SUMMARY
Succinate dehydrogenase (SDH) occupies a central place in cellular energy production, linking the tricarboxylic cycle with the electron transport chain. As a result, a subset of cancers and neuromuscular disorders result from mutations affecting any of the four SDH structural subunits or either of two known SDH assembly factors. Herein we characterize a novel evolutionarily conserved SDH assembly factor designated Sdh8/SDHAF4, using yeast, Drosophila, and mammalian cells. Sdh8 interacts specifically with the catalytic Sdh1 subunit in the mitochondrial matrix, facilitating its association with Sdh2 and the subsequent assembly of the SDH holocomplex. These roles for Sdh8 are critical for preventing motility defects and neurodegeneration in Drosophila as well as the excess ROS generated by free Sdh1. These studies provide insights into the mechanisms by which SDH is assembled and raise the possibility that some forms of neuromuscular disease may be associated with mutations that affect this SDH assembly factor.
Keywords: metabolism, mitochondria, electron transport chain, succinate dehydrogenase, neurodegenerative disease
INTRODUCTION
Mitochondria are dynamic organelles that are essential for many cellular processes including metabolism, signal transduction, and apoptosis. Consistent with these broad cellular functions, mitochondrial dysfunction is associated with a wide range of human diseases, including diabetes, neurodegeneration, and cancer. These important roles have led to comprehensive characterization of the mitochondrial proteome, identifying more than 1,000 nuclear-encoded proteins (Sickmann et al., 2003; Pagliarini et al., 2008). In spite of these efforts, however, many mitochondrial proteins remain uncharacterized, including proteins that are conserved throughout eukaryotic evolution. We are studying this subset of proteins as part of a systematic effort to comprehensively understand mitochondrial physiology. Herein, we describe the function of one of these evolutionarily conserved proteins and demonstrate that it is essential for proper succinate dehydrogenase (SDH) complex assembly and activity in yeast, flies, and mammalian cells.
SDH is unique in that it functions in both the tricarboxylic acid (TCA) cycle and the electron transport chain (ETC), wherein it is referred to as Complex II. SDH links the oxidation of succinate to fumarate in the TCA cycle with the reduction of ubiquinone to ubiquinol in the ETC, which contributes to the establishment of the mitochondrial membrane potential and ATP synthesis. SDH is a heterotetrameric protein complex that is embedded in the inner mitochondrial membrane (IMM), with its catalytic domain facing the mitochondrial matrix. The complex is anchored to the IMM by two integral membrane proteins, Sdh3 (SDHC in humans) and Sdh4 (SDHD). The Sdh3/Sdh4 dimer binds the peripheral membrane protein Sdh2 (SDHB), which tethers the catalytic Sdh1 (SDHA) subunit to the complex. Sdh1 harbors a covalently bound FAD cofactor that is required for the oxidation of succinate (Robinson et al., 1994). The two electrons that result from succinate oxidation are channeled through the three iron-sulfur clusters in Sdh2 to ubiquinone, which interacts with SDH via the Sdh3/Sdh4 membrane anchor (Rutter et al., 2010).
The assembly of complexes like SDH presents the cell with the problem of coordinating the synthesis and stepwise interactions of individual subunits to form an intricate membrane bound complex. This problem is exacerbated in the case of ETC complexes as individual subunits contain redox-active cofactors that can perform inappropriate and deleterious reactions when they are not properly secluded within the native complex. As a result, a number of dedicated factors assist the assembly of these complexes by facilitating cofactor insertion, preventing non-productive interactions, and stabilizing assembly intermediates. While the importance of assembly factors for Complexes I and IV are well established (Fernandez-Vizarra et al., 2009; Diaz et al., 2011), dedicated SDH assembly factors have only recently begun to emerge with the discovery of SDHAF1 (Sdh6 in yeast) and SDHAF2 (Sdh5; Ghezzi et al., 2009; Hao et al., 2009). Human patients with loss of function mutations in SDHAF1 or SDHAF2 display reduced SDH complex levels and activity and present with infantile leukoencephalopathy and neuroendocrine tumors, respectively (Ghezzi et al., 2009; Hao et al., 2009). This is consistent with the spectrum of diseases that associate with mutations affecting the core subunits of SDH – SDHA, SDHB, SDHC, and SDHD (Rutter et al., 2010) and with a series of additional diseases that are characterized by loss of SDH activity, but which lack mutations in known SDH genes (Jain-Ghai et al., 2013). Disruption of the yeast orthologs for SDHAF1 or SDHAF2 (SDH6 or SDH5) prevents respiratory-dependent growth, with a clear defect in SDH activity. While the precise mechanism by which Sdh6 supports SDH biogenesis has remained elusive, Sdh5 was shown to be required for the covalent attachment of the FAD cofactor to Sdh1 (Ghezzi et al., 2009; Hao et al., 2009). The current model proposes that Sdh1 is flavinated by Sdh5, binds iron-sulfur loaded Sdh2, which then docks onto the pre-assembled membrane bound Sdh3-Sdh4 dimer to form the holocomplex (Rutter et al., 2010). We hypothesized that the complexity of this assembly process likely necessitates the action of additional dedicated assembly factors and that the genes encoding these factors may contribute to the currently idiopathic SDH-associated genetic diseases.
With the goal of discovering new human disease genes and gaining a better understanding of SDH assembly and activity, we have focused on our ongoing functional elucidation of uncharacterized mitochondrial proteins. These studies led us to identify an evolutionarily conserved SDH assembly factor, Ybr269, which we have renamed Sdh8 in yeast and SDHAF4 in higher organisms. Utilizing yeast, flies and mammalian cells, we have defined the molecular function of the SDHAF4 protein family in SDH assembly and an important role for SDHAF4 in metazoan physiology.
RESULTS
Yeast Sdh8 is a conserved mitochondrial matrix protein required for maximal SDH activity
Sdh8 is a small protein of approximately 15 kDa that belongs to a pan-eukaryotic protein family, which we have named the SDHAF4 family (Figure S1A). N- and C-terminal Sdh8 fusion proteins were unable to complement the phenotype of an sdh8Δ mutant strain and were therefore deemed to be non-functional. Efficient rescue was seen, however, when either a tandem affinity tag (His6HA3) or GFP was fused into a predicted unstructured region within Sdh8, between residues N82 and S83. These constructs, which were subsequently named Sdh8-His6HA3 and Sdh8-GFP, respectively were expressed under the control of the native SDH8 promoter and terminator and determined to be fully functional (See Figures 1A and S1E). As expected, we found that Sdh8 localizes exclusively to mitochondria and determined it to be a soluble matrix protein (Figure S1B–D). To begin to assess the role of Sdh8 in mitochondrial function, an sdh8Δ mutant was established in two strains (BY4741 and W303), both of which were used for key experiments to control for genetic background. While sdh8Δ mutants grew normally in glucose medium, growth was impaired on glycerol and acetate, demonstrating a defect in mitochondrial respiration (Figure 1A and S1E). Interestingly, the growth impairment was exacerbated on acetate-containing medium, which is a hallmark of mutants that affect SDH activity. Consistent with this possibility, metabolomic analysis of sdh8Δ mutants revealed an apparent block in the TCA cycle at SDH, with an accumulation of succinate and depletion of the two TCA cycle intermediates downstream of SDH, fumarate and malate (Figure 1B and S1F-H). We also measured the enzymatic activity of SDH in mitochondria harvested from wild-type (WT) and sdh8Δ cells. SDH activity in sdh8Δ cells was decreased 60% relative to WT (Figure 1C), while malate dehydrogenase (MDH) activity was unaffected (Figure 1D). Finally, the steady state level of the SDH holocomplex was assessed by blue native polyacrylamide gel electrophoresis (BN-PAGE). Mitochondria purified from either WT or sdh8Δ cells were solubilized in digitonin, resolved by BN-PAGE, and probed by immunoblot. We observed a marked decrease in assembled SDH and demonstrated that re-expression of Sdh8-His6HA3 (see supplemental text) in sdh8Δ cells was sufficient to rescue the defect in SDH assembly (Figure 1E and S1I). Taken together, these data demonstrate that Sdh8 is required for the stability and activity of SDH.
Figure 1. Yeast Sdh8 is a conserved mitochondrial matrix protein required for maximal SDH activity and assembly.
(A) Ten-fold serial dilutions of WT yeast transformed with an empty vector (EV) or sdh8Δ mutants transformed with either EV or a plasmid expressing SDH8-His6HA3, all in a BY4741 background. (B) GC/MS was used to measure the abundance of metabolites in WT and sdh8Δ yeast (BY4741 background. N=6 biological replicates. ****p < 0.0001; **p < 0.005; *p < 0.05). (C) SDH and (D) MDH enzyme assays were performed on mitochondrial extracts of WT, sdh8Δ, and sdh1Δ strains, normalized to total protein (±SEM. N=3 biological replicates. **p < 0.005). (E) WT and sdh8Δ mitochondria were solubilized in digitonin, fractionated by BN-PAGE, and analyzed by immunoblotting to detect Atp2 (complex V), Cor1/2 (complex III), Cox3 (complex IV), Sdh1 (complex II), or Porin. (F) Ten-fold serial dilutions of the indicated strains were grown as in (A). SDHAF4 and dSdhaf4 are the human and Drosophila orthologs of SDH8, respectively, and were each expressed under the control of the endogenous SDH8 promoter and terminator.
As a first step toward determining if its function is conserved across eukaryotic species, the human and Drosophila orthologs of SDH8 (SDHAF4 and dSdhaf4, respectively) were expressed in the sdh8Δ mutant under the control of the native SDH8 promoter and terminator. Both orthologs complement the sdh8Δ growth defects (Figure 1F), suggesting that the role of Sdh8 in SDH assembly is an evolutionarily conserved feature of the SDHAF4 family.
Mammalian SDHAF4 is required for maximal SDH activity
To assess a possible role for mammalian SDHAF4 in SDH function, we transfected C2C12 mouse myoblasts with a non-targeting control siRNA (control) or either of two siRNAs targeting mouse SDHAF4 (si1 and si2). Knockdown of SDHAF4 mRNA was confirmed (Figure 2A) and mitochondria were harvested from cells transfected with control, si1, and si2 siRNAs. Immunoblot of the isolated mitochondria revealed that depletion of SDHAF4 did not affect the steady state abundance of SDHA or SDHB (Figure S2A). We detected, however, a decrease in SDH enzymatic activity in SDHAF4 knockdown cells (Figure 2B), with no significant change in MDH activity (Figure S2B). In addition, cells transfected with si1 or si2 showed a reproducible and specific decrease in steady-state SDH complexes as assayed by BN-PAGE, of a magnitude similar to that seen in SDH activity (Figure 2C). We conclude that SDHAF4 is required for the proper assembly and activity of SDH in both yeast and mammalian cells.
Figure 2. Mammalian SDHAF4 is required for maximal SDH activity.
(A) C2C12 cells were transfected with either a control non-targeting siRNA or either of two siRNAs directed against SDHAF4 (si1 and si2), and SDHAF4 mRNA abundance was assayed by quantitative RT-PCR and normalized to tubulin mRNA (±SEM. N=3 biological replicates. ***p < 0.0005). (B) SDH enzyme activity was measured in mitochondria harvested from C2C12 cells treated as in (A) (±SEM. N=3 biological replicates. ***p < 0.0005; *p < 0.05). (C) Mitochondria extracted C2C12 cells treated as in (A) were solubilized in digitonin, fractionated by BN-PAGE, and analyzed by immunoblotting for ATP5A (complex V monomers (CVI) or dimers (CVII), QCR2 (complex III), VDAC, or SDHA (complex II).
Drosophila Sdhaf4 is required for maximal succinate dehydrogenase activity
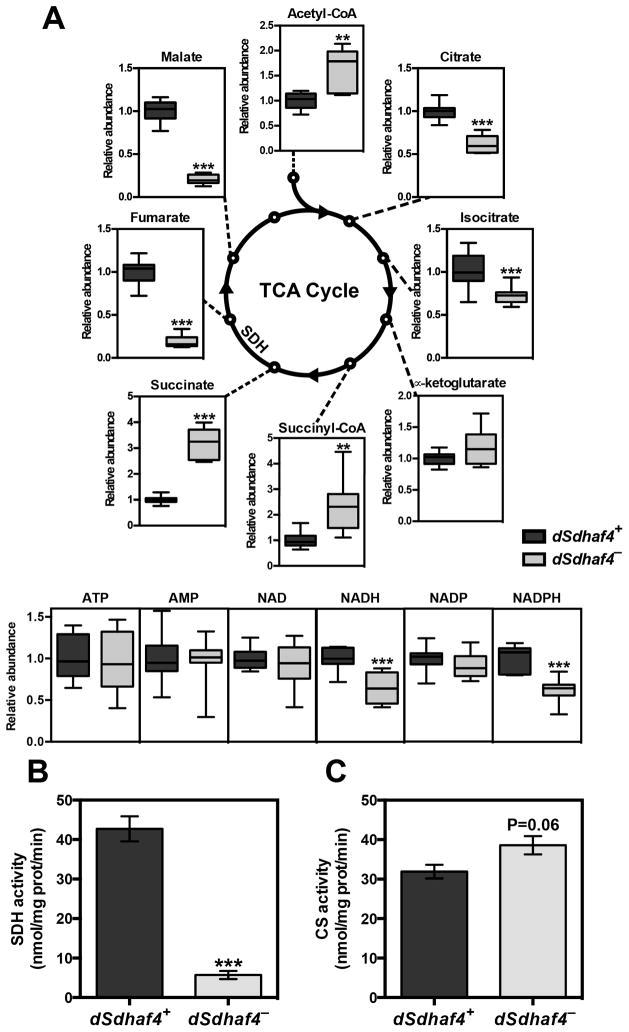
To further define the function of Sdh8 in a multicellular organism, we extended our analysis to Drosophila. Three null alleles were generated in the Drosophila ortholog of SDH8 (CG7224) that we have renamed Drosophila Sdhaf4 (dSdhaf4) (Figure S3A). Similar to our findings in yeast, metabolomic analysis of dSdhaf4 mutants compared to genetically matched controls revealed a significant accumulation of succinate and depletion of malate and fumarate (Figure 3A). Acetyl-CoA was also elevated, while citrate and isocitrate were reduced in the mutants (Figure 3A). These effects may be due to a reduction in oxaloacetate that is required for the formation of citrate from acetyl-CoA. Consistent with this, aspartate, which is interconvertible with oxaloacetate, is depleted in dSdhaf4 mutants (Figure S3C). Similar changes were observed upon metabolomic analysis of flies carrying a dSdhaf4 TALEN-induced allele over an independently derived imprecise excision, indicating that these results remain constant across genetic backgrounds (Figure S3B and S3C). Consistent with this metabolomic profile, dSdhaf4 mutant mitochondria display an 85% reduction in SDH activity (Figure 3B) while citrate synthase (CS) activity was unaffected (Figure 3C). Based on these data, we conclude that, like yeast sdh8Δ mutants, dSdhaf4 mutants have a specific impairment in SDH function.
Figure 3. Drosophila Sdhaf4 is required for SDH activity.
(A) Either LC/MS (acetyl-CoA, succinyl-CoA, ATP, AMP, NAD, NADH, NADP, and NADPH) or GC/MS (citrate, isocitrate, alpha-ketoglutarate, succinate, fumarate and malate) was used to measure the abundance of metabolites in trans-heterozygous dSdhaf41/2 (dSdhaf4−) mutants compared to genetically matched w1118 controls (dSdhaf4+). 8–12 biological replicates from two independent experiments were combined per genotype. (B) SDH or (C) citrate synthase enzymatic activity was measured in extracts from mitochondria isolated from either control or dSdhaf4 mutant flies (±SEM. N=4–6 biological replicates. **p < 0.01 and ***p <0.001). Two independent experiments were performed with similar results.
Drosophila Sdhaf4 is required for the stability of SdhA and SdhB
The decrease in SDH activity in dSdhaf4 mutants is greater than that seen in yeast sdh8Δ mutants, suggesting that there might be a more profound effect on the steady-state level of SDH. Consistent with this, the SDH holocomplex was undetectable by BN-PAGE, whereas the levels of Complex V were unaffected (Figure 4A). This lack of SDH holocomplex was accompanied by an apparent reduction in the stability of the SdhA and SdhB subunits in dSdhaf4 mutants (Figure 4B, S4A, and S4B). In contrast, dSdhaf4 heterozygous mutants displayed similar SDH activity to controls (Figure S4C), and dSdhaf4 overexpression had no effect on SDH activity or SDH subunit protein levels (Figure S4D and S4E).
Figure 4. Drosophila Sdhaf4 is required for SdhA and SdhB stability.
(A) Mitochondrial extracts from either w1118 controls (dSdhaf4+) or dSdhaf41/2 mutants (dSdhaf4) were fractionated by BN-PAGE and analyzed by immunoblotting to detect Atpα (Complex V) or SdhB (Complex II). (B) Total protein from mitochondria isolated from either control or dSdhaf4 mutant flies was resolved using SDS-PAGE, followed by immunoblotting to detect SdhA, SdhB or Atpα. (C) Flies carrying either control or dSdhaf4 mutant chromosomes over a balancer chromosome were crossed to flies carrying a strong loss-of-function allele for either SdhA or Cytochrome c oxidase Va subunit (CoVa) over a balancer. The resulting progeny were scored for the absence or presence of the marker linked to the balancer chromosome. The proportion of the expected progeny that eclosed, based on Mendelian ratios of the indicated genotypes, is shown. (±SEM. N = number of progeny assayed from each cross. ***p <0.001 (Student’s t test).
The reduced levels of SDH subunits in the dSdhaf4 mutant suggest that we might see genetic interactions between dSdhaf4 alleles and mutations in SDH subunit-encoding genes. Consistent with this, we found that dSdhaf4 mutant flies heterozygous for a strong loss-of-function mutation in SdhA or homozygous for a hypomorphic mutation in SdhB displayed reduced viability compared to controls (Figure 4C and S4F). This genetic interaction is specific for SDH subunit-encoding genes, as dSdhaf4 mutants heterozygous for a null allele of CoVa (Mandal et al., 2005) displayed normal viability (Figure 4C). Taken together, these biochemical and genetic studies indicate that dSdhaf4 is required for SDH assembly and activity.
dSdhaf4 mutants display neurodegeneration, early-adult lethality, and sensitivity to oxidative stress
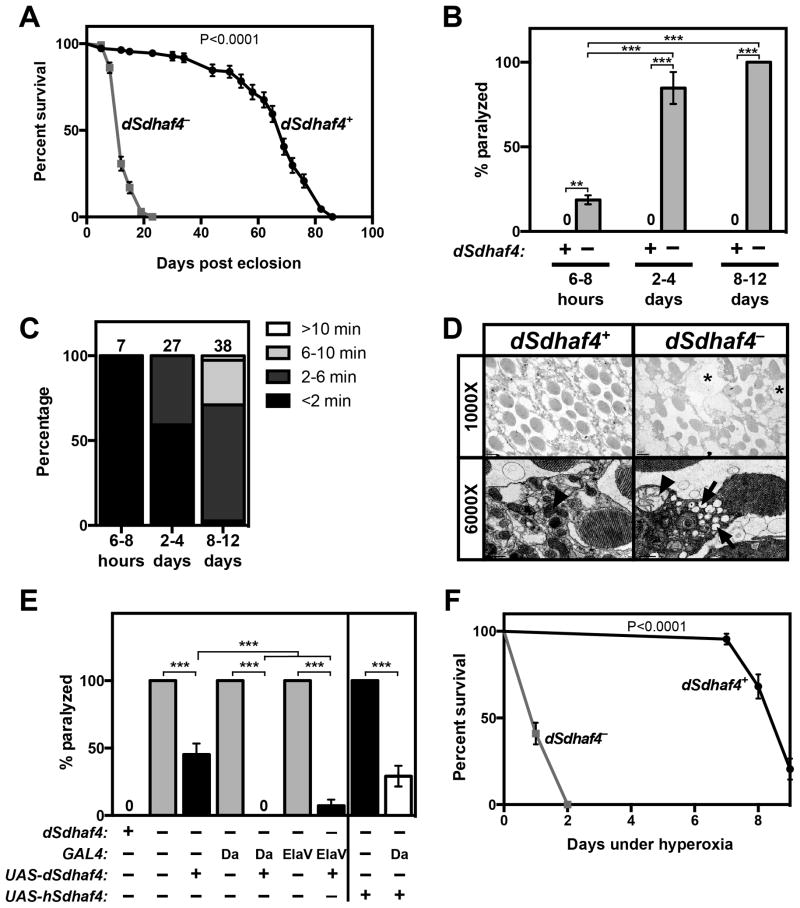
Consistent with their metabolic defects, dSdhaf4 mutants died significantly earlier than controls, with a median lifespan of 12 days compared to a median lifespan of 69 days for control flies (Figure 5A). Interestingly, these mutants also displayed two phenotypes that are hallmarks of mitochondrial dysfunction, bang sensitivity and erect wings (Figure 5B, 5C, S5A, and S5B; Greene et al., 2003; Fergestad et al., 2006). In addition, dSdhaf4 mutant retinas displayed a marked disorganization of retinal architecture (Figure 5D, asterisks), consistent with the known association between bang sensitivity and neurodegeneration (Celotto et al., 2006a; Gnerer et al., 2006; Liu et al., 2007). The photoreceptors were also highly vesicularized (Figure 5D, arrows; Kiselev et al., 2000), with aberrant mitochondrial morphology (Figure 5D, arrowheads). The bang sensitivity of dSdhaf4 mutants can be rescued by specific expression of wild-type dSdhaf4 in neurons, suggesting that it is associated with the neurodegeneration (Figure 5E, S5C, and S5D). Moreover, global expression of human Sdhaf4 in dSdhaf4 mutants partially rescued the bang-sensitivity of these animals, providing further evidence of a conserved function for Sdhaf4 (Figure 5E, S5C, and S5D). Taken together, these results demonstrate that dSdhaf4 is required in neurons to maintain tissue architecture and function.
Figure 5. Drosophila Sdhaf4 mutants display neurodegenerative phenotypes, early-adult lethality, and sensitivity to oxidative stress.
(A) The number of surviving w1118 control (dSdhaf4+) or dSdhaf41/2 mutant (dSdhaf4−) females maintained on standard laboratory food at normoxia was assayed at 2–4 day intervals. Each data point is the percentage of surviving flies±SEM, with at least 110 flies assayed per genotype. (B) Control or dSdhaf4 mutant females aged 6–8 hours, 2–4 days, or 8–12 days were vortexed for 15 seconds and the percentage of paralyzed flies was counted as described (Ganetzky and Wu, 1982). (±SEM. N>32 flies per age per genotype. **p <0.01 and ***p <0.001 (ANOVA)). (C) The length of time required for the dSdhaf4 mutant flies in (A) to recover from paralysis is presented as a bar graph, with the number of paralyzed dSdhaf4 mutants analyzed from each age group depicted above the bar. (D) Cross sections of compound eyes from 8–12 day old control or dSdhaf4 mutant flies visualized by transmission electron microscopy. Arrows point to aberrant vesicularization of dSdhaf4 mutant photoreceptors, arrowheads point to mitochondria, and asterisks denote vacuoles (E) Control or dSdhaf4 mutant flies at 8–12 days of age carrying either the ubiquitous Daughterless-Gal4 (Da-GAL4) driver, the pan-neuronal Elav-Gal4 driver, or no driver, in the presence or absence of either a UAS-dSdhaf4 transgene or a UAS-human Sdhaf4 (hSdhaf4) transgene, were vortexed as in (A). Note that UAS-dSdhaf4 alone has leaky dSdhaf4 activity since it partially rescues in the absence of a GAL4 driver. (±SEM. N=15–40. ***p <0.001 (ANOVA)). (F) Control or dSdhaf4 mutant females at 2–4 days of age were transferred to 100% oxygen and the number of surviving animals was counted each day. The median lifespan after transfer to hyperoxia is 9 days for controls and 1 day for dSdhaf4 mutants. Data is shown as the percentage of surviving flies±SEM. At least 40 flies were assayed per genotype. P values for both survival curves (panels A, F) were calculated using a log-rank test.
Defects in mitochondrial ATP production and reactive oxygen species (ROS) homeostasis have been previously shown to cause neurodegenerative phenotypes similar to those observed in dSdhaf4 mutants (Fergestad et al., 2006; Liu et al., 2007; Celotto et al., 2012). Since loss of dSdhaf4 does not appear to affect steady state ATP levels (Figure 3A and S3B), we hypothesized that dSdhaf4 may be required to protect against ROS toxicity. Consistent with this, dSdhaf4 mutants were highly sensitive to oxidative stress induced by hyperoxia (Figure 5F), but showed no sensitivity to starvation, which is unassociated with ROS production (Supplemental Figure 5E). These data suggest a connection between the role of dSdhaf4 in SDH assembly and oxidative stress resistance.
Yeast Sdh8 is a chaperone for soluble and covalently flavinated Sdh1
Data from three distinct eukaryotic systems indicate that the SDHAF4 protein family plays an evolutionarily conserved role in SDH assembly and activity. Turning back to yeast, we sought to understand the mechanism by which Sdh8 supports SDH biogenesis. To identify proteins interacting with Sdh8, we performed a large scale HA immunoprecipitation from either WT or Sdh8-His6HA3 containing mitochondria. The final eluates were resolved by SDS-PAGE, revealing a band of ~70 kDa that was present only in the Sdh8-His6HA3 immunoprecipitation. Using mass spectrometry, we found that this band contained Sdh1, the catalytic subunit of SDH (Figure S6A). The putative Sdh8/Sdh1 interaction was confirmed by immunoprecipitation of Sdh8 followed by identification of Sdh1 in the eluate using an Sdh1-specific antibody (Figure 6A, lanes 1 and 6 vs. 2 and 7). Interestingly, it appears that Sdh2 is not a member of this complex as it is undetectable in the eluate (Figure 6A).
Figure 6. Yeast Sdh8 is a co-chaperone for unbound and covalently flavinated Sdh1.
(A) Sdh8-His6HA3 was immunoprecipitated from digitonin-solubilized mitochondria isolated from the yeast strains described as follows. Lanes 1 and 6, WT cells; Lanes 2 and 7, sdh8Δ cells expressing Sdh8-His6HA3; Lanes 3 and 8, sdh2Δ sdh8Δ cells expressing Sdh8-His6HA3; Lanes 4 and 9 sdh1Δ sdh8Δ cells expressing Sdh8-His6HA3 and Sdh1H90A; Lanes 5 and 10, sdh1Δ sdh2Δ sdh8Δ cells expressing Sdh8-His6HA3 and Sdh1H90A. (B) Mitochondrial lysates isolated from the following strains were fractionated by SDS-PAGE. Lane 1, sdh8Δ cells expressing Sdh8-His6HA3; Lane 2, sdh1Δ sdh8Δ cells expressing Sdh8-His6HA3; Lane 3, sdh2Δ sdh8Δ cells expressing Sdh8-His6HA3; Lane 4, sdh8Δ cells; Lane 5, sdh2Δ sdh8Δ cells (BY4741 background). Steady state protein abundance was assessed by immunoblot using antibodies against Sdh1, Sdh2, and HA (Sdh8-His6HA3). (C) Mitochondria isolated from the following yeast strains were solubilized in digitonin and fractionated by BN-PAGE. Lane 1, sdh8Δ cells expressing Sdh8-His6HA3; Lane 2, sdh2Δ sdh8Δ cells expressing Sdh8-His6HA3; Lane 3, sdh1Δ sdh2Δ sdh8Δ cells expressing Sdh8-His6HA3; Lane 4, sdh2Δ sdh8Δ; Lane 5, sdh4Δ sdh8Δ cells expressing Sdh8-His6HA3; Lane 6, sdh1Δ sdh4Δ sdh8Δ cells expressing Sdh8-His6HA3, Lane 7, sdh4Δ sdh8Δ cells. Immunoblotting was used to detect Atp2 (complex V) as a control and complexes containing Sdh1, Sdh2, and HA (Sdh8-His6HA3). SDH complex (~232 kDa), Sdh1-Sdh8 sub-complex (~150 kDa).
Since Sdh8 binds Sdh1 independent of Sdh2, we hypothesized that Sdh8 may be acting as a chaperone for a free soluble pool of Sdh1 in the mitochondrial matrix. If this is true, then deletion of SDH2 might shift Sdh1 to a free soluble form, which would increase the demand for Sdh8 function. Consistent with this possibility, sdh2Δ mutant mitochondria exhibit an accumulation of Sdh8-His6HA3 (Figure 6B, lane 1 vs. 3), while sdh1Δ mutant mitochondria exhibit a depletion of Sdh8-His6HA3 (Figure 6B, lane 1 vs. 2). Furthermore, this accumulation of Sdh8-His6HA3 results in enhanced Sdh1-Sdh8 association in the sdh2Δ mutant (Figure 6A, lanes 2 and 7 vs 3 and 8) despite an approximately two-fold decrease in steady state Sdh1 levels (Figure 6B, lane 1 vs. 3).
Although the stability of Sdh8 depends on Sdh1, Sdh1 abundance is unaffected by the deletion of SDH8 (Figure 6B, lane 1 vs. 4). However, Sdh2 levels are reduced in an sdh8Δ mutant, likely due to decreased Sdh1 competence for interaction with Sdh2 (Figure 6B, lane 1 vs. 4). This is further supported by the observation that the formation of the Sdh1-Sdh2 soluble dimer, which is readily apparent in a mutant lacking the Sdh4 membrane anchor, is impaired in an sdh8Δ mutant (Figure S6F). Interpretation of this experiment is complicated by the decreased Sdh2 steady state level in the sdh8Δ mutant, but the data are most consistent with the hypothesis that Sdh8 is required to maintain Sdh1 in a state that is fully competent to bind Sdh2 and assemble in the SDH holocomplex.
Sdh1 is known to be covalently flavinated at H90 in a reaction that requires the activity of Sdh5 (Hao et al., 2009). Initially, we found that deletion of SDH8 does not impact the flavination of Sdh1 (Figure S6B). We reasoned, however, that Sdh8 might act specifically on the flavinated form of free Sdh1. Accordingly, we immunoprecipitated Sdh8-His6HA3 from mitochondria that express wild-type Sdh1, Sdh1H90A, or Sdh1H90S, which are Sdh1 mutants that cannot covalently bind FAD. Indeed, Sdh1H90A and Sdh1H90S both failed to co-purify with Sdh8-His6HA3 (Figure 6A, lanes 2 and 7 vs. 4 and 9, Figure S6C) even in the absence of Sdh2 (Figure 6A, lanes 3 and 8 vs. 5 and 10). In further support of this hypothesis, we found that Sdh1 failed to interact with Sdh8-His6HA3 in sdh5Δ mutant cells (Figure S6D), which lack covalent Sdh1 flavination (Figure S6B; Hao et al., 2009). Furthermore, Sdh8-His6HA3 was destabilized in the sdh5Δ mutant as was observed for the sdh1Δ mutant (Figure S6E, lane 1 vs 6).
Finally, Sdh8-His6HA3 is not present in the SDH holocomplex, as assayed by BN-PAGE (Figure 6C, lane 1). We also observed that deletion of SDH2 or SDH4, which ablates the SDH holocomplex, causes the emergence of a sub-complex of ~150 kDa, which lacks Sdh2 but clearly contains both Sdh1 and Sdh8 (Figure 6C, lanes 2 and 5). This is not coincidental co-migration as loss of either Sdh1 or Sdh8 causes this sub-complex to disappear from sdh2Δ and sdh4Δ mutant strains (Figure 6C, lanes 3, 4, 6, and 7). The Sdh1-Sdh8 subcomplex can also be detected in WT cells, albeit at lower steady state levels than in sdh2Δ and sdh4Δ mutants (Figure S6G). Taken together, these data demonstrate that Sdh8 interacts specifically with flavo-Sdh1, independent of other components of the SDH complex, and its stability depends upon this interaction.
Sdh8 protects the cell from oxidative stress
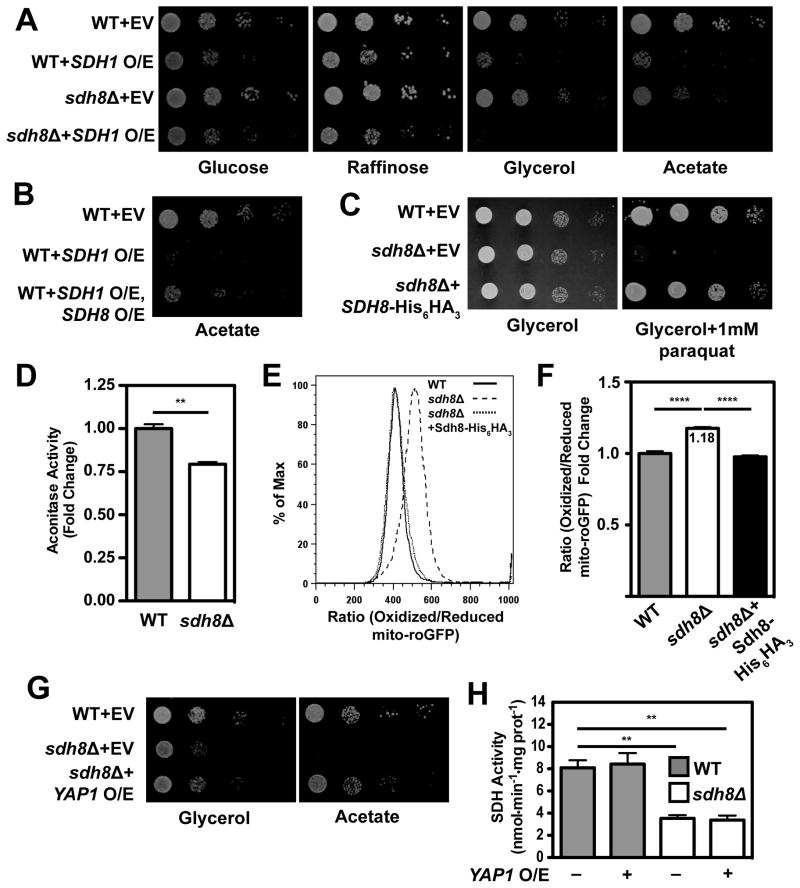
The data presented thus far demonstrate that Sdh8 acts as a chaperone for Sdh1 to promote formation of the Sdh1-Sdh2 dimer. We found that overexpression of Sdh1 does not rescue the growth defects of sdh8Δ mutants but, rather, is toxic to both WT and sdh8Δ cells on respiratory carbon sources, with the sdh8Δ mutant being particularly sensitive (Figure 7A and S7A). This observation raises the possibility that free Sdh1 is toxic and that Sdh8 may alleviate this toxicity. To test this, we co-overexpressed Sdh8 with Sdh1 and observed a significant rescue of the Sdh1 toxicity in WT cells (Figure 7B). We conclude that, in addition to promoting assembly of the Sdh1-Sdh2 dimer, Sdh8 also prevents the toxicity associated with free Sdh1.
Figure 7. Sdh8 protects the cell from oxidative stress.
(A) Ten-fold serial dilutions of WT and sdh8Δ yeast either transformed with an empty vector or over-expressing Sdh1 were plated on media containing the indicated carbon source (W303 background). (B) Ten-fold serial dilutions of WT yeast transformed with an empty vector, WT yeast overexpressing Sdh1, and WT yeast overexpressing Sdh1 and Sdh8-His6HA3 were plated on media containing acetate (W303 background). (C) Ten-fold serial dilutions of WT yeast transformed with an empty vector, sdh8Δ cells transformed with an empty vector, and sdh8Δ cells expressing Sdh8-His6HA3 were plated on media containing glycerol with and without 1mM paraquat and incubated at 30°C (W303 background) as in (A). (D) Aconitase activity was measured in WT and sdh8Δ cells grown to stationary phase in synthetic complete media containing 2% glucose and normalized to total protein (±SEM; N=4 biological replicates. **p < 0.005). (E) WT cells (solid line), sdh8Δ cells (dashed line), and sdh8Δ cells expressing Sdh8-His6HA3 (dotted line) transformed with mito-roGFP were plated on synthetic media containing 2% acetate, allowed to grow for 48 hours and analyzed by flow cytometry. Shown are representative histograms measuring the ratio of oxidized/reduced mito-roGFP in 30,000 cells (BY4741 background). (F) The median value of the ratio of oxidized/reduced mito-roGFP is plotted for WT cells, sdh8Δ cells, and sdh8Δ cells expressing Sdh8-His6HA3 (±SEM; N=4 biological replicates. ****p < 0.0001). (G) Ten-fold serial dilutions of WT yeast transformed with an empty vector, sdh8Δ cells transformed with an empty vector, and sdh8Δ cells over-expressing YAP1 were plated on media containing the indicated carbon source and incubated at 30°C (BY4741 background) as in (A). (H) SDH activity enzyme assays were performed using mitochondria harvested from WT and sdh8Δ cells carrying an empty vector or overexpressing YAP1. Activity was normalized to total protein (±SEM; N=3 biological replicates. **p < 0.005).
In considering possible mechanisms for toxicity of free Sdh1, we hypothesized that the exacerbated sdh8Δ phenotype on acetate compared to glycerol medium (Figure 1A) may be the result of increased oxidative stress. This is consistent with previous reports using acetate to trigger ROS-dependent cell death in yeast (Carmona-Gutierrez et al., 2010), as well as the sensitivity of dSdhaf4 mutant to hyperoxia (Figure 5F). In support of this hypothesis, we found that the sdh8Δ mutant is profoundly hypersensitive to paraquat in glycerol media relative to WT cells (Figure 7C). To test this hypothesis directly we sought to quantify the levels of ROS in the sdh8Δ mutant. We observed a significant decrease in aconitase activity in sdh8Δ cells, consistent with an accumulation of ROS in the mutant strain (Figure 7D). We also established a system for measuring the redox state of yeast mitochondria using a mitochondrially-targeted redox sensitive GFP construct (mito-roGFP; McFaline-Figueroa et al., 2011; Vevea et al., 2013). This construct has been engineered to contain two surface exposed cysteines that are capable of forming a disulfide bond. Mito-roGFP exhibits two excitation peaks at ~400nm and ~480nm with a single emission peak at ~510nm. Oxidation of the surface-exposed cysteines promotes excitation at 400nm while reduction of these residues favors excitation at 480nm (Figure S7B and S7C; McFaline-Figueroa et al., 2011; Vevea et al., 2013). Thus, the excitation ratio of mito-roGFP is a measure of the redox state of yeast mitochondria in live cells. To quantify the redox state of the matrix, we transformed WT cells, sdh8Δ cells, and sdh8Δ cells expressing Sdh8-His6HA3 with mito-roGFP (Figure S7D). Cells were grown in the presence of 2% acetate and analyzed by flow cytometry. Consistent with the aconitase activity data, we observed an increase in the ratio of oxidized/reduced mito-roGFP in the sdh8Δ strain that was fully rescued by re-expressing Sdh8-His6HA3 (Figure 7E and 7F). If this impaired growth of the sdh8Δ mutant is the result of the elevated ROS seen in these cells, then overexpression of genes involved in ROS detoxification might suppress the growth phenotype. Consistent with this, overexpression of YAP1, a transcription factor that stimulates the expression of many genes important for oxidative stress defense (including TRR1, TRX2, GSH1, and GLR1), as well as the mitochondrial superoxide dismutase SOD2, completely or partially rescued the growth phenotype of sdh8Δ cells (Figure 7G and S7E). Interestingly, despite a profound ability to rescue the growth phenotype associated with sdh8Δ cells, YAP1 overexpression has no effect on SDH activity (Figure 7H), SDH assembly (Figure S7F), or Sdh2 steady state levels (Figure S7G). Thus, we conclude that the ability of YAP1 to rescue the sdh8Δ mutant phenotype is not related to SDH activity and likely due to an amelioration of oxidative stress in the mutant cells.
DISCUSSION
The critical role of SDH in primary cellular metabolism is reflected by the variety of diseases associated with its dysfunction. Unlike other ETC complexes, which require multiple factors for their assembly, only two factors, SDHAF1 and SDHAF2, have been identified to date that are required for the assembly of the SDH holocomplex within the IMM (Ghezzi et al., 2009; Hao et al., 2009). Herein we use three distinct eukaryotic model systems, yeast, Drosophila, and mammalian cells, to demonstrate that the SDHAF4 protein family consists of evolutionarily conserved SDH assembly factors. We show that Sdh8 binds directly to flavinated Sdh1, blocking the generation of excess ROS and facilitating its interaction with Sdh2 and the subsequent assembly of the SDH holocomplex. These functions for SDHAF4 appear to be conserved in metazoans and suggest a role for this factor in disease, as Drosophila Sdhaf4 mutants display muscular and neuronal dysfunction as well as neurodegeneration.
Yeast sdh8Δ and Drosophila Sdhaf4 mutants exhibit the classic metabolomic phenotype of SDH deficiency, with an apparent block in the TCA cycle that results in an accumulation of succinate and a depletion of fumarate and malate, the two TCA cycle intermediates downstream of SDH. Consistent with these effects, the yeast and fly mutants display a reduction in the steady-state levels of the SDH holocomplex and reduced SDH enzyme activity, indicating an important role for SDHAF4 in SDH stability and activity. siRNA-mediated knockdown of SDHAF4 in mammalian cells leads to similar defects. Moreover, expression of either fly or human SDHAF4 is sufficient to rescue the growth defects of an sdh8Δ mutant yeast strain. Taken together, these results demonstrate that the central requirement for SDHAF4 in SDH assembly has been conserved through evolution, from yeast to mammals.
Our biochemical studies in yeast provide a mechanistic context for understanding the role of SDHAF4 in SDH assembly. An unbiased proteomic analysis of Sdh8-associated proteins revealed a physical interaction between Sdh8 and Sdh1, the catalytic subunit of SDH. We confirmed the specificity of this interaction by co-immunoprecipitation and demonstrated that an Sdh1-Sdh8 subcomplex of ~150 kDa can be detected in unstressed WT cells, albeit at very low levels. Increased levels of this subcomplex, however, become readily detectable upon genetic disruption of the SDH complex through deletion of either Sdh2 or Sdh4. Taken together, these results support the model that Sdh8 interacts with a soluble pool of Sdh1 prior to the formation of the Sdh1-Sdh2 soluble dimer and assembly of the holocomplex. Moreover, Sdh8 interacts specifically with the flavinated form of Sdh1, indicating that this association follows Sdh1 flavination by Sdh5/SDHAF2 (Hao et al., 2009). This model is consistent with the localization of Sdh8 to the matrix where it can readily access free flavo-Sdh1, which is subsequently tethered to the IMM through its association with Sdh2 and the membrane-bound Sdh3/Sdh4 dimer.
We observed that Sdh8 stability directly relates to its association with Sdh1, further supporting the centrality of its role as an Sdh1 chaperone. Thus, deletion of Sdh1 leads to a dramatic reduction in Sdh8 levels while deletion of Sdh2, which causes greatly increased availability of free Sdh1, leads to an increase in Sdh8. Although Sdh1 protein levels are unperturbed in sdh8Δ cells, we observed considerable destabilization of Sdh2, which is consistent with impaired formation of Sdh1-Sdh2 dimers. Therefore, we hypothesize that the occupation of free Sdh1 by Sdh8 facilitates the formation of Sdh1-Sdh2 dimers. This dimer fails to form efficiently in the absence of Sdh8, and Sdh2 is destabilized as it is in mutants lacking Sdh1 altogether.
Interestingly, the loss of SDHAF4 appears to have more severe consequences in Drosophila than in yeast or mammalian cells. dSdhaf4 mutants display a profound reduction in SDH enzyme activity, destabilization of both SdhA and SdhB, and a significant loss of SDH holocomplexes. In contrast, yeast sdh8Δ mutants and C2C12 mouse myoblasts treated with SDHAF4 siRNA maintain ~40–50% of wild-type SDH activity and preserve a similar fraction of assembled SDH holocomplexes. While yeast sdh8Δ mutants have relatively normal levels of Sdh1, dSdhaf4 mutants have greatly reduced levels of SdhA, which probably underlies the nearly complete loss of SDH holocomplexes. It is possible that yeast and mammalian cells express an additional SDHA chaperone that can function in the absence of SDHAF4, maintaining the levels of SDHA to support holocomplex assembly. It is also possible that growth in rich culture medium provides a buffer against oxidative stress in yeast that is not present in the more complex Drosophila system. Identification of the mechanism that underlies this discrepancy, however, will require further study of SDH assembly in both yeast and flies. Regardless, it is important to note that dSdhaf4 mutants retain some SDH activity (~15%) and that this is sufficient to support limited viability, albeit with both muscular (erect wings) and behavioral (neurodegeneration) dysfunction. In contrast, SdhA null mutants are lethal, demonstrating that a further reduction in SDH activity cannot be tolerated (Mast et al., 2008). Thus, while the SDHAF4 protein family is clearly involved in SDH stability and activity, it does not appear to be an absolute requirement as some level of SDH activity remains in the absence of the respective SDHAF4 ortholog in all three model systems.
Our demonstration that Sdh8 acts through Sdh1 to facilitate formation of the Sdh1/Sdh2 dimer and subsequently the SDH holocomplex further emphasizes the importance of dedicated chaperones for ETC complex assembly. We propose that this unique demand might relate to the need to protect the redox-active cofactors present in many ETC complex subunits (Messner and Imlay, 2002). Prior to assembly, these cofactors are exposed to solvent and, therefore, susceptible to damaging chemical reactions. The susceptibility of free cofactors to damaging chemical reactions is exemplified by our data suggesting that free flavinated Sdh1 is toxic and that this toxicity is alleviated by Sdh8. Overexpression of Sdh1 causes growth defects in WT cells, but this phenotype is exacerbated in sdh8Δ mutants and ameliorated by co-overexpression of Sdh8. Thus, we conclude that the chaperone activity of Sdh8 protects the cell from potentially damaging effects of unbound Sdh1, which are likely due to the generation of ROS enabled by solvent-accessible FAD. Sdh1, which contains a covalently bound, redox active, FAD cofactor is capable of oxidizing succinate to fumarate independent of the SDH complex. This oxidation of succinate leads to the reduction of FAD, which is then auto-oxidized by molecular oxygen to yield superoxide (Messner and Imlay, 2002; Guzy et al., 2008). The observation that Sdh8 interacts specifically with covalently flavinated Sdh1, but not with apo-Sdh1, is consistent with Sdh8 acting to prevent this mechanism of oxidative stress. Further, sdh8Δ mutant cells were unable to grow on respiratory medium in the presence of the superoxide producer paraquat and demonstrate a marked accumulation of ROS as determined by two independent assays. Similarly, dSdhaf4 mutant flies are highly sensitive to oxidative stress induced by hyperoxia. Importantly, overexpression of two genes involved in the detoxification of ROS — YAP1 and SOD2—rescued the sdh8Δ growth defects in yeast without affecting the SDH activity, SDH assembly, or steady state Sdh2 levels. We conclude that, in addition to promoting Sdh2 binding and holocomplex assembly, Sdh1 protects the redox-active FAD cofactor from potentially deleterious solvent interactions.
These biochemical aberrations resulted in profound physiological defects in Drosophila lacking dSdhaf4, all of which are hallmarks of mitochondrial dysfunction and which suggest possible links to human disease. Most notably, dSdhaf4 mutants display bang sensitivity, a phenotype characterized by a susceptibility to mechanically induced paralytic seizures, which is commonly elicited by defects in mitochondrial function (Fergestad et al., 2006; Liu et al., 2007). dSdhaf4 mutants also display neurodegeneration and reduced lifespan, which often accompany bang sensitivity (Fergestad et al., 2008). In addition, we observed abnormal wing posture in dSdhaf4 mutants, which is characteristic of muscular dysfunction (Greene et al., 2003). Similar phenotypes have been reported in flies harboring mutations in several known disease-causing genes that impact mitochondrial function. For example, Pink and Parkin mutants display abnormal wing posture, and mtATP6 mutants are bang sensitive (Celotto et al., 2006b; Fernandes and Rao, 2011). Human mutations in Pink/Parkin and mtATP6 are known to cause familial Parkinson’s disease and mitochondrial encephalopathy, respectively (Schapira, 2012). Moreover, as described in more detail below, some neurodegenerative diseases such as Leigh’s syndrome are known to be associated with a loss of SDH activity. Our studies of dSdhaf4 thus provide a genetic model for better understanding the pathophysiology of neurodegenerative disorders that are caused by SDH deficiency (Rutter et al., 2010; Hoekstra and Bayley, 2013).
Similar to dSdhaf4 mutants, SdhB hypomorphic mutants that retain ~40% of normal SDH activity are sensitive to oxidative stress and display a reduced lifespan (Walker et al., 2006). The SdhB hypomorphs, however, were not reported to undergo mechanically induced paralytic seizures or display neurodegenerative phenotypes (Walker et al., 2006). This lack of neuronal phenotypes in SdhB mutants could be due to the higher overall SDH activity seen in these partial loss-of-function mutants relative to a complete loss of dSdhaf4 activity. It is also possible, however, that the neurodegeneration phenotype is driven by increased ROS production rather than a defect in mitochondrial ATP production. In support of this model, clones generated using a lethal null allele of SdhA in the eye display retinal degeneration similar to that seen in dSdhaf4 mutants, which was shown to be specifically caused by increased ROS production (Mast et al., 2008). This is also consistent with our finding that dSdhaf4 mutants are not depleted of ATP, possibly enabled by metabolic compensation (Celotto et al., 2011). Thus, our study of dSdhaf4, and other work characterizing SdhA, SdhB, and dSdhaf3 mutants in Drosophila, suggests that tissue dysfunction induced by a loss of SDH appears to be largely driven by increased ROS production. This is consistent with our hypothesis that SDH assembly factors may have evolved as a mechanism to protect cells and organisms from ROS production during SDH assembly.
Mutations in all genes encoding previously published SDH subunits and assembly factors have been shown to cause human disease. These include neurodegenerative diseases (Leigh’s Syndrome, Leukodystrophy, and Leukoencephalopathy), which have been linked to mutations in SDHA, SDHB and SDHAF1 (Horvath et al., 2006; Ghezzi et al., 2009; Alston et al., 2012; Ohlenbusch et al., 2012). Mutations in SDHA, SDHB, SDHC, SDHD, and SDHAF2 have also been shown to cause paragangliomas and pheochromocytomas – two classes of neuroendocrine tumors (Astuti et al., 2001; Hao et al., 2009; Fishbein and Nathanson, 2012). Recently, several cases of wild-type gastrointestinal stromal tumors (WT GIST) have been linked to germline loss-of-function mutations in SDHA and SDHB (Janeway et al., 2011; Celestino et al., 2012; Italiano et al., 2012; Oudijk et al., 2013). This wealth of evidence linking SDH dysfunction to disease raises the possibility that damaging mutations in SDHAF4 may cause human disease. In fact, several cases of SDH-deficient diseases, including Leigh’s Syndrome and WT GIST, lack mutations in the genes encoding all known SDH subunits (Horvath et al., 2006; Janeway et al., 2011; Jain-Ghai et al., 2013). Our phenotypic studies raise the important possibility that SDHAF4 is a candidate gene for these and other diseases characterized by SDH deficiency. Our future efforts will focus on determining the incidence of these possible causative SDHAF4 mutations.
On the basis of the data presented in this report we conclude that the SDHAF4 gene family encodes a novel SDH assembly factor. Insights gained in our studies of this protein have allowed us to propose a more complete model of SDH assembly (Figure S7H). Upon import into the matrix by TOM and TIM apo-Sdh1 is flavinated in a reaction that requires Sdh5 (Hao et al., 2009). Free flavo-Sdh1 is then bound by Sdh8 to form a ~150 kDa sub-complex. This sub-complex occupies Sdh1, preventing auto-oxidation and supporting its ability to dimerize with Sdh2. The Sdh1-Sdh2 soluble dimer is then recruited to the IMM via interactions with Sdh3 and Sdh4 to form the SDH holocomplex. This model highlights the importance of subunit-specific chaperones in the process of assembly. Indeed, failure of SDHAF4 to occupy free SdhA prevents normal SDH assembly in three eukaryotic systems and has profound physiological consequences. The identification and characterization of the SDHAF4 protein family thus provides important new insights into the process of SDH assembly, the mechanisms by which SDH subunits are protected for generating ROS, and a new candidate gene to understand human disease associated with a loss of SDH activity.
EXPERIMENTAL PROCEDURES
Drosophila metabolomic analysis and enzyme activity assays
Adult male flies were aged 5–7 days, washed with PBS, and snap frozen in liquid nitrogen. Metabolites were extracted from frozen flies and GC/MS was performed as previously described (Bricker et al., 2012). Succinate dehydrogenase activity was measured in mitochondrial extracts as previously described, by examining the reduction of iodonitrotetrazolium at 500 nm in the presence of succinate (Celotto et al., 2011). Citrate synthase activity was measured in mitochondrial extracts by examining the reduction of 5,5′-5 dithiobis-2-nitrobenzoic acid in the presence of oxaloacetate and acetyl-CoA, as previously described (Tang et al., 2009). Enzyme activity per gram of mitochondrial protein was calculated.
Drosophila paralytic assays
Bang sensitivity was measured as previously described (Ganetzky and Wu, 1982). Flies were collected in groups of 5–8 females per vial and allowed to recover from CO2 anesthesia for at least 2 hours. Flies were then vortexed for 15 seconds and the time it takes to return to a standing position was assayed. At least 3 vials were assayed per condition in each experiment, and all experiments were repeated at least twice. Experiments on female flies are presented, but similar results were observed when assaying males (data not shown).
Electron microscopy
Heads were removed from 8–12 day old female flies, the proboscis was removed, and EM was performed as described in the supplemental experimental procedures.
Mito-roGFP and flow cytometry
The protocol used in this investigation has been adapted from Vevea et al., 2013. For each experiment an equal number of cells transformed with mito-roGFP were plated thinly on synthetic media containing 2% acetate or 2% glycerol. Cells were allowed to incubate for approximately 48 hours before being harvested and analyzed by flow cytometry. Data was collected at the University of Utah Flow Cytometry Core Facility. Cells were acquired on a BD FACSCanto II by collecting emission wavelengths of 530/30 from a violet (405 nm) and blue (488 nm) laser. Post acquisition analysis was accomplished in Flowjo (Treestar, Inc). After single cells were selected using FSC-W and FSC-H parameters, GFP+ cells were gated on for further analysis. The fluorescence intensity was expressed as a ratio of oxidized (405nm excitation) to reduced (488nm excitation) mito-roGFP.
Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
BN-PAGE was performed as described previously (Wittig et al., 2006). Mitochondria were resuspended in lysis buffer (50 mM NaCl, 5 mM 6-aminocaproic acid, 50 mM imidazole, 1 mM AEBSF, and protease inhibitor cocktail). Yeast and Drosophila mitochondria were solubilized with 1% digitonin and mammalian mitochondria with 2% digitonin. Lysates were resolved on a 3%–13% gradient native gel using a PROTEAN®II xi Cell gel running system (Bio-Rad). Western blot performed as described elsewhere using a Trans-blot transfer cell (Bio-Rad) and PVDF membranes.
Statistics
PRISM software was used to graph all quantitative data and perform statistical analyses. P values for pairwise comparisons were determined using a Student’s t test and p values for multiple comparisons were calculated using one-way analysis of variance (ANOVA) with a Bonferroni correction. Metabolomics data is presented in box-plot form, where the box represents the 25th–75th percentile, the line is the median, and the whiskers represent the range of the data. All other quantitative data is shown as the mean±SEM. P values for Kaplan-Meier survival curves in Figure 5 and Supplemental Figure 5 were calculated using a log-rank test.
Supplementary Material
Highlights.
The SDHAF4 gene family encodes an SDH assembly factor.
Drosophila Sdhaf4 mutants display neurodegeneration and attenuated lifespan.
Sdh8 interacts with flavinated Sdh1 and facilitates assembly of SDH holocomplexes.
Sdh8 protects the cell from toxicity associated with unbound Sdh1 in the matrix.
Acknowledgments
We thank Tim Dahlem at the University of Utah Mutation and Detection Core for generating the TALEN gene-targeting constructs, Linda Nikolova at the University of Utah Electron Microscopy core for assistance performing TEM, the Bloomington Stock Center for providing stocks, FlyBase for critical information that made these studies possible, Dennis Winge for providing antibodies and other reagents, James Marvin at the University of Utah Flow Cytometry core for help in establishing the mito-roGFP system, and Liza Pon (Columbia University) for providing the mito-roGFP construct. This research was supported by NIH grant NIH 1R01 GM094232 (JR and CST) and we acknowledge support of funds in conjunction with the grant P30CA042014 awarded to the Huntsman Cancer Institute. DKB was supported by the NIH Genetics Predoctoral Training Grant T32 GM007464. JGV was supported by the Huntsman Cancer Institute Multidisciplinary Cancer Research Training Program (T32 CA093247).
Footnotes
Author Contributions
JGV, DKB, CST, and JR designed the research and prepared the manuscript. JGV performed all experiments in yeast and mammalian cells and BN-PAGE experiments. DKB performed all the experiments in Drosophila. NP and SPG performed protein mass spectroscopy. JEC performed the metabolomics analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alston CL, Davison JE, Meloni F, van der Westhuizen FH, He L, Hornig-Do HT, Peet AC, Gissen P, Goffrini P, Ferrero I, et al. Recessive germline SDHA and SDHB mutations causing leukodystrophy and isolated mitochondrial complex II deficiency. J Med Genet. 2012;49:569–577. doi: 10.1136/jmedgenet-2012-101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- Celestino R, Lima J, Faustino A, Maximo V, Gouveia A, Vinagre J, Soares P, Lopes JM. A novel germline SDHB mutation in a gastrointestinal stromal tumor patient without bona fide features of the Carney-Stratakis dyad. Fam Cancer. 2012;11:189–194. doi: 10.1007/s10689-011-9499-x. [DOI] [PubMed] [Google Scholar]
- Celotto AM, Chiu WK, Van Voorhies W, Palladino MJ. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS One. 2011;6:e25823. doi: 10.1371/journal.pone.0025823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Frank AC, McGrath SW, Fergestad T, Van Voorhies WA, Buttle KF, Mannella CA, Palladino MJ. Mitochondrial encephalomyopathy in Drosophila. J Neurosci. 2006a;26:810–820. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Frank AC, Seigle JL, Palladino MJ. Drosophila model of human inherited triosephosphate isomerase deficiency glycolytic enzymopathy. Genetics. 2006b;174:1237–1246. doi: 10.1534/genetics.106.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Liu Z, Vandemark AP, Palladino MJ. A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav. 2012;2:424–434. doi: 10.1002/brb3.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Kotarsky H, Fellman V, Moraes CT. Mitochondrial disorders caused by mutations in respiratory chain assembly factors. Semin Fetal Neonatal Med. 2011;16:197–204. doi: 10.1016/j.siny.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Bostwick B, Ganetzky B. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics. 2006;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Olson L, Patel KP, Miller R, Palladino MJ, Ganetzky B. Neuropathology in Drosophila mutants with increased seizure susceptibility. Genetics. 2008;178:947–956. doi: 10.1534/genetics.107.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes C, Rao Y. Genome-wide screen for modifiers of Parkinson’s disease genes in Drosophila. Mol Brain. 2011;4:17. doi: 10.1186/1756-6606-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim Biophys Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Fishbein L, Nathanson KL. Pheochromocytoma and paraganglioma: understanding the complexities of the genetic background. Cancer Genet. 2012;205:1–11. doi: 10.1016/j.cancergen.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B, Wu CF. Indirect suppression involving behavioral mutants with altered nerve excitability in Drosophila melanogaster. Genetics. 1982;100:597–614. doi: 10.1093/genetics/100.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- Gnerer JP, Kreber RA, Ganetzky B. wasted away, a Drosophila mutation in triosephosphate isomerase, causes paralysis, neurodegeneration, and early death. Proc Natl Acad Sci U S A. 2006;103:14987–14993. doi: 10.1073/pnas.0606887103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra AS, Bayley JP. The role of complex II in disease. Biochim Biophys Acta. 2013;1827:543–551. doi: 10.1016/j.bbabio.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Horvath R, Abicht A, Holinski-Feder E, Laner A, Gempel K, Prokisch H, Lochmuller H, Klopstock T, Jaksch M. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA) J Neurol Neurosurg Psychiatry. 2006;77:74–76. doi: 10.1136/jnnp.2005.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Chen CL, Sung YS, Singer S, DeMatteo RP, LaQuaglia MP, Besmer P, Socci N, Antonescu CR. SDHA loss of function mutations in a subset of young adult wild-type gastrointestinal stromal tumors. BMC Cancer. 2012;12:408. doi: 10.1186/1471-2407-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain-Ghai S, Cameron JM, Al Maawali A, Blaser S, MacKay N, Robinson B, Raiman J. Complex II deficiency--a case report and review of the literature. Am J Med Genet A. 2013;161A:285–294. doi: 10.1002/ajmg.a.35714. [DOI] [PubMed] [Google Scholar]
- Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, Gaal J, Dahia PL, Liegl B, Ball ER, Raygada M, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci U S A. 2011;108:314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Liu W, Gnanasambandam R, Benjamin J, Kaur G, Getman PB, Siegel AJ, Shortridge RD, Singh S. Mutations in cytochrome c oxidase subunit VIa cause neurodegeneration and motor dysfunction in Drosophila. Genetics. 2007;176:937–946. doi: 10.1534/genetics.107.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Mast JD, Tomalty KM, Vogel H, Clandinin TR. Reactive oxygen species act remotely to cause synapse loss in a Drosophila model of developmental mitochondrial encephalopathy. Development. 2008;135:2669–2679. doi: 10.1242/dev.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Ohlenbusch A, Edvardson S, Skorpen J, Bjornstad A, Saada A, Elpeleg O, Gartner J, Brockmann K. Leukoencephalopathy with accumulated succinate is indicative of SDHAF1 related complex II deficiency. Orphanet J Rare Dis. 2012;7:69. doi: 10.1186/1750-1172-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudijk L, Gaal J, Korpershoek E, van Nederveen FH, Kelly L, Schiavon G, Verweij J, Mathijssen RH, den Bakker MA, Oldenburg RA, et al. SDHA mutations in adult and pediatric wild-type gastrointestinal stromal tumors. Mod Pathol. 2013;26:456–463. doi: 10.1038/modpathol.2012.186. [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea JD, Swayne TC, Zhou C, Liu C, Leung G, Boldough IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age assymetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl PL, Gibson KM, Cortez MA, Wu Y, Carter Snead O, 3rd, Knerr I, Forester K, Pettiford JM, Jakobs C, Theodore WH. Succinic semialdehyde dehydrogenase deficiency: lessons from mice and men. J Inherit Metab Dis. 2009;32:343–352. doi: 10.1007/s10545-009-1034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K, Rothery R, Weiner J, Lemire B. The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur J Biochem. 1994;222:983–990. doi: 10.1111/j.1432-1033.1994.tb18949.x. [DOI] [PubMed] [Google Scholar]
- Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion. 2010;10:393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial diseases. Lancet. 2012;379:1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Le PK, Tse S, Wallace DC, Huang T. Heterozygous mutation of Opa1 in Drosophila shortens lifespan mediated through increased reactive oxygen species production. PLoS One. 2009;4:e4492. doi: 10.1371/journal.pone.0004492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Alessi Wolken DM, Swayne TC, White AB, Pon LA. Ratiometric biosensors that measure mitochondrial redox state and ATP in living yeast cells. JOVE. 2013;77:e50633. doi: 10.3791/50633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DW, Hajek P, Muffat J, Knoepfle D, Cornelison S, Attardi G, Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc Natl Acad Sci U S A. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.