blocking drug efflux in order to increase susceptibility to azoles has attracted attention, particularly in Candida glabrata, a species in which azole efflux is the major mechanism of drug resistance (Bennett et al., 2004). Search strategies have centered around agents which increase azole inhibition of C. glabrata growth, that is, are synergistic with azoles. Milbemycin synergy with azoles was first reported by Lee and colleagues, who screened a library of 85000 microbial fermentation products for those which would decrease azole resistance in Candida albicans and C. glabrata. (Lee et al., 2001) They found that milbemycin α9 at 1 µg/ml decreased the fluconazole minimum inhibitory concentration (MIC) from an average of 8 µg/ml to 0.5 µg/ml in 50 strains of C. glabrata.(Lee et al., 2001). Milbemycin oximes were already known as Streptomyces hygroscopicus aureolacrimosus fementation products which have broad spectrum activity against nematode infection in animals, such as heart worm in dogs. Evidence that synergy was due to drug efflux in Candida was indirect and no direct correlation was made between amount of synergy and extent of drug resistance. Lamping and colleagues showed that milbemycin affected drug efflux when they reported that milbemycin β9 increased the fluconazole susceptibility of a Sacchaaromyces cerevisiae mutant overexpressing CgCDR1, the major azole transporter in C. glabrata.. There was a minor effect on azole susceptibility when PDH1, another azole transporter, was overexpressed in S. cerevisiae (Lamping et al., 2007). Silva and colleagues studied milbemycin A3, A4 and their oximes for fluconazole synergy in four strains of C. glabrata, (Silva et al., 2013). They reported that milbemycin blocked rhodamine 6G efflux, could have a cidal effect and increased azole efficacy in experimentally infected mice. They also performed microarray analysis of milbemycin A3 oxime’s effect on transcription (Silva et al., 2013). Induction of CgCDR1 and CgPDR1 transcription, reported here for strain NCCLS84, was not found. Other authors have reported milbemycin-azole synergy in Candida albicans (Holmes et al., 2008) and Candida krusei (Lamping et al., 2009). The current work extends these studies by showing that a four-fold reduction in voriconazole and fluconazole susceptibility by milbemycin A4 oxime held across a broad range of MIC’s in 28 isolates of C glabrata. Specificity of milbemycin A4 oxime effect for transporter and substrate is also detailed.
Materials and Methods
Candida glabrata was identified using API 20C Aux strips (BioMerieux Vitek Inc., Marcy l’Etoile, France). Isolates are listed in Table 1. Paired fluconazole susceptible and resistant isolates from 15 patients were chosen to provide a broad range of azole susceptibilities (2–128 µg/ml). Additional strains studied were a clinical resistant isolate (Cg40a) and a stock strain, NCCLS84 (ATCC90030), both isolates having a fluconazole minimum inhibitory concentration (MIC) of 256 µg/ml. Also studied were three mutants derived from NCCLS84: 84870, with the two major azole efflux transporters deleted, ΔCgpdr1, with the major transcriptional activator of azole efflux pumps deleted, and ΔCgsnq2 with the CgSNQ2 transporter deleted as described below. Isolates were incubated at 30° C in one of three media: YEPD (Difco Laboratories, Detroit, MI), containing 1% Bacto Yeast extract, 2% BactoPeptone, and 2% Dextrose, MIN, containing 0.67% yeast nitrogen base without amino acids (YNB, Difco) plus 2% dextrose, or YEPG, containing 1% Bacto yeast extract (Difco Laboratories), 1.8% Bactopeptone (Difco Laboratories), 0.9% ethanol and 2.7% glycerol,
Table 1.
Candida glabrata isolates used in this study
| Isolate | Genotype | Reference |
|---|---|---|
| 15 pairs of fluconazole susceptible and resistant clinical isolates | wild type | (Bennett et al., 2004) |
| Cg40a (clinical isolate) | wild type | (Miyazaki et al., 1998) |
| NCCLS84 | wild type | ATCC90030 |
| 84u | ura3 | (Izumikawa et al., 2003) |
| 84870 | cgcdr1 pdh1::ScURA3 | (Miyazaki et al., 1998) |
| Cgpdr1 | 84u pdr1Δ::ScURA3 | (Noble et al., 2013) |
| Cgsnq2 | 84u cgsnq2Δ::ScURA3 | this study |
Chemicals included fluconazole and voriconazole (both kind gifts of Pfizer, Sandwich, UK), milbemycin A-oxime (Sankyo Research Laboratories, Tokyo, Japan), 4-nitroquinoline 1-oxide (4NQO) (Supelco Analytical, St. Louis, MO), benomyl (Sigma-Aldrich, St. Louis, MO), oligomycin (USB, Cleveland, Ohio), and rhodamine 6G (Sigma, Steinheim, Germany). Benomyl was dissolved in dimethylsulfoxide. Oligomycin and rhodamine 6G were dissolved in ethanol. Solvent controls were included in all experiments.
Drug susceptibility was determined using a modification of the CLSI M27-A3 microdilution method with MIN broth and an endpoint of 80% reduction in optical density after 48 hours incubation (MIC80) (Clinical and Laboratory Standards Institute, 2008). As an exception, oligomycin was tested using YEPG broth. Interaction was evaluated by testing each chemical in the absence or presence of milbemycin in two fold dilutions ranging from 0.5 to 32 µg/ml plus additional concentrations 1.5 and 2.5 µg/ml. Concentrations of the other chemicals were 11 two-fold dilutions ranging down from the following: 256 µg/ml for fluconazole, 100 µg/ml for 4NQO, and 32 µg/ml for voriconazole and oligomycin.
[3H] Fluconazole accumulation
The effect of milbemycin A4 oxime upon the uptake of fluconazole was measured in the azole resistant strain, Cg40a, and the azole susceptible mutant, 84870, using our previously published method (Bennett et al., 2004). For the treatment of cells in the presence of milbemycin A4 oxime, a 2.5 µg/ml was used during the 2-hour incubation period preceding the addition of [3H] fluconazole.
Rhodamine 6G accumulation
Using our previously published method (Izumikawa et al., 2003) accumulation of rhodamine 6G was measured by flow cytometer in the presence and absence of milbemycin A4 oxime. Cells were treated with 5 µg/ml of the drug during the two-hour incubation period. Then, rhodamine 6G was added to the cells to a final concentration of 0.2 µg/ml. Cells were subsequently incubated for 4 hours at 30° C. After incubation, 0.05ml aliquots of the milbemycin A4 oxime-treated and non-treated cells were added to 0.9 ml cold phosphate buffered saline (pH 7.0). Cells were incubated on ice for 5 minutes before being analyzed by flow cytometry.
CgSNQ2 deletion
The targeted deletion of CgSNQ2 (CAGL0I04862g) in 84u was performed by the homologous recombination of a deletion cassette containing the two 120bp regions flanking the CgSNQ2 open reading frame (ORF), using the Saccharomyces cerevisiae URA3 open reading frame as a selection marker. Homologous recombination via a double crossover resulted in the replacement of the native CgSNQ2 ORF by the deletion cassette. Deletion of the CgSNQ2 locus was confirmed by PCR and Southern blot (data not shown).
RT-qPCR
RNA was isolated from log phase cultures of the wild-type strain, NCCLS84 and the CgPDR1 deleted strain, ΔCgpdr1. Cells were grown in MIN with or without milbemycin A4 oxime 4 µg/ml, fluconazole 32 µg/ml or both drugs together for two hours. RNA was prepared from the cell pellet using TRIzol (Invitrogen, Carlsbad, CA) and lysing matrix C with FastPrep-24 (MP Biomedicals) and purified with the RNeasy MinElute cleanup kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). The reaction took place in a thermal cycler (T3 Thermocycler; Biometra, Goettingen, Germany) with a single cycle and incubation periods of 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. Quantitative real-time PCR (RT-qPCR) was utilized to determine the expression level of CgPGK1, CgCDR1, and CgPDR1. The sequences of forward and reverse primers are listed in Table 2. RT-qPCR for reference gene RNA transcription was performed by SYBR Green chemistry (SYBR Green PCR Master Mix; Applied Biosystems). The increase in fluorescence of the SYBR Green dye was monitored using a 7500 Real-time PCR System (Applied Biosystems). Data analysis was performed by the threshold cycle (ΔΔCT) method. Threshold cycles of the target were normalized to the CT of the CgPGK1gene (ΔCT =CT Target -CT CgPGK1). (Li et al., 2012). Therefore, the negative ΔΔCT values are equivalent to log2 relative fold changes.
TABLE 2.
Primers Used in RT-qPCR
| Primer | Sequence (5’-3’) |
|---|---|
| CgPGK1-F | CAAACGGTGAAAGAAACGAGAA |
| CgPGK1-R | CCGACACAGTCGTTCAAGAAAG |
| CgCDR1-F | AGATGTGTTGGTTCTGTCTCAAAGAC |
| CgCDR1-R | CCGGAATACATTGACAAACCAAG |
| CgPDR1-F | AACGATTATTCAATTGCAACAACG |
| CgPDR1-R | CCTCACAATAAGGAAAGTCTGCG |
Statistics
The Spearman correlation coefficient and p value were calculated using Prism 5 (GraphPad software, Inc. San Diego, CA).
RESULTS
Effect on susceptibility
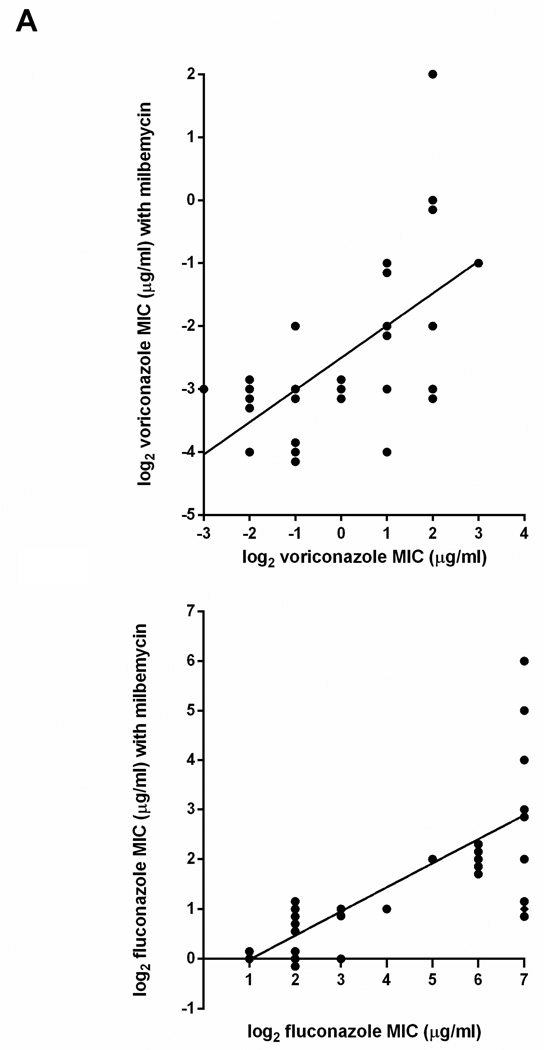
Milbemycin A4 oxime 2.5 µg/ml decreased susceptibility to fluconazole and voriconazole in 28 clinical isolates of C. glabrata (Fig. 1). Four additional isolates could not be evaluated because milbemycin A4 oxime alone at 2.5 µg/ml decreased growth to a small extent. The concentration of milbemycin needed for 80% inhibition (MIC80) was greater than 32 µg/ml for all isolates in this study except for NCCLS84, which had an MIC of 16 µg/ml. At a concentration of 2.5 µg/ml, milbemycin A4 oxime caused a decrease in fluconazole MIC that was in direct proportion to the amount of fluconazole resistance (Spearman correlation p<0.0001). As shown in Fig 1. the slopes of best-fit lines were the same (0.5) for both fluconazole and voriconazole. This calculates to a four-fold reduction in MIC of both fluconazole and voriconazole in the presence of milbemycin A4 oxime. With fluconazole, that proportionality was the least evident at the highest fluconazole MIC of 128 µg/ml (Fig. 1B), suggesting that azole resistance at the highest level may have involved more than one mechanism. With this caveat, the results indicated that milbemycin A4 oxime was blocking the main mechanism of resistance to these azoles.
Fig. 1.
MIC of voriconazole (A) and fluconazole (B) in presence and absence of milbemycin 2.5 µg/ml in 28 clinical isolates of C. glabrata
Efflux activity
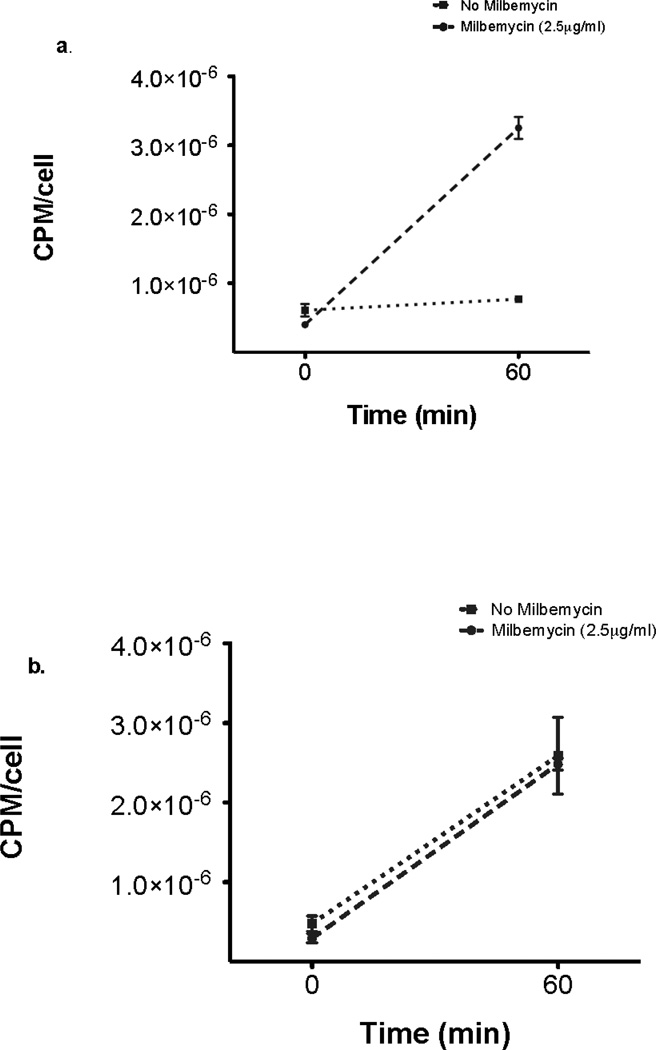
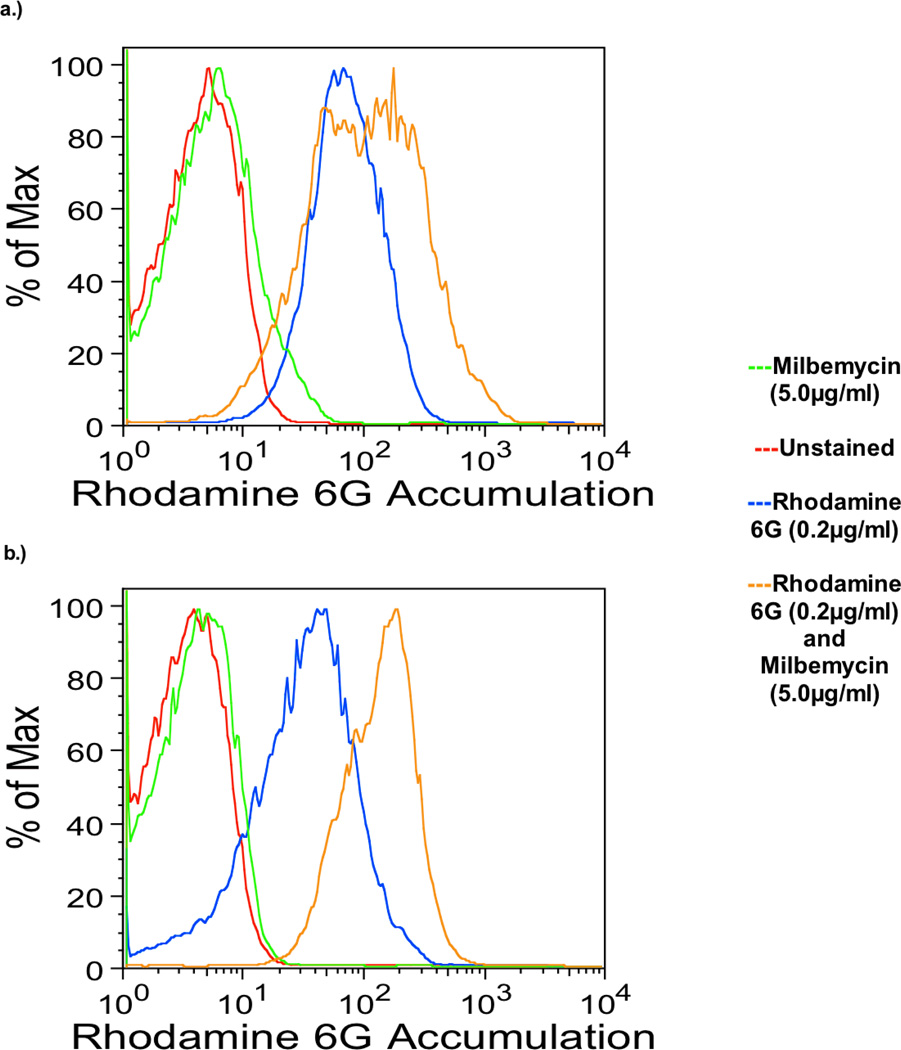
Because drug efflux is a major mechanism of azole resistance in C. glabrata, the effect of milbemycin on drug efflux was measured directly with radiolabeled fluconazole and also by rhodamine 6G content using flow cytometry. Rhodamine 6G is known to be effluxed by the same mechanism as fluconazole (Parkinson et al., 1995). As shown in Fig. 2, milbemycin A4 oxime 2.5 µg/ml blocked the efflux of radiolabeled fluconazole in the azole resistant strain Cg40a but not in the mutant 84870 with deletion of the efflux pumps CgCDR1 and PDH1. Milbemycin A4 oxime 5 µg/ml also blocked efflux of rhodamine 6G in strain 40a but not in 84870 (Fig 3). These results indicated that the major effect of milbemycin A4 oxime may be to block drug efflux by these transporters.
Fig. 2.
3H-fluconazole accumulation as counts per minute (CPM) per fungal cell in Cg40a (A) and Candida glabrata 84870 (B) Mean +/− SD of triplicate experiments
Fig. 3.
Rhodamine 6G accumulation in C. glabrata strain 84870 (upper) and Cg40a (lower) in the presence and absence of milbemycin 5.0 µg/ml.
Gene Expression
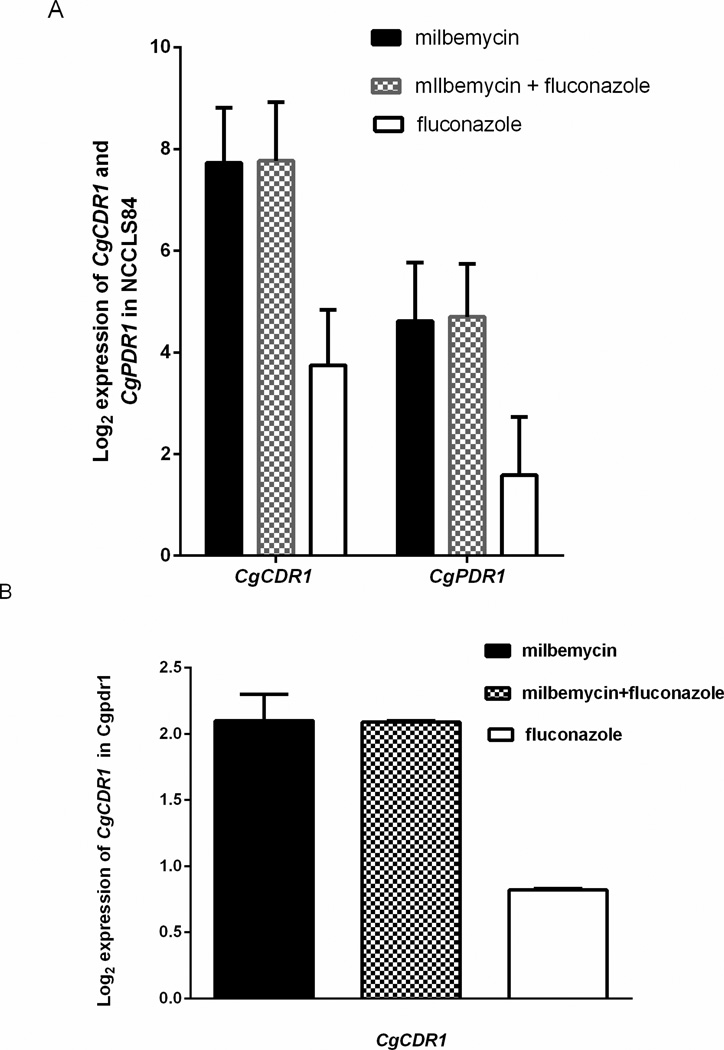
Milbemycin A4 oxime did not block expression of CgCDR1, the major azole transporter, or CgPDR1, the major transcriptional activator of CgCDR1 (Tsai et al., 2006). Rather, milbemycin A4 oxime strongly increased transcription of these genes, independent of the much weaker effect of fluconazole on gene transcription.(Fig. 4A) In a strain in which CgPDR1 had been deleted, milbemycin still increased CgCDR1 transcription, though the level of activation was reduced (Fig. 4B). Likely, milbemycin A4 oxime was affecting efflux function, resulting in a strong feedback pathway that increased transcription, using a pathway that included but was not exclusive to CgPDR1.
Fig. 4.
Expression of CgCDR1 and CgPDR1 in NCCLS84 (A) and expression of CgCDR1 in the deletant, Cgpdr1. Cells were treated for 2 hours with fluconazole 32 µg/ml, milbemycin 4 µg/ml, or both. Mean +/−SD are the log2 ratio of drug to no drug.
Drug specificity
Effect of milbemycin A4 oxime on susceptibility of NCCLS84 to different drugs was explored, Benomyl was studied because it is effluxed by CgFLR1, a member of the major facilitator superfamily, not an ABC transporter (Chen, et al., 2007). Oligomycin was selected because it is transported in S. cerevisiae by the ABC transporter, YOR1 (Katzmann et al., 1995). We found that oligomycin susceptibility was not enhanced by milbemycin A4 oxime, nor was benomyl susceptibility altered (Table 3). 4NQO was studied because resistance to this DNA damaging agent is reduced when CgSNQ2, an ABC transporter, is deleted (Torelli, et al, 2008). Susceptibility to 4NQO was increased 8 fold by milbemycin A4 oxime. This result indicates that a 4NQO transporter is a target of milbemycin A4 oxime. As discussed in the next paragraph, that transporter was not CgSNQ2.
Table 3.
Milbemycin A4 oxime effect on MIC using different drugs and transporter deletions.
| C. glabrata strain | Drug | MIC without milbemycin (µg/ml) |
MIC with milbemycin 4 µg/ml (µg/ml) |
|---|---|---|---|
| NCCLS84 | Fluconazole | 256 | 32 |
| 4-nitroquinoline 1-oxide | 3.12 | 0.39 | |
| Oligomycin | 0.5 | 0.5 | |
| Benomyl | 62.5 | 62.5 | |
| ΔCgsnq2 | Fluconazole | 256 | 64 |
| 4-nitroquinoline-1-oxide | 0.78 | 0.09 | |
| Oligomycin | 0.5 | 0.5 | |
| 84870 | Fluconazole | 8 | 8 |
| 4-nitroquinoline-1-oxide | 3 | 1.5 | |
| Oligomycin | 0.25 | 0.25 | |
| Benomyl | 62.5 | 62.5 |
Deletion of CgCDR1 and PDH1 (Table 3)
Deletion of CgCDR1 and PDH1 in 84870 ablated the effect of milbemycin A4 oxime on fluconazole, consistent with the results of those deletions on rhodamine 6G accumulation (Fig. 2) and H3-fluconazole efflux (Fig 3.) This confirmed that these transporters are a target of milbemycin A4 oxime. In 84870, the effect of milbemycin A4 oxime on 4NQO susceptibility was repeatedly marginal.
Deletion of CgSNQ2 (Table 3)
Deletion of CgSNQ2 increased susceptibility of 4NQO four-fold, as expected (Torelli, et al., 2008) but did not decrease the effect of milbemycin A4 oxime on fluconazole or 4NQO susceptibility. These results fail to clarify the 4NQO transporter(s) which milbemycin A4 oxime is inhibiting, though excluding CgSNQ2.
DISCUSSION
Milbemycin caused a four-fold reduction in the MIC of fluconazole and voriconazole in 28 clinical isolates, compatible with blocking the major mechanism of azole resistance in these isolates, with the possible exception of a few of the most resistant isolates (Fig. 1). In order to examine the mechanism by which the blockage occurred, we studied the effect of milbemycin A4 oxime on resistance to different drugs and the effect of deleting transporter genes. This approach is limited by the broad range of drug substrates which a single transporter can efflux, as well as the ability of some substrates to be effluxed by more than one transporter (Cannon, et al, 2009). In C. glabrata, azoles are transported largely by two ABC transporters, CgCDR1 and, to a lesser extent, by PDH1 (also called CgCDR2).(Tsai, et al. 2006) As anticipated from the data in Fig. 1, when both CgCDR1 and PDH1 were deleted in an azole resistant strain, milbemycin A4 oxime no longer blocked efflux of rhodamine 6G or H3-fluconazole.(Fig 2 and 3).. It was also anticipated that milbemycin A4 oxime would not affect susceptibility to benomyl, which is transported by CgFLR1, a member of a different family of efflux pumps (Table 3). However it could not be predicted that milbemycin A4 oxime would have no effect on oligomycin. YOR1, which transports oligomycin. YOR1 is an ABC transporter which shares with CgCDR1 highly conserved ATP binding sites, that is, the N- and C-terminal Walker A, Walker B and Signature sequences (Miyazaki et al., 1998). These results are consistent with the concept that milbemycin A4 oxime is blocking and likely binding to discrete domains in CgCDR1 and possibly PDH1. Specificity and activity of homologous ABC transporters in S. cerevisiae and C. albicans have been shown to be altered by mutations in the transmembrane, cytosolic and extracellular regions (Tutulan-Cunita et al., 2005),(Ernst et al., 2008),(Rawal et al., 2013). It is possible the milbemycin binds to one of these critical domains in one or more transporters. Results with 4NQO were not sufficiently definitive to show activity of milbemycin A4 oxime against other transporters so that possibility remains unresolved
Expression of CgCDR1 was studied to determine whether milbemycin A4 oxime increased azole susceptibility by interfering with transcription of the major transporter, CgCDR1 or its transcriptional activator CgPDR1. Instead, we found that CgCDR1 expression was increased 212-fold and CgPDR1 increased 25-fold by milbemycin A4 oxime (Fig. 4A). When CgPDR1 was deleted, the upregulation of CgCDR1 by milbemycin A4 oxime was only 4-fold, indicating that upregulation largely required CgPDR1 (Fig. 4B). It is possible that decreased function of the transporter lead to a feed-back loop that included CgPDR1 and its many targets. Silva and colleagues studied the effect of milbemycin A3 oxime 10 µg/ml on transcription in an azole susceptible C. glabrata, DSY562 (Silva, et al.,2013). They did not find upregulation of either CgCDR1 or PDR1 There appears to be a strain specificity for this effect.
In hopes that compounds with structural similarity to milbemycin A4 oxime might be synergistic with azoles in C. glabrata, we searched but did not find fluconazole synergy with ivermectin, a drug used to treat nematode infections in humans, or with the veterinary drug, moxidectin (data not provided). There are multiple classes of drug efflux inhibitors, as discussed elsewhere, leaving open many pathways to pursue azole synergism further (Cannon et al., 2009).
Acknowledgements
The work was supported by the Division of Intramural Research, NIAID.
Footnotes
The authors have no conflicts of interest with this work.
References
- Bennett JE, Izumikawa K, Marr KA. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrobial agents and chemotherapy. 2004;48:1773–1777. doi: 10.1128/AAC.48.5.1773-1777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. Efflux-mediated antifungal drug resistance. Clinical microbiology reviews. 2009;22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Miyazaki T, Tsai HF, Bennett JE. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene. 2007;386:63–72. doi: 10.1016/j.gene.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference method for broth dilution antifunal susceptibility testing of yeasts. 2008. Approved standard, 3rd ed. M27-A3. [Google Scholar]
- Ernst R, Kueppers P, Klein CM, Schwarzmueller T, Kuchler K, Schmitt L. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, Lin YH, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrobial agents and chemotherapy. 2008;52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa K, Kakeya H, Tsai HF, Grimberg B, Bennett JE. Function of Candida glabrata ABC transporter gene, PDH1. Yeast. 2003;20:249–261. doi: 10.1002/yea.962. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Hallstrom TC, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley WS. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Molecular and cellular biology. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E, Ranchod A, Nakamura K, Tyndall JD, Niimi K, Holmes AR, Niimi M, Cannon RD. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrobial agents and chemotherapy. 2009;53:354–369. doi: 10.1128/AAC.01095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E, Monk BC, Niimi K, Holmes AR, Tsao S, Tanabe K, Niimi M, Uehara Y, Cannon RD. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryotic cell. 2007;6:1150–1165. doi: 10.1128/EC.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD, Galazzo JL, Staley AL, Lee JC, Warren MS, Fuernkranz H, Chamberland S, Lomovskaya O, Miller GH. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco. 2001;56:81–85. doi: 10.1016/s0014-827x(01)01002-3. [DOI] [PubMed] [Google Scholar]
- Li QQ, Skinner J, Bennett JE. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC molecular biology. 2012;13:22. doi: 10.1186/1471-2199-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer DJ, Ward DJ, Marsden K, Bennett JE. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrobial agents and chemotherapy. 1998;42:1695–1701. doi: 10.1128/aac.42.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Tsai HF, Suffis SD, Su Q, Myers TG, Bennett JE. STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrobial agents and chemotherapy. 2013;57:959–967. doi: 10.1128/AAC.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson T, Falconer DJ, Hitchcock CA. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrobial agents and chemotherapy. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal MK, Khan MF, Kapoor K, et al. Insight into pleiotropic drug resistance ATP-binding cassette pump drug transport through mutagenesis of Cdr1p transmembrane domains. The Journal of biological chemistry. 2013;288:24480–24493. doi: 10.1074/jbc.M113.488353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LV, Sanguinetti M, Vandeputte P, Torelli R, Rochat B, Sanglard D. Milbemycins: more than efflux inhibitors for fungal pathogens. Antimicrobial agents and chemotherapy. 2013;57:873–886. doi: 10.1128/AAC.02040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Molecular microbiology. 2008;68:186–201. doi: 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- Tsai HF, Krol AA, Sarti KE, Bennett JE. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrobial agents and chemotherapy. 2006;50:1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutulan-Cunita AC, Mikoshi M, Mizunuma M, Hirata D, Miyakawa T. Mutational analysis of the yeast multidrug resistance ABC transporter Pdr5p with altered drug specificity. Genes to cells : devoted to molecular & cellular mechanisms. 2005;10:409–420. doi: 10.1111/j.1365-2443.2005.00847.x. [DOI] [PubMed] [Google Scholar]