Abstract
Elmo1 and Elmo2 are highly homologous cytoplasmic adapter proteins that interact with Dock family guanine nucleotide exchange factors to promote activation of the small GTPase Rac. In T lymphocytes, Dock2 is essential for CCR7- and CXCR4-dependent Rac activation and chemotaxis, but the role of Elmo proteins in regulating Dock2 function in primary T cells is not known. Here we show that endogenous Elmo1 but not Elmo2 interacts constitutively with Dock2 in mouse and human primary T cells. CD4+ T cells from Elmo1−/− mice were profoundly impaired in polarization, Rac activation and chemotaxis in response to CCR7 and CXCR4 stimulation. Transfection of full-length Elmo1, but not Elmo2 or a Dock2-binding mutant of Elmo1, rescued defective migration of Elmo1−/− T cells. Interestingly, Dock2 protein levels were reduced by four-fold in Elmo1−/− lymphocytes despite normal levels of Dock2 mRNA. Dock2 polyubiquitination was increased in Elmo1−/− T cells, and treatment with proteasome inhibitors partially restored Dock2 levels in Elmo1−/− T cells. Finally, we show that Dock2 is directly ubiquitinated in CD4+ T cells and that Elmo1 expression in heterologous cells inhibits ubiquitination of Dock2. Taken together, these findings reveal a previously unknown, non-redundant role for Elmo1 in controlling Dock2 levels and Dock2-dependent T cell migration in primary lymphocytes. Inhibition of Dock2 has therapeutic potential as a means to control recruitment of pathogenic lymphocytes in diseased tissues. This work provides valuable insights into the molecular regulation of Dock2 by Elmo1 that can be used to design improved inhibitors that target the Elmo-Dock-Rac signaling complex.
Keywords: CD4+ T cell, chemokine signaling, GTPase, ubiquitination
INTRODUCTION
Chemokine signaling is an integral component of lymphocyte trafficking, activation and survival. Rac is a member of the Rho family of GTPases that are central drivers of actin cytoskeleton dynamics downstream of most chemokine receptors (1-4). Rac cycles between and inactive (GDP-bound) and active (GTP-bound) states owing largely to the action of guanine nucleotide exchange factors (GEFs). Dock2 is a ~200kDa Rac-GEF restricted to hematopoietic cells in mice and humans (5). Through the use of Dock2−/− mice, it is now well established that Dock2 is essential for Rac activation and chemotaxis in lymphocytes downstream of multiple chemokine receptors, including CCR7 and CXCR4 (1, 2, 6). Dock2-deficient lymphocytes show greatly reduced entry, egress and interstitial motility in lymphoid and non-lymphoid tissues. In murine models of cardiac allograft rejection and diabetes, deletion of Dock2 in lymphocytes was found to be protective, pointing to a role for Dock2 in migration of pathogenic T cells (7, 8). Over the past decade, Dock2 has also been found to regulate a range of Rac-dependent functions in neutrophils, dendritic cells and NKT cells (9-12). However the molecular regulation of Dock2 is poorly understood, particularly in primary cells and in human lymphocytes.
Elmo1 (75kDa) is a cytoplasmic adapter protein that physically associates with members of the Dock-A family of Rac-GEFs, of which Dock1 and Dock2 are the best characterized (5, 13, 14). Extensive structure-function analyses by a number of groups have shown that Elmo binding enhances Dock1 signaling by increasing its Rac-GEF activity, membrane localization and protein stability (13, 15-21). Studies in invertebrate models and mammalian cell lines have revealed an evolutionarily conserved role for Elmo1 in regulating Dock-Rac signaling in numerous cellular functions, including morphology, motility and phagocytosis (13, 18, 22-25). Elmo1 has also been shown to interact with Dock2 to promote Rac activation and migration in rodent cell lines (22, 26). More recently, studies in Elmo1−/− mice revealed a critical role for Elmo1 in apoptotic cell clearance during spermatogenesis and hippocampal neurogenesis (27, 28). However, as most of our insights into Elmo function stem from studies of Elmo1 and Dock1 in non-hematopoietic cells, the mechanisms and outcomes of Elmo1-dependent regulation of Dock proteins in leukocytes remain largely unknown.
Elmo1 and Elmo2 are 87% similar at the amino acid (a.a.) level, are widely expressed and, based on Dock1 studies, have largely been considered to be functionally redundant (13). Both proteins contain pleckstrin homology (PH) and proline-rich/PxxP domains located in the C-terminal 100a.a. (15, 29, 30). These C-terminal regions mediate multiple associations with the N-termini of Dock1 and Dock2 as revealed through crystallographic and biochemical analyses (30, 31). Dock1 and Dock2 contain an N-terminal Src homology 3 (SH3) domain that mediates interaction with the C-terminal polyproline regions of Elmo1 and Elmo2. Interestingly, this PxxP-SH3 association is essential for Elmo1 interaction with Dock2 but not Dock1 (31). The PxxP motif is conserved between mouse and human Elmo1 and Elmo2 (PKEP, Elmo1714-717), but whether Elmo2 can interact with or regulate Dock2 has not been reported.
In this study we used a number of approaches to address the function of Elmo1 and Elmo2 in regulating Dock2 in primary mammalian lymphocytes. Using Elmo1−/− mice and primary human T cells, we demonstrate a previously unknown, non-redundant role for Elmo1 in regulation of endogenous Dock2 that may provide insight toward the development of Dock2-targeting therapeutics.
MATERIALS AND METHODS
Mice
Animal experiments were approved by the University of Rochester Animal Care and Use Committee. Elmo1-deficient mice have been described elsewhere (27). All mice were 6-12 weeks of age and on a C57BL/6J background of at least 10 generations.
Reagents
Commercial reagents were purchased as follows: All chemicals unless noted otherwise were purchased from Sigma-Aldrich; Chemokines (PeproTech); ICAM-1 (R&D); Protein A/G agarose beads (Santa Cruz); 24 well transwell chambers (Corning); Taqman qRT-PCR probes (LifeTech); Cell culture media (Cellgro); CFSE and TAMRA (Invitrogen). BSA was purchased from Sigma-Aldrich (A4503).
Antibodies
Flow cytometry antibodies used in this study: CD3ε (17A2), CD4 (GK1.5), CD8α (53-6.7), CD11b (M1/70), CD16/32 (93), CD19 (6D5), CD45 (Ly-5), B220 (RA3-6B2), CXCR4 (2B11), CCR7 (4B12), F4/80 (BM8). Commercial antibodies for immunoprecipitation and immunoblotting: β-actin (Sigma-Aldrich); Dock2 (Millipore); Dock1 (Santa Cruz); GAPDH, Rac, pERK1/2 (9109), ERK1/2 (9102), phospho-Ser473 AKT (D9E), pan-AKT (40D4), HA, K48 polyubiquitin (D9D5), DYKDDDDK ‘flag’ tag (Cell Signaling Technology). Elmo1 and Elmo2 antibodies were provided by K.S. Ravichandran (15, 27), and were further tested in this study to confirm specificity (see Supplemental Figure 3).
Cell isolation
Mice were euthanized by CO2 asphyxiation. Spleen and lymph nodes were disaggregated through a 70μm mesh filter and RBC lysed using ACK lysis buffer (Sigma-Aldrich). Human leukocytes were obtained from healthy de-identified donors (New York Blood Center) and PBMCs isolated by Accu-Prep (Accurate Chemical). PBMC were first depleted of CD14+ cells by positive magnetic selection using MACS separation CD14 MicroBeads (Miltenyi Biotech). Mouse and human CD4 T cells were isolated by negative magnetic selection using the CD4+ T cell Isolation Kit II (Miltenyi Biotec). For isolation of murine macrophages, peritoneal cavities were lavaged with 5mL cation-free PBS twice and immediately stained and FACS sorted.
Cell culture
Primary murine CD4 T cells were cultured at 37°C/5%CO2 in RPMI1640, 20% FBS, 40U/mL rIL-2, 10mM HEPES, 1% pen-strep/L-glutamine. Jurkat T cells (clone E6.1) were grown at 37°C/5%CO2 in RPMI1640, 10% FBS, 10mM HEPES, 1% pen-strep/L-glutamine.
Cell staining for flow cytometry
For flow cytometry, cells were resuspended in cold FACS buffer (cation-free PBS, 0.5% BSA, 0.05% NaN3), incubated with 1:100 Fc receptor blocking antibodies on ice for 10min before addition of fluorescently labeled antibodies for 25min on ice. Staining with CXCR4 and CCR7 antibodies was carried out at 25°C. Cells were washed once and resuspended in FACS buffer before analysis or sorting.
Immunoprecipitation and immunoblotting
Cell lysates were prepared using lysis buffer (20mM Tris-HCl pH7.5, 1% Triton X-100, 150mM NaCl, 1mM MgCl2, 1x final protease inhibitor cocktail III (Calbiochem)) or Laemmli buffer. Protein quantification of lysates was done using BCA reagent (Pierce). Lysates were boiled 10min and protein separation carried out on 4-15% SDS-PAGE mini gel (BioRad). After transfer to PVDF at 100V for 1hr, membranes were blocked for 1hr with 5% non-fat dry milk/TBST before overnight incubation with indicated antibodies at 4°C. SuperSignal Pico or Dura ECL reagents were used per manufacturer's instructions (Pierce). For IP, cell pellets were resuspended in lysis buffer and rotated at 4°C for 10min prior to centrifugation at 12,000g, 5min, 4°C. Cleared lysates were incubated with anti-Dock2 (1:250), normal rabbit IgG (1:250) or anti-Elmo1 (1:100), in total volume of 500μL and rotated 18hr at 4°C. 25μL protein A/G beads were then added to each sample and rotated for 2hr at 4°C. Beads were washed four times in lysis buffer and boiled in Laemmli buffer. For co-IP analysis of endogenous Elmo1 and Elmo2, IPs were split equally after boiling, loaded in duplicate lanes and blotted with anti-Elmo1 or anti-Elmo2 antibodies separately. For co-IP of Elmo-Flag with Dock2 in 293T cells, all of each anti-Dock2 IP was loaded in a single lane and blotted for Flag using HRP-conjugated anti-DYKDDDDK. For detection of K48-ubiquitinated Dock2 under denaturing conditions, anti-Dock2 IP's from WT CD4+ T cells were boiled 5 minutes in 50μL of denaturing buffer (50mM Tris pH 7.5, 70mM β-mercaptoethanol, 1% SDS) followed by addition of 350μL IP lysis buffer and a second round of IP with anti-Dock2 for 2 hours at 4°C before IB analysis.
Quantitative RT-PCR
RNA was isolated using DNase I-treated RNeasy columns (Qiagen), and cDNA synthesized from 10-100ng RNA using iScript (BioRad). qRT-PCR was performed on a 7300 Real Time Thermocycler (Life Technologies) using SensiFast Probe Hi-ROX polymerase (Bioline) and the following gene-specific TaqMan probes from Life Technologies: Elmo1, (Mm00519109_m1); Elmo2 (Mm00475454_m1); Dock2 (Mm00473720_m1); Actb (Mm00607939_s1). Values were obtained using a relative standard method. In brief, a two-fold dilution standard curve of total cDNA was used to determine expression levels of each gene for each specimen. Expression levels were then normalized to Actb levels. For comparisons across genes, a calibrator sample was used to account for varying relative levels of each gene in the standard curve sample.
Time-lapse video microscopy
T cell motility experiments were carried out on Delta T dishes (Bioptechs) coated first with Protein A (10ug/mL, Invitrogen) then ICAM-1 Fc (10ug/mL, R&D) and 4ug/ml of CCL21 or CXCL12. Splenic CD4+ T cells were labeled with either 0.5μM CFSE or 1μM TAMRA-SE (Invitrogen) for 1hr at 37°C/5%CO2. Cells were washed and resuspended at 5×105/mL in Leibovitz's L-15 media supplemented with glucose (2mg/mL) and cultured at 37°C for 20min prior to being added to the microscopy dish. Dish was secured on a heated stage and imaging done with an epifluorescence Nikon Eclipse Ti microscope. Images were acquired every 15s for 15 or 30min using a 20X objective.
Migration assays
Transwell chemotaxis assays were performed using 24 well plates with 5μm pore size inserts (Corning). Cells were equilibrated at 37°C/5%CO2 in migration medium (RPMI1640, 1% BSA, 10mM HEPES, 1% pen-strep/L-glutamine) at 1×106 cells/mL for 30min before use. A total of 500μL of chemoattractant in migration medium was applied to the lower chamber and 100μL cells applied to the upper chamber. After 1hr at 37°C/5%CO2 inserts were discarded and 50μL Accucount beads (5.1μm diameter, Spherotech) were added to each lower chamber and input samples (100μL cells plus 400μL medium) for quantitation by flow cytometry. For post-migration antibody staining, 250μl cells from the lower chamber were removed prior to adding beads and stained with indicated antibodies. Percent migration was determined by: 100 × [(cell events in lower chamber/bead events in lower chamber)/(input cell events/input bead events)]. Staining and quantitation was carried with 2-3 replicates per condition.
Determination of Rac-GTP, phospho-AKT and phospho-ERK levels
Pulldown of active Rac was determined using GST-PAK beads (Cytoskeleton) according to manufacturer's instructions, with the following modifications. CD4+ cells were incubated in migration medium at 1×106/mL for 30min at 37°C/5%CO2. Cells were pelleted and resuspended at 2-3×106 cells per 200μl stimulation medium (RPMI1640, 10mM HEPES, 1% Pen-Strep/L-glutamine). Cells were incubated for 10min in 37°C water bath and stimulated by addition of 200μL of 500ng/mL chemokine in stimulation medium for 30sec. After stimulation, cells were immediately place on ice and 400μL ice-cold TBST added to each sample. Cells were then pelleted at 4,000g, 1min, 4°C and lysed in 165μL recommended lysis buffer and lysates cleared at 10,000g, 1min, 4°C. Cleared lysates were transferred to fresh tubes containing 15-30μg of GST-PAK beads and samples rotated for 1hr at 4°C. Beads were washed 2-3 times with recommended wash solution and pellets boiled 10min in Laemmli buffer, separated on 12% SDS-PAGE and analyzed by immunoblotting. For phospho protein analysis, T cells were stimulated as above except and immediately lysed in 1x Laemmli buffer before SDS-PAGE and immunoblotting.
Transfection
Jurkat T cells were transfected as previously described using the ECM 830 Square Wave Electroporation system (BTX) (32). The following SMARTpool ON-TARGET Plus siRNA duplexes were purchased from Thermo Scientific: non-targeting pool (D-001810-10-05) and human Elmo1 (L-012851-00-0005). HEK 293T cells were transfected with 1μg empty pEBB-Flag (vector) or Dock2-Flag (from M. Matsuda(5)) plus 4μg pEBB-Elmo-Flag plasmids by calcium phosphate (Profection, Promega). Primary T cells were transfected using the Mouse T cell Nucleofector Kit (Amaxa), with 2×106 CD4+ T cells, 1μg pMAX-GFP (Amaxa) and 4μg expression vectors. Elmo expression plasmids the pEBB-Flag backbone were provided by K.S. Ravichandran. MT123-HA-ubiquitin vector provided by Dirk Bohmann(33).
Data quantitation and statistical analysis
Densitometry values were determined by area under the curve analysis on ImageJ software (NIH). Quantitative analysis of time-lapse images was carried out using Nikon software (Nikon). Flow cytometry data was analyzed using FlowJo software (Treestar). Data shown are the average ±SEM. Statistical significance was determined using the two-tailed Student's t-test, where p<0.05 was considered significant.
RESULTS
Impaired migration of Elmo1−/− lymphocytes to CCR7 and CXCR4 ligands
We first analyzed Elmo1 levels in wild-type (WT) and Elmo1−/− mice by immunoblotting (IB). Splenocytes were FACS-sorted based on expression of B220 and CD3 surface markers and cell lysates analyzed by IB with anti-Elmo1. Elmo1 protein was readily detected in WT but not Elmo1−/− CD3+ (T cells) and B220+ (B cells) splenocytes (Figure 1A and 1B). CD3-/B220-splenocytes, which are comprised largely of CD11b+ myeloid cells, also expressed Elmo1, albeit at reduced levels compared to lymphocytes (Figure 1B). The frequency and total numbers of T and B cells in the spleen and lymph nodes of Elmo1−/− mice was not significantly different from WT (Figure 1A and Supplemental Figure 1). Also, the frequency of naïve splenic T cells (CD62Lhi/CD44lo) was similar between WT and Elmo1−/− mice (Supplemental Figure 1C).
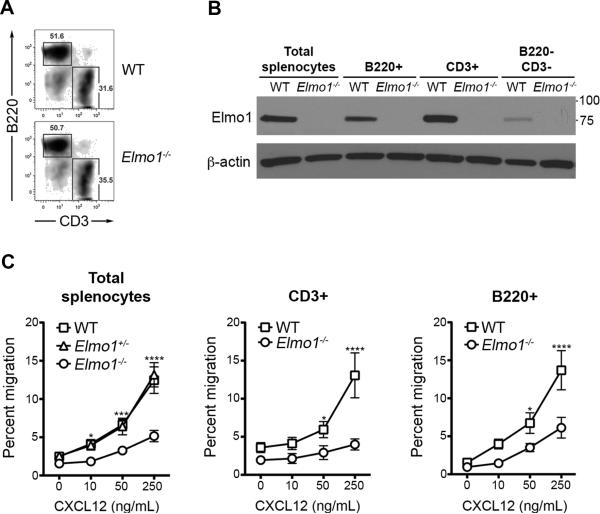
Figure 1. Impaired migration of Elmo1−/−splenocytes.
(A) Splenocytes from WT and Elmo1−/− mice were labeled with anti-B220 and anti-CD3 and analyzed by flow cytometry. The percentage of B220+ and CD3+ cells among all live splenocytes (7-AAD−) is shown. (B) Splenocytes were labeled as in A, FACS-sorted and analyzed by IB with antibodies indicated (left). Relative molecular weights are shown (right). One representative experiment of five is shown for A and B. (C) Transwell migration of splenocytes from WT, Elmo1+/− and Elmo1−/− mice to indicated concentrations of CXCL12. The percent of CD3+ and B220+ splenocytes that migrated to the lower chamber was determined by flow cytometry (n = 6 mice/genotype, ±SEM).
Splenocytes from WT, Elmo1+/− and Elmo1−/− were tested for migration to a range of CXCL12 concentrations in transwell chambers. At all doses tested, WT and Elmo1+/− splenocytes migrated similarly, whereas Elmo1−/− splenocytes showed significantly reduced migration (Figure 1C). Using flow cytometry we calculated the fraction of total B220+ (B cells) and CD3+ (T cells) splenocytes that migrated through the transwell. We observed a near complete loss of T cell migration to CXCL12 by Elmo1−/− T cells, while Elmo1−/− B cells showed an intermediate but significant decrease in migration compared to WT (Figure 1C). The ex vivo survival of unstimulated lymphocytes was similar between WT and Elmo1−/− after 24 hours in culture (data not shown). Thus the loss of Elmo1 in primary lymphocytes results in defective migratory responses to CXCL12.
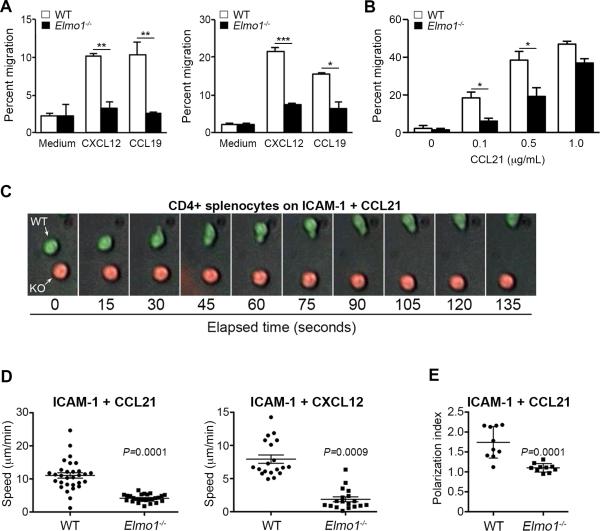
Based on the strong effect of Elmo1 deletion on T cell migration, we further examined migration responses of CD4+ T cells from WT and Elmo1−/− mice. CD4+ T cells were isolated from lymph nodes and spleen and analyzed for transwell migration to CXCL12, CCL19 and CCL21. Elmo1−/− CD4+ T cells from both tissues were significantly reduced in their capacity to migrate to these chemokines although some migration of Elmo1−/− T cells was observed, particularly at higher chemokine doses (Figure 2A and 2B). To determine if loss of Elmo1 specifically affected T cell polarization and migration, we used time-lapse microscopy to measure motility patterns of CD4+ T cells plated on ICAM-1 plus CCL21 or CXCL12. CD4+ T cells isolated from WT and Elmo1−/− spleens were labeled with CFSE or TAMRA-SE cell permeable dyes, mixed in equal numbers and plated for microscopic analysis. T cells from WT and Elmo1−/− mice adhered to ICAM-1, but while the majority of WT cells adopted a polarized morphology and migrated along the surface, Elmo1-deficient cells largely failed to do so (Figures 2C-E and Supplemental Videos 1 and 2). Together these results show that Elmo1 is required for normal T cell motility responses to CCR7- and CXCR4-dependent chemokines.
Figure 2. Defective chemotaxis of Elmo1−/− T cells to CCR7 and CXCR4 ligands.
(A) CD4+ T cells isolated from lymph nodes (mediastinal and axillary, left) or spleen (right) were analyzed for transwell migration to CXCL12 (25ng/mL) and CCL19 (10ng/mL). One representative experiment of three is shown. (B) Transwell migration of CD4+ T cells from WT and Elmo1−/− mice to CCL21. (C-E) CD4+ splenic T cells labeled with CFSE (WT, green) or TAMRA (Elmo1−/−, red) were plated on a glass dish coated with rmICAM-1 (μg/mL) and rmCCL21 (μg/mL). Epifluorescence images were acquired every 15 seconds with 20X objective. A representative series of time-lapse images from a single field containing one WT and one Elmo1−/− cell is shown. The speed (C) and polarization (D) of individual cells was calculated for all cells in one field over 30 minutes. One representative experiment of 2-3 for each condition is shown.
T cells require Elmo1 for CCR7- and CXCR4-mediated Rac activation
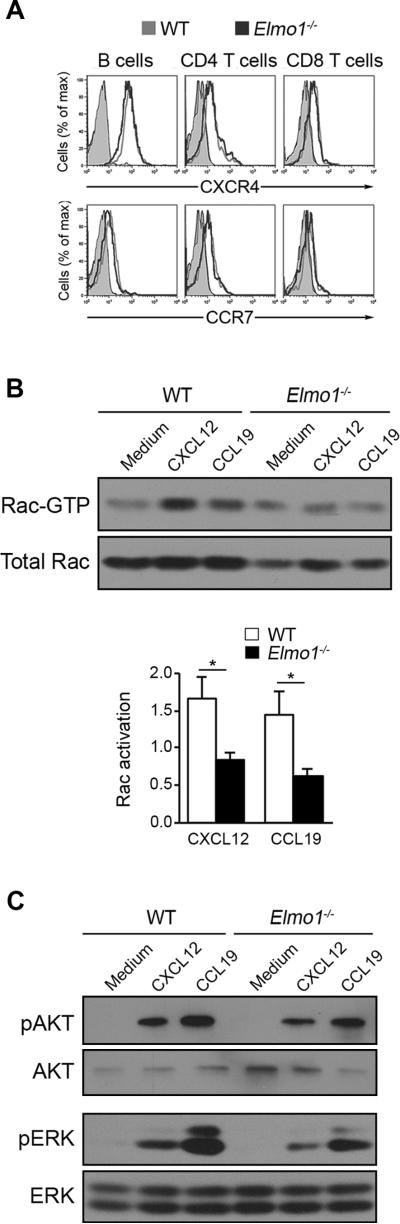
The failure of Elmo1−/− T cells to polarize and migrate suggested a requirement for Elmo1 in chemokine signaling. Since we found that the surface expression of CCR7 and CXCR4 on splenic lymphocytes was comparable between WT and Elmo1−/− (Figure 3A), we focused on activation of key molecular pathways downstream of CCR7 and CXCR4. The GTPase Rac is rapidly activated upon chemokine stimulation and is critically required for actin polymerization and polarization during T cell migration (4). To determine if loss of Elmo1 affects chemokine-induced Rac activation, we measured Rac-GTP levels in CD4+ T cells by GST-PAK pulldown and anti-Rac IB of lysates from WT and Elmo1−/− T cells following CXCL12 or CCL19 stimulation. In WT cells, Rac was clearly activated by both chemokines, while Elmo1−/− T cells showed no Rac activation (Figure 3B). Under the same stimulation conditions, we observed robust phosphorylation of AKT (Ser473) and ERK1/2 (Thr202/Tyr204) in both WT and Elmo1−/− T cells (Figure 3C). These results indicate that Elmo1 is specifically required for CXCR4 and CCR7 activation of Rac but not PI3K and Ras/Raf/MEK pathways.
Figure 3. CCR7 and CXCR4 signaling in Elmo1−/− T cells.
(A) CXCR4 and CCR7 surface levels on splenic lymphocyte populations from WT and Elmo1−/− mice were determined by flow cytometry. One representative experiment of four is shown. (B, C) CD4+ T cells isolated from WT and Elmo1−/− spleens were stimulated 15 seconds with 250ng/mL CXCL12 or CCL19 and lysed. (B) Lysates were analyzed for Rac-GTP levels by GST-PAK pulldown and anti-Rac IB. Below, graph showing Rac-GTP levels normalized to total Rac and expressed as relative to medium control for n = 4 mice/genotype ±SEM. (C) phospho-Ser473-AKT and phospho-Thr202/Tyr204-ERK1/2 levels were determined by IB. Blots were stripped and re-probed for total AKT and ERK1/2. One representative experiment of three is shown.
Elmo1 but not Elmo2 interacts with Dock2 in primary lymphocytes
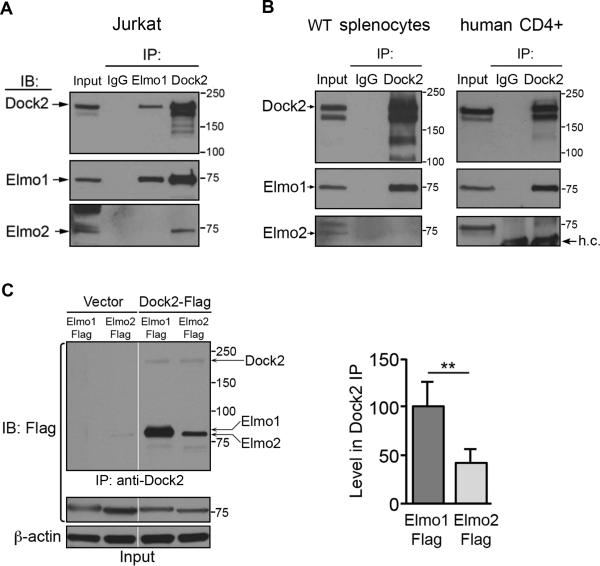
Dock2 is the primary GEF responsible for Rac activation downstream of CCR7 and CXCR4 in T cells (1, 2). Previous work has shown that Elmo1 and Dock2 can interact upon overexpression in cell lines, but the relevance of this interaction in primary lymphocytes is not known (26, 31). We tested for endogenous Elmo-Dock2 complexes in T cells by co-immunoprecipitation (co-IP). Elmo1 and Elmo2 were present in anti-Dock2 IP's of Jurkat lysates, while Dock2 but not Elmo2 was present in anti-Elmo1 IP's of these cells (Figure 4A). This confirmed that our co-IP approach was sufficient to detect native Elmo1-Dock2 and Elmo2-Dock2 complexes and also indicated that endogenous Dock2 may interact exclusively with either Elmo1 or Elmo2. Using this approach, we tested for Elmo-Dock2 complexes in normal mouse splenocytes and primary human CD4+ T cells. Surprisingly, we could detect Elmo1 but not Elmo2 in Dock2 IP's from these primary cells (Figure 4B). We then tested whether Dock2 may bind more readily to Elmo1 than Elmo2. Differences in the sensitivities of our anti-Elmo antibodies prevented us from testing this with endogenous proteins, so we expressed 1x Flag-tagged Elmo1 or Elmo2 along with Dock2 in 293T cells and examined Elmo-Dock2 complex formation by anti-Dock2 IP and anti-Flag IB. As shown in Figure 4C, Elmo1 and Elmo2 were expressed at equivalent levels in 293T cells and, as was seen in Jurkat cells (Figure 4A), both were detectable in anti-Dock2 IP's. However, significantly more Elmo1 co-precipitated with Dock2 (~2.5-fold) compared to Elmo2 (Figure 4C). We attempted to directly compare binding of Elmo1 and Elmo2 to Dock2 in the same cells, but we were unable to resolve distinct bands due to their similar molecular weights (Figure 4C). Together these data show that Dock2 can interact with either Elmo1 or Elmo2, but that in normal lymphocytes Elmo1-Dock2 complexes are the most prevalent. This difference may, in part, explain the failure of residual Elmo2 to fully compensate for loss of Elmo1 in T cell migration.
Figure 4. Dock2 interaction with Elmo1 and Elmo2.
(A) IP of Jurkat lysates with control rabbit IgG or anti-Dock2 followed by IB with antibodies indicated left. IP's were divided equally and loaded in separate wells for blotting with either anti-Elmo1 or anti-Elmo2. (B) IP of lysates from WT mouse splenocytes (left) and primary human CD4+ T cells (right) as in (A). One representative experiment of three for A and B is shown. h.c., heavy chain of IgG. (C) 293T cells were transfected with vector, Dock2-Flag, Elmo1-Flag or Elmo2-Flag plasmids followed by lysis and IP with anti-Dock2 or control IgG. IP's and total lysates (input) were analyzed by IB with anti-Flag-HRP. Right, the level of Elmo1-Flag and Elmo2-Flag present in anti-Dock2 IP's was normalized to input levels and the relative level of each in Dock2 IP's was calculated. The average of n = 3 ±SEM is shown right. Relative molecular weights (kDa) are indicated to right of blots.
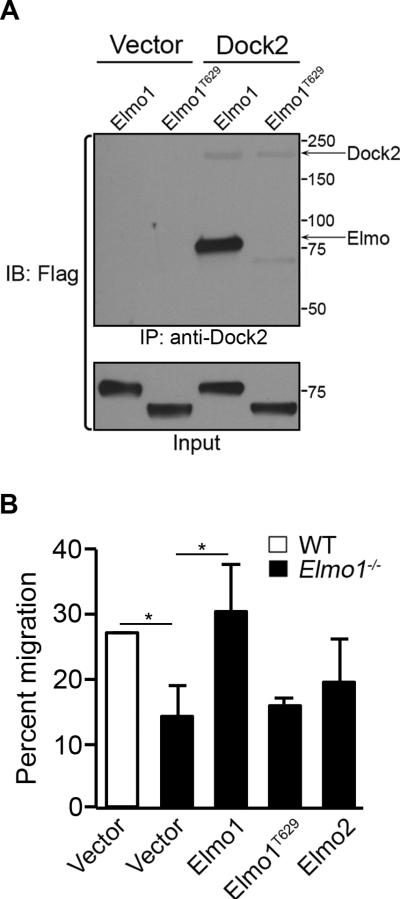
C-terminus of Elmo1 is essential for T cell migration
The data above suggested that Dock2 may preferentially associate with Elmo1 to promote T cell migration. To determine if Elmo1 interaction with Dock2 is required for Dock2-dependent migration, we attempted to rescue defective migration of Elmo1−/− T cells by transient transfection using full-length Elmo1 (a.a. 1-727) or C-terminal truncation mutant of Elmo1 (Elmo1T629, a.a. 1-629). In accord with a previous study we found that the C-terminal tail of Elmo1 is essential for interaction with Dock2 (Figure 5A and ref. 26). CD4+ splenic T cells from Elmo1−/− mice were then co-transfected with Elmo plasmids and a GFP reporter plasmid and tested for transwell migration to CXCL12. The number of GFP+ transfected cells migrating to the lower chamber was quantified by flow cytometry and compared to the total number of GFP+ cells (Supplemental Figure 2). Transfection with full-length Elmo1, but not Elmo1T629, restored migration of Elmo1−/− T cells to WT levels (Figure 5B). In the same experiments Elmo2 was unable to rescue migration of Elmo1−/− T cells (Figure 5B). These results show that Elmo1 interaction with Dock2 is required for Dock2-dependent T cell migration.
Figure 5. Elmo1-dependent CD4+ T cell migration requires interaction with Dock2.
(A) Lysates from 293T cells transfected with vector, Dock2-Flag, Elmo1-Flag, or mutant Elmo1T629-Flag were IP'd with anti-Dock2 and analyzed by IB with anti-Flag. One representative experiment of three is shown. Relative molecular weights (kDa) are indicated to right. (B) Splenic CD4+ T cells from WT and Elmo1−/− mice were nucleofected with GFP plasmid along with either empty vector, Elmo1, Elmo1T629 or Elmo2 plasmids and assayed for transwell migration to CXCL12 (50ng/mL). The number of GFP+ cells migrating to the lower chamber was determined by flow cytometry and normalized to the total number of GFP+ cells for each transfected population and used to calculate the percent migration for each transfected populations (n = 3, ±SEM).
Elmo1 selectively regulates Dock2 levels
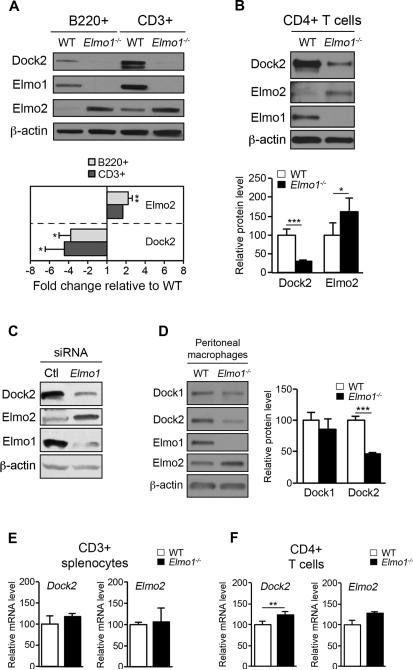
To gain mechanistic insight into the migration defect of Elmo1−/− T cells, we examined expression of Elmo1, Elmo2 and Dock2 levels in WT and Elmo1−/− lymphocytes. Surprisingly, we found that CD3+, CD4+ and B220+ lymphocytes from Elmo1−/− mice showed a ~4-fold reduction in Dock2 levels compared to WT (Figures 6A and 6B). Lymphocytes from all Elmo1−/− mice tested to date (>25) have shown a similar reduction in Dock2 levels compared to WT controls (data not shown). Interestingly, Elmo2 levels were increased ~2-fold in Elmo1-deficient lymphocytes (Figures 6A and 6B). Similar results were seen upon acute depletion of Elmo1 in human Jurkat cells by siRNA, further supporting the regulation of Dock2 and Elmo2 levels as a conserved function of Elmo1 in lymphocytes (Figure 6C). To determine if Elmo1 deficiency similarly affects Dock1 levels, F4/80hi resident peritoneal macrophages were FACS-sorted from WT and Elmo1−/− mice and tested for Dock1 and Dock2 levels by IB. Similar to lymphocytes, Dock2 levels were significantly reduced in Elmo1−/− F4/80hi macrophages compared to WT while the level of Dock1 in these macrophages was not significantly different (Figure 6D). These results show that Elmo1 plays a non-redundant and specific role in regulating the level of endogenous Dock2 protein.
Figure 6. Elmo1 control of Dock2 levels in leukocytes.
(A) Splenocytes from WT and Elmo1−/− mice were FACS-sorted by anti-B220 and anti-CD3 staining, lysed and analyzed by IB with the antibodies indicated to the left. Graph (below) shows fold change in Dock2 and Elmo2 levels in Elmo1−/− cells relative to WT as determined by IB for n = 3 mice/genotype, ±SEM. (B) Splenic CD4+ T cells were analyzed by IB as in A. Graph (below) is average of n = 8 mice/genotype, ±SEM. (C) Jurkat cells transfected with a non-targeting control (Ctl) or Elmo1 targeting siRNA were analyzed by IB with indicated antibodies. (D) F4/80hi resident peritoneal macrophages were FACS-sorted from WT and Elmo1−/− mice and analyzed by IB with indicated antibodies. Graph to the right shows the relative levels of Dock1 and Dock2 in F4/80hi RPM for n = 4 mice/genotype, ±SEM. (E, F) qRT-PCR analysis of splenic CD3+ (E) and CD4+ T cells (F) for n = 3-5 mice/genotype, ±SEM.
Evidence for in vivo posttranslational regulation of Dock2 by Elmo1
Elmo1 has been shown to control Dock1 levels through negative regulation of Dock1 ubiquitination and proteasomal degradation in cell lines, but Dock2 ubiquitination has not been studied (19-21). To address this, we first measured Dock2 mRNA levels in WT and Elmo1−/− lymphocytes by qRT-PCR. As shown in Figures 6E and 6F, steady-state levels of Dock2 mRNA were comparable between WT and Elmo1−/− splenic CD3+ and CD4+ T cells, although the latter showed a slight but significant increase in Dock2 mRNA. Likewise, levels of Elmo2 mRNA were not significantly different between WT and Elmo1−/− T cells (Figures 6E and 6F).
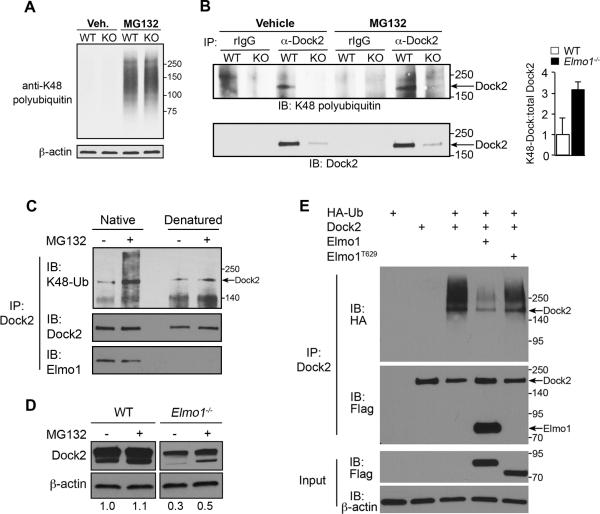
We next examined in vivo ubiquitination of Dock2 in CD4+ T cells. Lysine 48 (K48) of ubiquitin serves as a substrate for the formation of covalently attached polyubiquitin chains to lysine residues of target proteins. K48-linked polyubiquitinated proteins are subsequently targeted to the 26S proteasome for degradation (34). Using an antibody specific for K48-linked polyubiquitin, we measured in vivo levels of ubiquitinated endogenous Dock2 in WT CD4+ T cells. Cells were first treated with the proteasome inhibitor MG132 to allow for accumulation of ubiquitinated proteins (Figure 7A), followed by anti-Dock2 IP of CD4+ T cell lysates and IB with anti-K48. We observed one band corresponding to the molecular weight of Dock2 (~200kDa) in the anti-Dock2 but not control IgG IP's of WT T cells before and after MG132 treatment (Figure 7B). To determine whether the K48-ubiquitin band in Dock2 IP's is due to direct ubiquitination of Dock2 or association of Dock2 with ubiquitinated proteins, Dock2 IP's from WT CD4+ T cells were heat-denatured and subjected to a second round of anti-Dock2 IP prior to anti-K48-ubiquitin IB. As shown in Figure 7C, these denaturing conditions successfully disrupted Dock2-Elmo1 association but did not abrogate the K48-ubiquitin Dock2 band. These data show that Dock2 undergoes constitutive polyubiquitination in primary T cells.
Figure 7. Posttranslational regulation of Dock2 in T cells.
(A) Splenic CD4+ T cells from WT and Elmo1−/− mice were treated with the proteasome inhibitor MG132 for 3 hours, lysed and analyzed by IB using an antibody that recognizes K48-linked lysines of polyubiquitin chains or β-actin. (B) Splenic CD4+ T cells from WT and Elmo1−/− mice were treated as in A, followed by IP with anti-Dock2 or control rabbit IgG. IP's were analyzed by IB with anti-K48 ubiquitin (above) and anti-Dock2 (above). Arrows indicate position of Dock2 bands. Graph right shows relative levels of K48-Dock2 normalized to total Dock2 in IP. n = 2 mice/genotype, ±SEM. (C) WT splenic CD4+ T cells were treated 3 hours with 10μM MG132 followed by lysis and anti-Dock2 IP. Half of each IP was then denatured (5 minutes at 95°C) followed by a second round of anti-Dock2 IP before the denatured and nondenatured (“native”) fractions were analyzed by anti-K48 ubiquitin IB. (D) Splenic CD4+ T cells from WT and Elmo1−/− mice were treated with MG132 for 6 hours and total cell lysates were analyzed by IB using the indicated antibodies. Values below indicate the relative levels of Dock2 protein normalized to β-actin in each sample as determined by densitometry. Results are representative of three independent experiments. (E) 293T cells were transfected with indicated plasmids for 24 hours followed by anti-Dock2 IP of lysates and IB with indicated antibodies. Results are representative of three independent experiments. Relative molecular weights (kDa) are indicated to right of blots.
We next addressed the role of Elmo1 in regulating Dock2 ubiquitination. Unlike WT, K48-ubiquitinated Dock2 was only detectable in Elmo1−/− CD4+ T cells after MG132 treatment (Figure 7B). When normalized to the total amount of Dock2 present in each IP, the level of K48-ubiquitin Dock2 in Elmo1−/− cells was 3-fold higher than WT (Figure 7B, graph). MG132 treatment of Elmo1−/− T cells partially restored total Dock2 levels (Figure 7D), while treatment with bafilomycin, an inhibitor of lysosome-mediated proteolysis, had no effect on Dock2 levels in Elmo1−/− cells (data not shown). In time course experiments, we found that Dock2 levels in Elmo1−/− cells peaked 6 hours after MG132 treatment of Elmo1−/− T cells, and that neither extended incubation with MG132 (up to 24 hours) nor activation of the T cells (with plate-bound anti-CD3/anti-CD28 or PMA/ionomycin) enhanced Dock2 levels above that seen at 6 hours (data not shown). Finally, to determine if Elmo1 inhibits Dock2 ubiquitination, 293T cells were cotransfected with Dock2 and HA-tagged ubiquitin along with Elmo1 or Elmo1T629 followed by Dock2 IP and anti-HA IB. Expression of Elmo1, but not Elmo1T629, markedly inhibited ubiquitination of Dock2 (Figure 7E). Together, these data indicate that Elmo1 controls Dock2 protein levels, at least in part, through inhibition of Dock2 ubiquitination and proteasomal degradation.
DISCUSSION
Elmo1 and Elmo2 are expressed in most human and mouse tissues including central and peripheral lymphoid organs (13, 35). Both have been shown to promote actin-dependent phagocytosis and cell migration in a wide range of immortalized mammalian cell lines and invertebrate models. This report is the first to examine the function of endogenous Elmo1 in primary lymphocytes. Here we show that Elmo1 is essential for normal CCR7- and CXCR4-dependent migration of primary T and B lymphocytes in vitro. Although Elmo1−/− lymphocytes expressed normal surface levels of these chemokine receptors and were able to adhere to chemokine-infused ICAM-1 substrates, Elmo1-deficient T cells failed to effectively polarize and migrate in response to CCR7 and CXCR4 ligands. Rac activation in response to such stimulation was abrogated in Elmo1−/− T cells although AKT and ERK1/2 phosphorylation were similar to WT. The requirement for Elmo1 in Rac but not PI3K activation downstream of CCR signaling is strikingly similar to that seen in Dock2−/− T cells (1, 2). It is interesting to note that while Elmo1 and Dock2 are dispensable for PI3K activation downstream of CCR activation in lymphocytes, there are conditions where the Elmo-Dock module is important for Rac-dependent PI3K activation. Notably, recent findings from Fritsch et al show that the Elmo1-Dock1 complex is required for normal PI3K activation in fibroblasts stimulated with lysophosphatidic acid (LPA) or sphingosine 1-phosphate (S1P) (36). In this study Elmo1 was found to interact with the Gβγ complex upon LPA and S1P receptor activation and to promote Rac-dependent activation of the p110β type I PI3K(36). In another study using breast cancer cell lines, Elmo1 interaction with the Gβi subunit of CXCR4 was shown to be necessary for Dock1-dependent Rac activation, although CXCR4-dependent AKT phosphorylation was not affected by Elmo-Dock depletion (18). Interestingly, while PI3K activation downstream of CXCR4 appears to be Elmo1- and Dock2-independent in lymphocytes, Dock2 is required for S1P receptor-mediated AKT phosphorylation in lymphocytes(6). Thus while Elmo is clearly important for recruitment of Dock proteins to multiple GPCRs (18, 23, 36), it appears that the specific mechanisms of Elmo-GPCR association can regulate the pathways, including PI3K, that are activated by Dock-Rac signaling. Nevertheless, the data presented here show that in lymphocytes a principal function of Elmo1 in CCR signaling is the regulation of Dock2 levels.
Elmo binding has been shown to regulate Dock1 localization, relief of autoinhibition and protein stability (18, 23). The relative contribution and/or integration of these distinct mechanisms in optimal Dock1 signaling remains unsettled and is likely dependent on the specific tissue and function being studied. Although it was previously shown that Elmo1 interaction inhibits Dock1 ubiquitination and proteasomal degradation, our data show for the first time that endogenous Dock2 undergoes K48-linked polyubiquitination in normal primary T cells and that Elmo1 plays an important role in controlling overall Dock2 levels through regulation of Dock2 polyubiquitination. However, it is important to note that despite a residual pool of Dock2 in Elmo1−/− T cells, Rac activation and migration were almost completely impaired. This may indicate a threshold requirement for Dock2 levels in T cells, but may also reflect a requirement for Elmo1 in proper targeting and activation of Dock2 in response to chemokine stimulation. Indeed, Elmo1 was recently reported to be required for localization of Dock1 at the CXCR4 receptor in migrating human breast cancer cells (18). A better understanding of the molecular regulation of Dock2 ubiquitination by Elmo1, most notably the identity of lysines in Dock2 that are targeted for ubiquitination, the E3 ligase complex responsible for ubiquitination and how Elmo1 binding controls Dock2 modifications will be necessary to distinguish these modes of Elmo1-dependent Dock regulation.
Despite the homology and presumed redundancy of Elmo1 and Elmo2, we present several lines of evidence that show Elmo1 is a key regulator of Dock2-dependent migration of primary lymphocytes (13, 15). Despite being upregulated 2-fold in Elmo1−/− T cells, Elmo2 failed to compensate for Elmo1 in maintaining normal migration and Dock2 levels in vitro. Certainly this outcome could be a consequence of inadequate expression of Elmo2. Based on qRT-PCR analyses, Elmo2 mRNA levels were 3-fold lower that Elmo1 in WT CD4+ T cells (data not shown), which is in agreement with published microarray analyses (35). However, Elmo2 mRNA levels in Elmo1−/− T cells were comparable to WT, indicating that mRNA levels may not accurately reflect relative levels of Elmo proteins. Moreover, transfection of Elmo1 but not Elmo2 rescued defective migration of Elmo1−/− T cells, further suggesting that inadequate Elmo2 expression was unlikely to fully account for the lack of compensation. We then focused on potential differences in Dock2 binding. Quite surprisingly we found that endogenous Elmo1-Dock2 but not Elmo2-Dock2 complexes were present in primary mouse and human lymphocytes. In a direct comparison of Dock2 binding in 293T cells, Elmo1 showed a 2.5-fold greater ability to complex with Dock2 than did Elmo2. Our finding that Dock2 but not Dock1 levels were significantly reduced in Elmo1−/− peritoneal macrophages indicates that Elmo1 is an essential regulator of Dock2 levels while Elmo1 and Elmo2 may function redundantly in regulating Dock1 levels. Along these lines, it is interesting that peripheral lymphocyte populations of Elmo1−/− mice appear normal up to 12 weeks of age. This is in contrast to Dock2−/−mice which display a dramatic reduction in peripheral T and B cell populations due to the inability of Dock2-deficient lymphocytes to properly home to peripheral lymphoid organs(1, 2, 6). Dock2−/− T cells show a near-complete loss of in vitro migration to CXCL12 and CCL21 even at high concentrations of ligand(1), while in our study Elmo1−/− T cells showed some residual ability to migrate to these chemokines. Taken together these data suggest that under homeostatic conditions the residual pool of Dock2 protein in Elmo1−/− T cells is sufficient to allow for the normal accumulation of peripheral T cells. Whether this homeostatic migration occurs through Elmo2-dependent or Elmo-independent mechanisms remains to be determined. Moreover, the requirement for Elmo1 in regulating Dock2-dependent migration in non-lymphoid tissues or under non-homeostatic conditions is currently being investigated. Thus, it appears that Elmo1 plays a preferential role in interacting with and regulating Dock2 levels, but that as yet unidentified compensatory mechanisms exist to allow for normal homeostatic migration in vivo.
Recruitment of activated lymphocytes is a major cause of tissue pathology associated with many disease states including asthma, allograft rejection, chronic inflammation and autoimmunity. Chemokines play an integral role in lymphocyte trafficking and thus targeting these signaling pathways holds promise for curbing lymphocyte-mediated tissue damage. However, complex expression patterns and ligand-receptor promiscuity have hampered attempts to directly target chemokines or their receptors (37). Dock2 inhibition is an attractive approach for controlling T cell migration in vivo for a number of reasons. First, Dock2 regulates in vivo lymphocyte migration downstream of multiple receptors (1, 2, 6). Second, Dock2 inhibition does not appear to directly affect survival of peripheral T cells (1). Finally, Dock2 expression is largely restricted to hematopoietic cells (5). For these reasons, targeted inhibition of Dock2 could be used to specifically and reversibly block migration of lymphocytes to a broad range of chemokines in tissues without broadly depleting the repertoire of antigen-specific lymphocytes. Toward this goal, Fukui and colleagues recently reported the development of a small molecule inhibitor of Dock2 that acts at the Rac exchange domain to block Rac activation and T cell migration (38). Unfortunately this inhibitor was also found to strongly inhibit Dock1 function probably due to the high degree of conservation of the Rac exchange domains of Dock-A proteins (38). Because Dock1 is widely-expressed and essential for life, any strategy to disrupt Dock2 must avoid Dock1 inhibition (5, 39). In this present study, we describe important and previously unknown features of Elmo-Dock2 interaction in primary cells that can guide efforts to design more effective strategies to selectively disrupt Dock2 function and T cell migration.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jim Miller and Edward Stites for reviewing the manuscript, Taeg Kim for helpful discussions, Nathan Laniewski and the URMC Flow Cytometry Core for technical assistance.
This work was supported by the Ellison Medical Foundation (MRE) and the National Institutes of Health: AI027767-24 (to MRE through a CNIHR award from the University of Alabama – Birmingham Center for AIDS Research), HL087088 (MK) and T32AI007285 (PSM, TC).
Footnotes
DISCLOSURES
The authors have declared no conflict of interest related to this work.
REFERENCES
- 1.Fukui Y, Hashimoto O, Sanui T, Oono T, Koga H, Abe M, Inayoshi A, Noda M, Oike M, Shirai T, Sasazuki T. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 2.Nombela-Arrieta C, Lacalle RA, Montoya MC, Kunisaki Y, Megías D, Marqués M, Carrera AC, Mañes S, Fukui Y, Martínez-A C, Stein JV. Differential requirements for DOCK2 and phosphoinositide-3-kinase gamma during T and B lymphocyte homing. Immunity. 2004;21:429–441. doi: 10.1016/j.immuni.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Tybulewicz VLJ, Henderson RB. Rho family GTPases and their regulators in lymphocytes. Nat. Rev. Immunol. 2009;9:630–644. doi: 10.1038/nri2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faroudi M, Hons M, Zachacz A, Dumont C, Lyck R, Stein JV, Tybulewicz VLJ. Critical roles for Rac GTPases in T-cell migration to and within lymph nodes. Blood. 2010;116:5536–5547. doi: 10.1182/blood-2010-08-299438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishihara H, Kobayashi S, Hashimoto Y, Ohba F, Mochizuki N, Kurata T, Nagashima K, Matsuda M. Non-adherent cell-specific expression of DOCK2, a member of the human CDM-family proteins. Biochim. Biophys. Acta. 1999;1452:179–187. doi: 10.1016/s0167-4889(99)00133-0. [DOI] [PubMed] [Google Scholar]
- 6.Nombela-Arrieta C, Mempel TR, Soriano SF, Mazo I, Wymann MP, Hirsch E, Martínez-A C, Fukui Y, Von Andrian UH, Stein JV. A central role for DOCK2 during interstitial lymphocyte motility and sphingosine-1-phosphate-mediated egress. J. Exp. Med. 2007;204:497–510. doi: 10.1084/jem.20061780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Pan F, Erickson LM, Jang M-S, Sanui T, Kunisaki Y, Sasazuki T, Kobayashi M, Fukui Y. Deletion of DOCK2, a regulator of the actin cytoskeleton in lymphocytes, suppresses cardiac allograft rejection. J. Exp. Med. 2005;202:1121–1130. doi: 10.1084/jem.20050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikimi A, Kukimoto-Niino M, Yokoyama S, Fukui Y. Immune regulatory functions of DOCK family proteins in health and disease. Experimental Cell Research. 2013 doi: 10.1016/j.yexcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Kunisaki Y, Nishikimi A, Tanaka Y, Takii R, Noda M, Inayoshi A, Watanabe K-I, Sanematsu F, Sasazuki T, Sasaki T, Fukui Y. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 2006;174:647–652. doi: 10.1083/jcb.200602142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunisaki Y, Tanaka Y, Sanui T, Inayoshi A, Noda M, Nakayama T, Harada M, Taniguchi M, Sasazuki T, Fukui Y. DOCK2 is required in T cell precursors for development of Valpha14 NK T cells. J. Immunol. 2006;176:4640–4645. doi: 10.4049/jimmunol.176.8.4640. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh K, Tanaka Y, Nishikimi A, Inayoshi A, Enjoji M, Takayanagi R, Sasazuki T, Fukui Y. Differential requirement for DOCK2 in migration of plasmacytoid dendritic cells versus myeloid dendritic cells. Blood. 2008;111:2973–2976. doi: 10.1182/blood-2007-09-112169. [DOI] [PubMed] [Google Scholar]
- 12.Ippagunta SK, Malireddi RKS, Shaw PJ, Neale GA, Walle LV, Green DR, Fukui Y, Lamkanfi M, Kanneganti T-D. The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nature Immunology. 2011:1–9. doi: 10.1038/ni.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 14.Côté J-F, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 16.Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, Hengartner MO, Yajnik V, Ravichandran KS. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 17.deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, Sondek J, Hengartner MO, Wu YC, Ravichandran KS. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr. Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun. 2013;4:1706. doi: 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino Y, Tsuda M, Ichihara S, Watanabe T, Sakai M, Sawa H, Nagashima K, Hatakeyama S, Tanaka S. Elmo1 inhibits ubiquitylation of Dock180. J. Cell. Sci. 2006;119:923–932. doi: 10.1242/jcs.02797. [DOI] [PubMed] [Google Scholar]
- 20.Sanematsu F, Hirashima M, Laurin M, Takii R, Nishikimi A, Kitajima K, Ding G, Noda M, Murata Y, Tanaka Y, Masuko S, Suda T, Meno C, Cote JF, Nagasawa T, Fukui Y. DOCK180 Is a Rac Activator That Regulates Cardiovascular Development by Acting Downstream of CXCR4. Circ. Res. 2010;107:1102–1105. doi: 10.1161/CIRCRESAHA.110.223388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R, Cheng SY. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Research. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M, Chen Q, Klingele D, Hengartner MO, Ravichandran KS. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- 23.Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 24.Van Goethem E, Silva EA, Xiao H, Franc NC. The Drosophila TRPP Cation Channel, PKD2 and Dmel/Ced-12 Act in Genetically Distinct Pathways during Apoptotic Cell Clearance. PLoS ONE. 2012;7:e31488. doi: 10.1371/journal.pone.0031488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Ham TJ, Kokel D, Peterson RT. Apoptotic Cells Are Cleared by Directional Migration and elmo1-Dependent Macrophage Engulfment. Curr. Biol. 2012 doi: 10.1016/j.cub.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanui T, Inayoshi A, Noda M, Iwata E, Stein JV, Sasazuki T, Fukui Y. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood. 2003;102:2948–2950. doi: 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- 27.Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, Kinchen JM, Zhang J, Lysiak JJ, Ravichandran KS. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2010;467:333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Z, Elliott MR, Chen Y, Walsh JT, Klibanov AL, Ravichandran KS, Kipnis J. Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nature Cell Biology. 2011;13:1–9. doi: 10.1038/ncb2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Kinchen JM, Rossman KL, Grimsley C, deBakker C, Brugnera E, Tosello-Trampont A-C, Haney LB, Klingele D, Sondek J, Hengartner MO, Ravichandran KS. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat Struct Mol Biol. 2004;11:756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- 30.Komander D, Patel M, Laurin M, Fradet N, Pelletier A, Barford D, Côté J-F. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell. 2008;19:4837–4851. doi: 10.1091/mbc.E08-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanawa-Suetsugu K, Kukimoto-Niino M, Mishima-Tsumagari C, Akasaka R, Ohsawa N, Sekine S-I, Ito T, Tochio N, Koshiba S, Kigawa T, Terada T, Shirouzu M, Nishikimi A, Uruno T, Katakai T, Kinashi T, Kohda D, Fukui Y, Yokoyama S. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1113512109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, Lysiak JJ, Harden TK, Leitinger N, Ravichandran KS. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature. 2009;461:282–286. doi: 10.1038/nature08296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 34.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng TSP, Painter MW, Immunological Genome Project Consortium The Immunological Genome Project: networks of gene expression in immune cells. Nature Immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 36.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO Families of GTPases Directly Regulate Distinct Phosphoinositide 3-Kinase Isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schall TJ, Proudfoot AEI. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat. Rev. Immunol. 2011;11:355–363. doi: 10.1038/nri2972. [DOI] [PubMed] [Google Scholar]
- 38.Nishikimi A, Uruno T, Duan X, Cao Q, Okamura Y, Saitoh T, Saito N, Sakaoka S, Du Y, Suenaga A, Kukimoto-Niino M, Miyano K, Gotoh K, Okabe T, Sanematsu F, Tanaka Y, Sumimoto H, Honma T, Yokoyama S, Nagano T, Kohda D, Kanai M, Fukui Y. Blockade of Inflammatory Responses by a Small-Molecule Inhibitor of the Rac Activator DOCK2. Chem. Biol. 2012;19:488–497. doi: 10.1016/j.chembiol.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Côté J-F. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proceedings of the National Academy of Sciences. 2008;105:15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.