Abstract
Background
Surgical site infections (SSIs) remain a common and widespread problem contributing to a significant morbidity and mortality, attributed partly by the increase in antimicrobial resistance among the etiological agents. This study was done to determine the spectrum of bacterial isolates and their susceptibility patterns causing SSIs at Muhimbili National Hospital, Tanzania.
Methods
This descriptive cross sectional study was conducted between September, 2011 and February, 2012. Pus swabs or pus were cultured on blood agar (Oxoid, UK) and MacConkey agar (Oxoid, UK) and incubated aerobically at 37°C for 18–24 hours. Bacterial identification was done using API 20E and VITEK and antimicrobial susceptibility was determined by Kirby Bauer disc diffusion.
Results
Of the 100 patients, from whom wound swabs were collected, 90 (90%) had positive aerobic bacterial growth. A total of 147 pathogenic bacteria were isolated, including 114 (77.5%) gram negative and 33(22.5%) gram positive organisms. The most prevalent bacterial species were Pseudomonas aeruginosa (16.3%), followed by Staphylococcus aureus (12.2%) and Klebsiella pneumoniae (10.8%). Of the 18 S. aureus , 8 (44%) were methicillin resistant Staphylococcus aureus (MRSA) and three of them (17%) were carrying both MRSA and induced clindamycin resistance (ICR). Extended spectrum beta-lactamase (ESBL) producing Enterobacteriaceae were observed in 23 (79.3%) of the 29 isolates tested. Majority of Escherichia coli 12 (92.3%) and K. pneumoniae 11 (69%) isolates were ESBL producers. About 63% (93/147) were multiple-drug resistance (MDR) isolates, and the overall MDR among Gram positive and Gram negative bacteria was 60.6% (20/33) and 61.4%, (73/114), respectively. The prevalence of MDR for E. coli, A. baumannii and P. stuartii was 100% each. Majority (97%) of the Gram negative bacteria were resistant to more than four categories (classes) of antibiotics.
Conclusion
A high proportion (63%) of the isolates causing SSIs in this tertiary hospital were MDR, of which (90%) were resistant to more than four classes of antibiotics. In the light of these findings, an urgent and significant change in antibiotic prescription policy is required at this National hospital.
Keywords: Multidrug resistance, Surgical site infections, Tanzania
Background
Surgical site infections (SSI) remain a common and widespread problem that contributes to significant morbidity and mortality, prolongs hospital stay and consequently increasing health care cost [1]. Globally, SSI is reported to be the third most common nosocomial infection preceded only by urinary tract infections and pneumonias [1,2]. The outcome of SSI is partly attributed to increase in antimicrobial resistant bacterial pathogens [3], which make the choice of empirical therapy more difficult. Globally, the incidence of SSIs may range from 1.5 – 20% depending on the diversity and complexity of procedures performed, age of patients, pre-operative hospitalization, length of surgery as well as geographical locations [4,5].
SSI remains the major cause of nosocomial infections in Tanzania that imposes substantial burdens on healthcare resources [6]. Recent studies reported increase of SSI ranging from 19.4% to 36.5% [7-10], being higher at Muhimbili National Hospital (MNH) compared to other referral hospitals [10]. In these studies Staphylococcus aureus was reported to be a predominant pathogen followed by Escherichia coli, and Klebsiella species with varying antimicrobial susceptibility patterns. A study at Bugando Medical Centre (BMC) reported resistance to ciprofloxacin of 86%, 80% and 54% for E. coli, Klebsiella pneumoniae and S. aureus respectively. Of the E. coli and K. pneumoniae involved in SSI at Bugando Medical Centre 65% and 80% were ESBL producers, respectively [7]. Rural settings have not been spared of the antibacterial drug resistance problem as shown by a study done (2004) at a remote district hospital in Tanzania which reported more than 95% of S. aureus isolates being resistant to penicillin with 1(0.8%) being resistant to methicillin [8]. However, recent data from the same geographical location has revealed significant increase of methicillin resistant S. aureus (MRSA) (18.8%) causing SSI [7].
Previous studies conducted in Tanzania have shown persistently high levels of antimicrobial resistance in bacteria isolated from urine and bloodstream infections [11,12]. However, there is rather limited data regarding the magnitude and pattern of antimicrobial resistance among pathogens causing SSI. We undertook this study at MNH, which is the national referral hospital, and where antimicrobial use is probably the highest to determine antimicrobial resistance pattern, including multidrug resistance, among bacteria causing SSI.
Methods
Study design and study setting
This was a descriptive cross sectional study performed between September, 2011 and February, 2012 at the Muhimbili National hospital (MNH). Located in Dar es Salaam, MNH is a 1400-bed facility and the largest hospital in Tanzania, which attends 1000 to 1200 outpatients a week and serves approximately four million people living in the Dar es Salaam region. MNH is the main referral and teaching hospital.
Patient recruitment
All patients with clinical evidence of SSI as defined by the Centers for disease control and prevention (CDC) [13] were eligible for the study. Recruitment was done after obtaining an informed consent. A total of 100 patients were enrolled consecutively from general surgery, obstetrics and gynecology, and orthopedic wards.
Data collection and laboratory procedures
Structured standard questionnaires were used to obtain social-demographic and clinical data from patients and the patient’s case notes. Thereafter two pus swabs or pus were collected aseptically from the depth of the wound and transported immediately to the laboratory in Amies transport media (Oxoid, UK) for processing.
Gram’s stain was performed from the first swab while the second pus swab was inoculated into blood agar (Oxoid, UK) and MacConkey agar (Oxoid, UK); and incubated aerobically at 37°C for 18–24 hours. Identification was based on microscopic characteristics, colonial characteristics, and Biochemical tests as described by Murray et al. [14], including VITEX (BioMerieux, France) and API 20E (BioMerieux, France). Gram negative organisms were identified by oxidase, Triple sugar Iron (TSI), sulphur indole and motility (SIM), urease, citrate test, VP and Methyl red test. Whereas Gram positive organisms were catalase reaction, coagulase test, DNase test and bile esculin test.
Antimicrobial susceptibility testing
This was performed by Kirby Bauer disc diffusion method as described by Clinical and Laboratory Standards Institute [CLSI] [15]. Briefly test and control organisms (listed below) were suspended in normal saline to McFarland 0.5 standard and inoculated on Mueller-Hinton agar [Oxoid, UK]. Appropriate discs were placed onto the media then incubated at 37°C for 24 hrs. For Gram positive organisms discs included penicillin (10 units), ampicillin (10 μg), amoxicillin/clavulanate (20/10 μg), ceftriaxone (30 μg), gentamicin (10 μg), erythromycin (15 μg), tetracycline (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), trimethoprim/sulphamethaxazole (1.25/23.75 μg) and chloramphenicol (30 μg). The Gram negative organisms were tested against ampicillin (10 μg), amoxicillin/clavulanate (20/10 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cefotaxime (30 μg) gentamicin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), trimethoprim/sulphamethaxazole (1.25/23.75 μg), chloramphenicol (30 μg) and meropenem (10 μg). For P. aeruginosa discs that were used included ceftriaxone (30 μg), ceftazidime (30 μg), gentamicin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg) and meropenem (10 μg).
MRSA was determined by disc diffusion test using cefoxitin (30 μg) disc on Mueller –Hinton Agar, incubated and maintained at 33-35°C for 24 hours, zone of inhibition ≤ 21 mm was considered a positive result for MRSA strain [15]. Induced clindamycin resistance among S. aureus was detected by disc diffusion method on Mueller-Hinton Agar, where 15 μg of erythromycin disc and 2 μg of clindamycin disc were spaced 15 mm apart, incubated at 35 ± 2°C for 16–18 hours. Flattening of the zone of inhibition adjacent to erythromycin (D-zone) was interpreted as ICR [15]. ESBLs production was screened by disc diffusion on Mueller-Hinton Agar with ceftazidime (30 μg) or cefotaxime (30 μg) and confirmed by double disc approximation method [15,16]. Quality of media, antibiotic disc as well as performance of a person carrying the tests was controlled by reference strains: Escherichia coli ATCC 25922, K. pneumoniae ATCC 700603 for ESBL, S.aureus ATCC 25923, and S.aureus ATCC 29213 for MRSA.
Multidrug resistant (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [17]. Antimicrobial categories tested were Penicillin class (ampicllin, penicillin); Cephalosporin class (ceftazidime, cefotaxime, ceftriaxone); Aminoglycosides class (gentamicin); Tetracycline class (Tetracycline); fluorquinolones class (ciprofloxacin); folate pathway inhibitors class (co-trimoxazole); phenicols class (chloramphenicol); macrolides class (erythromycin) and lincosamides class (clindamycin).
Data analysis
Data were entered and analyzed using Epidata Entry version 3.1 and STATA version 11.2, and expressed in a descriptive analysis. Categorical variables were compared using Chi-square test or fisher exact test, p values of < 0.05 was considered significant.
Ethical approval
Ethical approval was obtained from the Senate Research and Publications Committee of Muhimbili University of Health and Allied Sciences, Dar es Salaam and a written informed consent was obtained from each patient/caretaker.
Results
Demographical – clinical characteristics
We enrolled a total of 100 patients with clinical SSI, aged between 18 and 80 years. Most of the patients (53%) were males and nearly half (46%) were from orthopedic and trauma wards. Regarding surgical procedures, 27% of the patients had surgical debridement and external fixation, 25% had laparotomy while 15% had caesarian section (see Additional file 1). Almost all patients (95%) had documentation of antimicrobial exposure within a month, with ceftriaxone, cloxacillin and amoxicillin-clavulanic acid the most frequently used, being prescribed to 80 patients each, followed by gentamicin (19), ciprofloxacin (17) and metronidazole (15). Notably, three-fourth had a history of previous admission within 6 months.
Etiology of SSI
Of the 100 wound swabs collected, 90% had bacterial growth. More than half (52.2%, 47/90) had pure bacterial growth (mono isolate). Gram negative organisms were more prevalent than gram positive bacteria accounting for 77.5% (114/147) of all isolates. The most predominant Gram negative organism was P. aeruginosa comprising 16.3% (24/147) of all bacterial isolates. K. pneumoniae 10.8% (16/147) and Proteus mirabilis 10.8% (16/147) were the next two common Gram negative organisms [Figure 1]. Most (92.3%) of the 13 Escherichia coli and 11/16 (69%) of K. pneumoniae isolates were ESBLs producing strains. Of the Gram positive isolates, S.aureus was the leading cause of SSIs accounting 12.2% (18/147) of all isolates, followed by coagulase negative Staphylococci (6.8%) and Enterococcus faecalis (3.4%). Of the 18 S. aureus isolates, 44.4% (8/18) were MRSA strains and three of them had induced clindamycin resistance (ICR).
Figure 1.
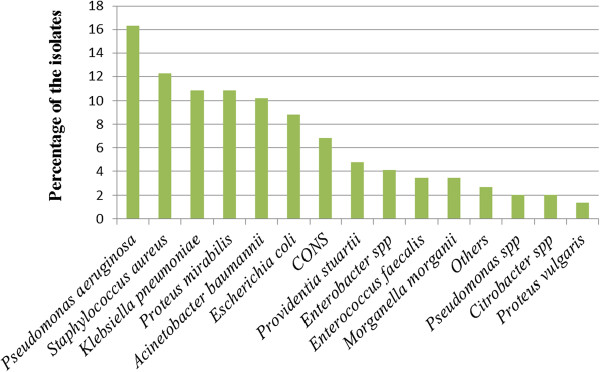
Isolation frequency of pathogenic bacterial isolates from post-operative wound infections (N = 147).
Antimicrobial resistance pattern
Most of S. aureus isolates were highly resistant (83%) to penicillins group of antibiotic; some had low to moderate resistance (17% to 33%) to erythromycin, gentamicin and ciprofloxacin. All Enterobacteriaceae displayed high magnitude of resistance (69% to 100%) to multiple antibiotics tested but all were sensitive to meropenem; and 20-56% were resistant to ciprofloxacin. Surprisingly, 92% of 24 P. aeruginosa isolates were sensitive to both gentamicin and ciprofloxacin. Majority of A. baumannii isolates were highly resistant (73% to 100%) to most antimicrobial agents; however 60% of them appeared to be moderately sensitive to imipenem [Table 1]. The magnitude of resistance to gentamicin, ciprofloxacin, co-trimoxazole and tetracycline was extremely higher in organisms carrying MRSA (44.4%) and ESBL (79.3%) combined than in non-carrying isolates [p < 0.001]. Of all isolates 63% (93/147) were MDR strains, the overall prevalence of MDR strains was 60.6% among Gram positive and 61.4% among the Gram negatives. MDR status was displayed in all Escherichiacoli, A. baumannii and P. stuartii, and 87.5% of Klebsiella spp. Surprisingly, only one isolate of the 24 P. aeruginosa displayed MDR [Table 2].
Table 1.
Antimicrobial resistance pattern of bacteria isolated from post-operative SSIs
| Bacteria isolates (N) |
Antimicrobial agents resisted (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | AP | AMC | CRO | CAZ | CTX | C | TS | TE | E | CD | CN | CP | MEM | |
|
E. coli
(n = 13) |
NA |
100 |
92 |
92 |
92 |
92 |
42 |
85 |
85 |
NA |
NA |
92 |
58 |
0 |
|
K. pneumonia
(n = 16) |
NA |
100 |
94 |
81 |
88 |
88 |
54 |
94 |
56 |
NA |
NA |
67 |
56 |
0 |
|
P. mirabilis (
n = 16) |
NA |
73 |
87 |
69 |
67 |
100 |
69 |
69 |
94 |
NA |
NA |
63 |
56 |
0 |
|
A. baumannii
(n = 15) |
NA |
100 |
100 |
100 |
86 |
100 |
100 |
77 |
73 |
NA |
NA |
86 |
47 |
40 |
|
P. aeruginosa
(n = 24) |
NA |
NA |
NA |
88 |
21 |
NA |
NA |
NA |
NA |
NA |
NA |
8 |
8 |
0 |
|
Other G – (n = 30) |
NA |
90 |
83 |
83 |
57 |
76 |
73 |
77 |
65 |
NA |
NA |
38 |
47 |
0 |
|
S. aureus
(n = 18) |
83 |
92 |
73 |
47 |
NA |
NA |
20 |
35 |
39 |
17 |
6 |
33 |
29 |
NA |
| Other G + (n = 15) | 67 | 40 | 67 | 50 | NA | NA | 50 | 42 | 47 | 79 | 25 | 62 | 65 | NA |
G-: Gram negative bacteria [P.stuartii, M.morgannii, P.vulgaris, Enterobacter spp, Citrobacter spp), G+: Gram positive cocci [CoNS, E.faecalis]. P = Penicillin, AP = Ampicillin, AMC = Amoxycillin/clavulanic acid, CRO = Ceftriaxone, CAZ = Ceftazidime, CTX = Cefotaxime, C = Chloramphenicol, TS = Co-trimoxazole, TE = Tetracycline, E = Erythromycin, CD = Clindamycin, CN = Gentamicin, CIP = Ciprofloxacin, MEM = Meropenem.
Table 2.
Multi-drug resistant bacteria isolated from surgical site infections
| Bacteria |
Classes of antimicrobial resisted to N(%) |
Average N(%) | |||||
|---|---|---|---|---|---|---|---|
| R3 | R4 | R5 | R6 | R7 | R8 | ||
|
K. pneumoniae
|
- |
1(6.2) |
2(12.5) |
3(18.8) |
8(50) |
- |
14(87.5) |
|
E. coli
|
- |
1(7.1) |
1(7.1) |
2(15.4) |
9(62.9) |
- |
13(100) |
|
P. mirabilis
|
- |
- |
1(6.3) |
2(12.5) |
9(56.2) |
- |
12(75) |
|
A. baumannii
|
1(6.6) |
1(6.6) |
- |
2(13.3) |
11(73.3) |
- |
15(100) |
|
P. aeruginosa
|
1(4) |
- |
- |
- |
- |
- |
1(4) |
|
P. stuartii
|
- |
2(28.6) |
- |
4(57.1) |
1(14.3) |
- |
7(100) |
|
Other GNR |
- |
- |
2(12.5) |
3(18.7) |
6(37.5) |
- |
11(36.6) |
|
S. aureus
|
1(5.5) |
- |
2(11.1) |
3(16.6) |
- |
2(11.1) |
8(44.4) |
|
CONS |
2(20) |
4(40) |
- |
- |
- |
2(20) |
8(80) |
|
E. faecalis
|
1(20) |
- |
2(40) |
1(20) |
- |
- |
4(80) |
| Total | 6(4) | 9(6.1) | 9(6.1) | 20(13.6) | 16(10.8) | 33(22.4) | 93(63) |
Key: R3- R7 = resistant to 3, 4, 5, 6 or 7 classes of antimicrobials tested. Other GNR = (M.morgannii, P.vulgaris, Enterobacter spp, Citrobacter spp).
Discussion
This report on antimicrobial resistance from the national hospital in Tanzania demonstrates the predominance of gram negative bacterial isolates in SSIs (77.5%). Predominately Pseudomonas aeruginosa being the commonest followed by K. pneumoniae (10.8%) and Proteus mirabilis (10.8%). On the other hand S.aureus which was the commonest Gram positive organism accounted for (12.2%). This observation is in contrast to finding in previous studies from the same setting which reported Gram positive predominantly S. aureus as the most common SSI bacterial pathogen [7,18] (Figure 2).
Figure 2.
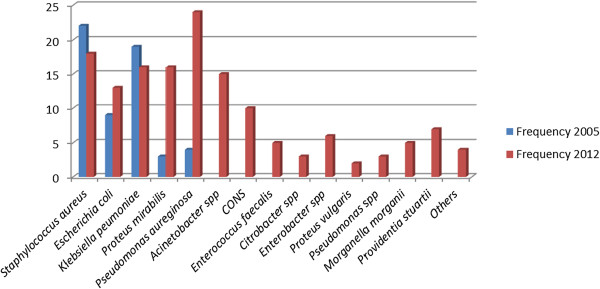
Trend of pathogenic bacterial isolated from SSI in 2005 and 2012.
With regard to antimicrobial resistance we found, among Gram negative bacteria, very high levels of resistance (73 -100%) to commonly prescribed antibiotics like ampicillin, amoxicillin/clavulanic acid, co-trimoxazole and tetracycline, which is in keeping results (32 – 100%) reported in the same hospital and in Kenya [19,20]. The level of resistance to ciprofloxacin was 20 – 56% in Enterobacteriaceae and Acinetobacter baumannii, showing an increase in resistance compared to (6.7 – 30%) previously reported at this hospital in 2004/2010, [11,19]. Resistance to ciprofloxacin is an early warning sign since fluoroquinolones are effective agents for treatment of gram negative bacterial infections [21].
With regard to third generation cephalosporins we found very high levels of resistance (67 -100%) among Gram negative bacteria, with most of E coli and K. pneumoniae being ESBL producing strains. These finding is in keeping with recent studies from Northern-Western Tanzania and Uganda that shows (61 – 92%) [7,22]. We noted that about 80% of patients received ceftriaxone as prophylaxis, thus favoring the emergence of resistant bacteria.
Significantly, we note an increasing trend of resistance to cotrimoxazole, gentamicin, ceftazidime and ciprofloxacin over a period from 2004 to 2012 (Figures 3 and 4) [11,19]. In addition to the high rate of resistance to individual antibiotics, we also found significant number of MDR isolates. The overall proportion of MDR among Gram negative bacteria isolates was high (64%), this finding is in agreement with recent study from Uganda [22]. We noted that, all Escherichia coli, Acinetobacter baumannii and Providentia stuartii were MDR strains.
Figure 3.
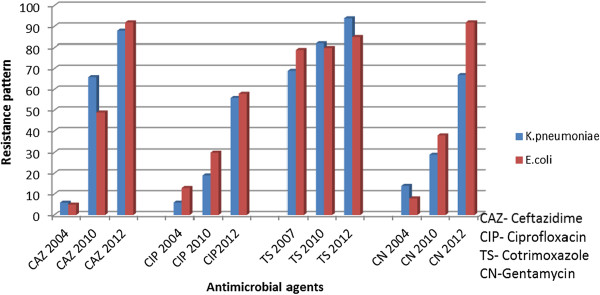
Increasing trend of resistance among Gram negative bacteria isolated from clinical isolates 2004, 2010 and 2012.
Figure 4.
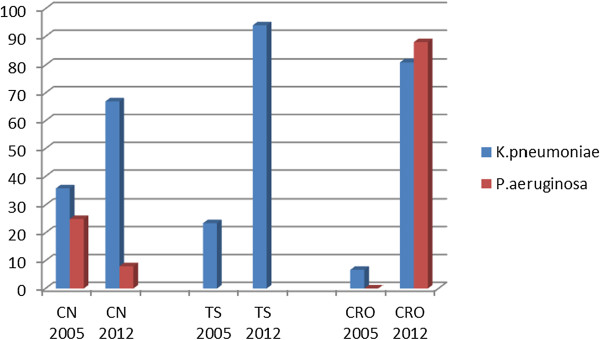
Increasing pattern of resistance to isolates from SSI at Muhimbili National Hospital 2005 and 2012.
The very high levels of antibiotics resistance seen in this study could be attributed, at least in part, to inappropriate use of antimicrobial agents affects both the cost and efficacy, thus favoring the emergence of resistant bacteria.
One of the limitations of our study is cross-sectional design of the study. Also there was no patient follow up after discharge up to 30 days which is required according to CDC definition of SSI and hence some cases of SSI after discharge from hospital may be missed.
Conclusion
The very high levels of antimicrobial resistance among bacteria causing SSIs, including resistant to more than four categories (classes), complicates the management of these infections at MNH, and could be associated with higher SSI rate and prolongation of hospital stay and increased number of drugs usage. There is a very urgent need of revising the perioperative antimicrobial prophylaxis at this national referral Hospital to provide clinicians with standardized approach to the rational, safe and effective use of antibiotics management of SSI.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JM participated in conception, design, and collection of data, analysis, interpretation and drafting of the manuscript. MIM helped to draft the manuscript. MM and SEM participated in critically revising the manuscript. SM participated in analysis and interpretation of data. EFL participated in conception, design of the study and critically revising the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Demographic and clinical characteristics of patients with post-operative wound infections.
Contributor Information
Joel Manyahi, Email: manyahijoel@yahoo.com.
Mecky I Matee, Email: mateemecky@yahoo.com.
Mtebe Majigo, Email: mmajigo@gmail.com.
Sabrina Moyo, Email: sabmoyo26@yahoo.com.
Stephen E Mshana, Email: mshana72@yahoo.com.
Eligius F Lyamuya, Email: eligius_lyamuya@yahoo.com.
Acknowledgements
We wish to thank Prof. S.C Yongolo for his valuable clinical assistance throughout the different stages of this work. We also thank Dr Mabula Kasubi and Mary Martin for their technical assistance, Dr Francis Mhimbira for statistical inputs, and all members of the department of Microbiology/Immunology for their support. This work was supported by Belgium Technical Cooperation (Tanzania).
References
- Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR. Guideline for prevention of surgical site infection, 1999. Centers for disease control and prevention (CDC) hospital infection control practices advisory committee. Am J Infect Control. 1999;27:97–132. [PubMed] [Google Scholar]
- Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt JA, Lipsky BA, Tabak YP, Derby KG, Kim M, Gupta V. Surgical site infections: causative pathogens and associated outcomes. Am J Infect Control. 2010;38:112–120. doi: 10.1016/j.ajic.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Leaper DJ, van Goor H, Reilly J, Petrosillo N, Geiss HK, Torres AJ, Berger A. Surgical site infection – a European perspective of incidence and economic burden. Int Wound J. 2004;1:247–273. doi: 10.1111/j.1742-4801.2004.00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TA, Sohn AH, Nguyen PT, Vo TC, Vo VN, Tran Nguyen TH, Ewald B, Dibley M. Microbiology of surgical site infections and associated antimicrobial use among Vietnamese orthopedic and neurosurgical patients. Infect Control Hosp Epidemiol. 2006;27:855–862. doi: 10.1086/506400. [DOI] [PubMed] [Google Scholar]
- Gosling R, Mbatia R, Savage A, Mulligan JA, Reyburn H. Prevalence of hospital-acquired infections in a tertiary referral hospital in northern Tanzania. Ann Trop Med Parasitol. 2003;97:69–73. doi: 10.1179/000349803125002724. [DOI] [PubMed] [Google Scholar]
- Mawalla B, Mshana SE, Chalya PL, Imirzalioglu C, Mahalu W. Predictors of surgical site infections among patients undergoing major surgery at Bugando Medical Centre in Northwestern Tanzania. BMC Surg. 2011;11:21. doi: 10.1186/1471-2482-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr J, Hatz C, Soka I, Kibatala P, Urassa H, Battegay M, Jeffrey Z, Smith T, Mshinda H, Frei R, Widmer AF. Antimicrobial prophylaxis to prevent surgical site infections in a rural sub-Saharan hospital. Clin Microbiol Infect. 2006;12:1224–1227. doi: 10.1111/j.1469-0691.2006.01551.x. [DOI] [PubMed] [Google Scholar]
- Eriksen HM, Chugulu S, Kondo S, Lingaas E. Surgical-site infections at Kilimanjaro Christian Medical Center. J Hosp Infect. 2003;55:14–20. doi: 10.1016/s0195-6701(03)00225-1. [DOI] [PubMed] [Google Scholar]
- Akoko LA, Mwanga HA, Fredrick F, Mbembati NM. Risk factors of surgical site infection at Muhimbili national hospital, Dar es Salaam, Tanzania. East Cent Afr J surg. 2012;17(3):12–17. [Google Scholar]
- Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res Notes. 2010;3:348. doi: 10.1186/1756-0500-3-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana SE, Kamugisha E, Mirambo M, Chakraborty T, Lyamuya EF. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC Res Notes. 2009;2:49. doi: 10.1186/1756-0500-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan TC, Gaynes RP, Martone WJ, Jarvis WR, Emori TG. CDC definitions of nosocomial surgical site infections, 1992: a modification of CDC definitions of surgical wound infections. Infect Control Hosp Epidemiol. 1992;13:606–608. [PubMed] [Google Scholar]
- Murray PR, Baron E, Pfaller MA, Tenovar FC, Yolken RH. Manual of Clinical Microbiology. 7. Washington, DC: American Society for Microbiology; 1999. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement CLSI Document M100–S20. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. [Google Scholar]
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Ussiri EV, Mkony C, Azizi MR. Surgical wound infection in clean-contaminated and contaminated laparotomy wounds at Muhimbili national hospital. East Cent Afr J Surg. 2005;10:19–23. [Google Scholar]
- Blomberg B, Mwakagile DS, Urassa WK, Maselle SY, Mashurano M, Digranes A, Harthug S, Langeland N. Surveillance of antimicrobial resistance at a tertiary hospital in Tanzania. BMC Public Health. 2004;4:45. doi: 10.1186/1471-2458-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andhoga J, Macharia AG, Maikuma IR, Wanyonyi ZS, Ayumba BR, Kakai R. Aerobic pathogenic bacteria in post-operative wounds at Moi Teaching and Referral Hospital. East Afr Med J. 2002;79:640–644. doi: 10.4314/eamj.v79i12.8671. [DOI] [PubMed] [Google Scholar]
- Masterton RG. Ciprofloxacin resistance –‘early-warning’ signs from the MYSTIC surveillance programme? J Antimicrob Chemother. 2002;49(1):218–220. doi: 10.1093/jac/49.1.218. [DOI] [PubMed] [Google Scholar]
- Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, Kapesa A, Bwanga F. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC Res Notes. 2013;6:298. doi: 10.1186/1756-0500-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic and clinical characteristics of patients with post-operative wound infections.


