Abstract
Cellular engineering of bacteria, fungi, insect cells and mammalian cells is a promising methodology to improve recombinant protein production for structural, biochemical, and commercial applications. Increased understanding of the host organism biology has suggested engineering strategies targeting bottlenecks in transcription, translation, protein processing and secretory pathways, as well as cell growth and survival. A combination of metabolic engineering and synthetic biology has been used to improve the properties of cells for protein production, which has resulted in enhanced yields of multiple protein classes.
Introduction
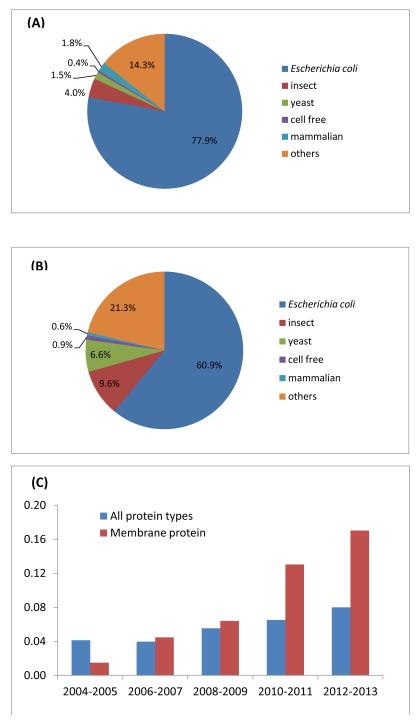
Expression of recombinant mammalian proteins is at the heart of structural studies in the biomedical field and is also important to commercial biotherapeutics, particularly the expression of antibodies. Although cloning, expression and production methods are available for many hosts [1-3], there is an ongoing effort to improve expression through traditional bioprocess optimization and cell re-engineering, especially for low yielding targets such as membrane proteins, large protein complexes and post-translationally modified proteins [4,5]. The first step in any strategy to over-produce proteins is the selection of the expression host. According to statistics from the Protein Data Bank (http://www.rcsb.org) and the ‘Membrane Proteins of Known Structure Database’ (http://blanco.biomol.uci.edu/mpstruc/), of all the proteins that had their structures determined between 2004 and 2013, 78% were expressed in Escherichia coli and only 4% in insect cells (Fig.1A). However, for the overexpression of membrane proteins, E. coli was utilized on average less (61%) and eukaryotic expression systems were used comparatively more (Fig. 1B). Notably, there is an increasing trend in the use of more complex eukaryotic hosts (insect and mammalian cells, Fig. 1C), which reflects an increase in the number of mammalian membrane proteins being crystallized, particularly G protein-coupled receptors (GPCRs) [6].
Fig. 1.
Summary of host cell line usage for production of recombinant proteins in structural studies between 2004 and 2013. (A) Break down of leading host cell choices for the expression of all types of proteins (B) Break down of leading expression organisms for integral membrane protein production. (C) Increasing application of higher eukaryotes (insect and mammalian cells) for recombinant protein production.
Despite many successes in the production of sufficient protein for crystallization and structure determination, proteins from, for example, mammalian sources and/or are integral membrane proteins are often difficult to overexpress[4,7]. Host cell engineering has emerged as one effective strategy for improving recombinant protein yields (Table 1) that can overcome bottlenecks in different steps along the protein production process. Here we discuss some recent successful cases that target potential bottlenecks in protein production, using strategies focused on optimizing transcription/translation, engineering the folding and secretory pathways, mutating the target protein sequence, and enhancing cell proliferation and/or survival.
Table 1.
Improvements in Protein Expression Levels for Different Cell Engineering Strategies
| Protein | Location | Expression host | Fold increase in protein production |
Reference |
|---|---|---|---|---|
| Strategy 1: optimizing transcription and enhancing translation | ||||
| Luciferase | intracellular | CHO-K1, HepG2, HEK-293, COS-7 |
3 | [8] |
| D-amino acid oxidase | intracellular | E.coli | 20 | [9] |
| Glutaryl-7-aminocephalosporanic acid acylase | intracellular | E.coli | 2 | [9] |
| N-carbamyl-D- amino acid amidohydrolase | intracellular | E.coli | 1.3 | [9] |
| Secreted alkaline phosphatase | extracellular | Insect cells | significant | [10] |
| Deltarhodopsin | membrane | E.coli | 5 | [11] |
| Sensory rhodopsin II | membrane | E.coli | 5 | [11] |
| 14 different membrane proteins | membrane | E.coli | significant | [12] |
| Cyclooxygenase-1 | extracellular | HEK293T | significant | [14] |
| Antibody | extracellular | COLO 320DM CHO DG44 |
>8 >20 |
[15] [15] |
| Tumor progression locus 2 complex | intracellular | HEK-293 | 10 | [16] |
| TBK1 | intracellular | HEK-293 | n.r. | [16] |
| Lck | membrane | HEK-293 | n.r. | [16] |
| CD40 | membrane | HEK-293 | n.r. | [16] |
| Bcl-2 | membrane | HEK-293 | n.r. | [16] |
| SEAP- EGFP fusion protein | extracellular | Insect cells | 2 | [17] |
| Strategy 2: Folding and secretory pathway engineering | ||||
| Secretory alkaline phosphatase- EGFP fusion protein |
extracellular | Insect cells | 2 | [17] |
| Human papillomavirus 16 E7 oncoprotein fused to C-terminus of Tobacco mosaic virus coat protein |
intracellular | E.coli | n.r. | [19] |
| Aldehyde dehydrogenase 3A1 | intracellular | E.coli | 4.9 | [20] |
| Serotonin transporter | membrane | Insect cells | 3 | [21] |
| Glycoprotein of rabies virus (truncated) | extracellular | H. polymorpha | n.r. | [22] |
| ZraS | membrane | E.coli | 3.6 | [26] |
| Deltarhodopsin | membrane | E.coli | 3.6 | [26] |
| Sensory rhodopsin II | membrane | E.coli | 3.4 | [26] |
| SEAP | extracellular | CHO-K1 | 2 | [27] |
| Antibody | extracellular | 3 | [27] | |
| α-amylase | extracellular | S. cerevisiae | 1.68 | [28] |
| Insulin precursor | extracellular | S. cerevisiae | 1.3 | [28] |
| t-PA | extracellular | CHO | 1.35 | [29] |
| HSA | extracellular | CHO | 1.6 | [30] |
| Antibodies | extracellular | CHO | 1.26 | [30] |
| Interleukin-1 receptor antagonist - HSA | extracellular | P. pastoris | 3.7 | [23] |
| HSA- human growth hormone | extracellular | P. pastoris | 4 | [23] |
| Antibody | extracellular | CHO | 4-7 | [24] |
| Strategy 3: Protein sequence mutagenesis | ||||
| Benzenediol- oxygen oxidoreductase | intracellular | E.coli | 3.14 | [31] |
| Coagulation Factor VIII | extracellular | COS-1 CHO |
1.3 1.6 |
[32] [32] |
| Neurotensin receptor | membrane | E.coli | 10 | [33] |
| Signal sequence of β-lactamase | intracellular | E.coli | 5.5 | [34] |
| YFP-Bcl-xL | membrane | Ramos B- cell CHO |
n.r. n.r. |
[35] [35] |
| Strategy 4: Cell proliferation and survival engineering | ||||
| Epidermal growth factor receptor | membrane | CHO | significant | [37] |
| Fibroblast growth factor receptor 3 | membrane | CHO | significant | [37] |
| Receptor tyrosine kinases proteins | membrane | CHO | significant | [37] |
| Antibody | extracellular | CHO-K1 | 4 | [38] |
| Secreted alkaline phosphatase | extracellular | CHO | 1.43 | [39] |
| Glycerol transport facilitator Fpsl | membrane | S. cerevisiae | 2 | [40] |
| A2a adenosine receptor | membrane | S. cerevisiae | 5 | [40] |
| Cannainoid receptor 2 | membrane | S. cerevisiae | 4.5 | [40] |
| Antibody | extracellular | CHO | 4 | [41] |
| SEAP | extracellular | CHO | 3-7 | [41] |
| Secreted α- amylase | extracellular | CHO | 3-7 | [41] |
| Epo-Fc | extracellular | CHO | n.r. | [42] |
| Other strategies | ||||
| lysozyme | intracellular | E.coli | 3000 | [44] |
n.r., not recorded
Optimizing transcription and enhancing translation
One of the most important choices in planning a strategy for overexpression of proteins is the type of promoter to use, and it is often the case that the strongest promoter will be the best for producing large amounts of correctly folded protein. Thus the most commonly used promoters are the T7 promoter in E. coli, the polyhedrin promoter in the baculovirus expression system and the CMV promoter in mammalian cells. If transcription is the rate limiting step in protein production, even after choosing a strong promoter, then increasing further the strength of the promoter may be effective. For example, Quilici et al. constructed a strong CMV promoter variant through introducing a 200-nucleotide deletion of intron A that increased luciferase expression up to 2 fold in mammalian cells [8]. However, recent studies have shown that increasing the amount of mRNA encoding the protein of interest does not necessarily lead to improved protein production in E.coli[9] or insect cells [10]. In these instances, it is possible that the rate limiting step is protein folding, perhaps due to limitations in host cell factors, such as molecular chaperones. Enhancements in protein expression can be achieved through reducing the rate of transcription, either by substituting a strong promoter with a weaker one [9,10], or by weakening a strong promoter by introducing a point mutation[11]. An alternative approach is to reduce the levels of polymerase in the host cell. For example, the levels of the T7 DNA polymerase expressed in E. coli can be modulated by altering the expression levels of the natural inhibitor T7 lysozyme, which is under the control of a tightly regulated inducible promoter, hence fine-tuning the rates of transcription. Wagner et al. improved expression of 14 membrane proteins using this methodology [12].
Even when strong promoters are used, host cell factors can result in low rates of transcription. For example, during the construction of stable mammalian cell lines with the gene of interest expressed from the CMV promoter, poor expression could result from epigenetic silencing of the promoter. This can be alleviated by engineering the nuclear matrix attachment region (MAR) [13] or by combining a MAR with a mammalian replication initiation region (IR) [14,15], consequently improving recombinant protein production in mammalian cell lines .
Translation of the gene of interest may also be inhibited by host cell silencing processes during protein production. For example, eukaryotic translation initiation factor 2 may become phosphorylated after DNA plasmid transfection or upon virus transduction, which will inhibit translation and thus decrease protein expression. However, viruses have evolved mechanisms to circumvent this. Gantke et al. co-expressed the Ebola virus protein 35, which is a viral protein that prevents translational silencing, and increased recombinant protein production by 10-fold [16]. An alternative approach to circumvent translational silencing in insect cells following baculovirus infection is to co-express eIF4E, which resulted in a 2-fold increase in the production of a secreted alkaline phosphatase (SEAP)-EGFP fusion protein (SEFP)[17].
Folding and secretory pathway engineering
Molecular chaperones have been applied to improve protein production in various systems, where they act to preserve nascent proteins in a folding-competent conformation and prevent aggregation [18]. The most extensively used chaperone systems that have facilitated protein production in E.coli are DnaK-DnaJ-GrpE and GroEL-GroES[19,20]. In insect cells, host protein biosynthesis shuts down as a result of infection by the recombinant baculovirus, which can adversely affect levels of molecular chaperones important for the folding of secreted proteins and membrane proteins in the endoplasmic reticulum (ER), particularly in relation to the high levels of protein synthesis resulting from high mRNA levels produced from the polyhedrin promoter. Hence, co-expression of the membrane-bound molecular chaperone calnexin enhanced the expression of functional serotonin transporter (SERT) by nearly 3 fold [21], and co-expression of the soluble molecular chaperone calreticulin increased secretion of SEFP in insect cells [17]. Whether a lack of appropriate molecular chaperones in heterologous systems contributes to low levels of functional protein sometimes is difficult to assess. However, overproduction of mammalian calnexin in the yeast Hansenula polymorpha did increase production of the truncated glycoprotein of rabies virus[22], suggesting that at least in this case the folding environment in the yeast ER was not optimal for folding large amounts of glycoprotein.
However, co-expression of molecular chaperones is not a panacea and does not often give a 10-fold or more improvement in expression levels. Part of the problem is that overexpression of ER resident chaperones such as calreticulin might burden the ER and activate an unfolded protein response [17]. Another more challenging issue is that molecular chaperones may act in a concerted fashion to promote protein folding in a poorly understood process, suggesting that it may be best to overexpress multiple chaperones simultaneously. However, expression levels will need to be tightly controlled to prevent overwhelming the cells protein production resource and also the stoichiometry between chaperones will have to be regulated. Another problem associated with engineering the chaperone and secretory pathway is that it can be protein and host specific. For example, co-expression of protein disulfide isomerase increased yields of albumin fusion proteins in the yeast Pichia pastoris[23] but did not improve functional SERT expression in insect cells [21]. Similarly, SRP 14 overexpression led to a substantial improvement of IgG production in CHO cells, but the strategy was ineffective in human cell lines producing alkaline phosphatase [24,25].
An alternative strategy to overexpressing molecular chaperones is to delete endogenous competing chaperones in order to channel the nascent peptide chain to the desired signal recognition particle (SRP) secretory pathway. Indeed, Nannenga et al. showed that membrane protein insertion in E. coli improved and expression levels increased through eliminating competition between trigger factor (TF) and the Signal Recognition Particle (SRP)for the nascent polypeptide chain [11,26].
Another strategy to improve secretion is to improve vesicular trafficking from the ER to the cell surface. Co-expression of secretory proteins which modulate vesicle trafficking, such as soluble NSF receptor (SNARE) proteins (SNAP-23 or VAMP8), improved production of SEAP and monoclonal antibodies by 2-3 fold in mammalian CHO-K1 cells [27]. Likewise, overexpression of SNARE-interacting Sec1p and Sly1p proteins improved expression of α-amylase and human insulin precursor in Saccharomyces cerevisiae[28]. In addition, the ceramide transfer protein S132A mutant improved production of tissue-plasminogen activator (t-PA) [29], human serum albumin (HSA) and monoclonal antibodies in CHO [30].
Protein sequence mutagenesis
Mutating the sequence of the protein target can also improve expression levels of the target protein. Sometimes this may be achieved through rational approaches such as analyzing the structure of the protein, as in the D500G mutation of laccase in E.coli [31] and the cysteine mutation of coagulation factor VIII [32]. However, in many instances there is insufficient evidence to suggest why a protein does not overexpress, so high-throughput mutagenic strategies can be used. For example, directed evolution coupled with random mutagenesis, followed by screening and selection was used by Sarkar et al. to evolve a GPCR, the rat neurotensin receptor (NTR) in E. coli. A mutant with 14 nucleotide substitutions retained the biochemical properties of the wild type receptor together with a 10-fold increase in functional expression and slightly increased thermostability [33]. Similarly, Heggeset et al. applied combinatorial mutagenesis and selection based on ampicillin tolerance in E.coli to evolve the signal sequence of β-lactamase and improved SEAP production up to 8-fold [34].
In theory, a more elegant and simple strategy would be to use in vivo mutagenesis coupled to screening or selection to improve expression. This approach was used by Majors et al. to evolve an anti-apoptotic gene Bcl-xL in a mammalian expression system by harnessing the somatic hypermutation capability of human Ramos B-cell line. The Bcl-xL gene, coupled to the YFP reporter, was mutated “in situ” and subjected to rounds of staurosporine treatment to identify mutants with reduced apoptosis activation and higher YFP-Bcl-xL expression levels [35].
Cell proliferation and survival engineering
The delay or prevention of the apoptosis cascade activation has been successful in preventing cell death and improving protein production in CHO cells under stress conditions [36]. Co-expression of the anti-apoptotic protein Bcl-xL in CHO cells improved the expression of epidermal growth factor receptor, fibroblast growth factor receptor 3 and receptor tyrosine kinases proteins [37]. Knock-out of the genes encoding the pro-apoptotic factors Bax and Bak in a CHO-K1 cell line improved cell viability, reduced levels of transfection-induced apoptosis and led to up to 4 fold higher antibody titers [38]. Similarly, knock-out of the pro-apoptotic microRNA mmu-miR-466h-5p in CHO cells delayed the onset of apoptosis, increased the maximum viable cell density and enhanced expression of SEAP [39].
Enhanced cell proliferation represents another potential approach to increase biomass and obtain higher volumetric yield during large scale production processes. For example, a metabolically engineered respiratory strain of S. cerevisiae (TM6*) doubled volumetric yield of Fps1 and at least quadrupled the yield of two human GPCRs (A2aR and CNR2)[40]. Overexpression of the mammalian target of rapamycin (mTOR) simultaneously improved cell growth, proliferation, viability and specific productivity of antibody, SEAP and secreted α-amylase in CHO cells [41]. Similarly, overexpression of miR-7 in CHO cells enhanced cell proliferation, leading to higher Epo-Fc titer [42]. However, accumulated biomass does not always lead to increased production as demonstrated by chemical inhibition of autophagy in CHO cells, which led to decreased cell concentration but a 2.8 fold increase in t-PA [43].
Other strategies
In cases where the heterologous proteins are toxic to the host cells, the presence of inhibitors can protect the host by sequestering proteins and keeping them in an inactive state. For example, co-expression of lysozyme together with its inhibitor Ivy, repressed lysozyme lytic activity in cytoplasm, and, along with transcription enhancement and chaperone co-expression, remarkably improved soluble lysozyme production in E.coli[44].
Conclusion
Recombinant protein expression has facilitated biochemical and structural studies of thousands of naturally low abundance proteins. Methodologies that improve expression levels can be particularly advantageous for many difficult-to-produce proteins or if the protein is being produced for therapeutic or industrial purposes. To improve expression levels further through cell engineering requires an understanding of both the host organism and the biology of protein expression. Considerable effort has been focused on engineering E.coli and yeast strains, and now there is an expanding effort to engineer insect and mammalian hosts such as HEK293 and CHO cell lines [45-47], especially for functional expression of mammalian membrane proteins that include particularly complex folding, assembly, and processing pathways [48-50].However, in many instances there is only limited information on the factors that affect expression of any particular protein, so current strategies are often piecemeal and focus on only one or two aspects of the protein production process. A goal for the future is robust cell factories generated through a holistic approach that considers all the bottlenecks in the protein expression process such as transcription, translation, protein folding, secretion and cell viability and engineer these through an integrative process to enable high-level expression of a wide spectrum of target proteins.
Highlights.
Cell engineering of bacteria, fungi, insect cells and mammalian cells is a successful strategy to improve protein expression.
Strategies have focused on optimizing transcription, translation, folding and secretion.
Improved protein expression has been observed particularly for complexes, membrane proteins and secreted proteins
Further work to combine engineering strategies in a holistic approach to improving protein expression are needed.
Acknowledgements
Funding was provided by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health and NIH grant [5R01GM095685-03].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Francis DM, Page R. Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci. 2011;Chapter 5:Unit 5 24:21–29. doi: 10.1002/0471140864.ps0524s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. doi: 10.1016/S0076-6879(09)63014-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv. 2012;30:1158–1170. doi: 10.1016/j.biotechadv.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Andrell J, Tate CG. Overexpression of membrane proteins in mammalian cells for structural studies. Mol Membr Biol. 2013;30:52–63. doi: 10.3109/09687688.2012.703703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bill RM, Henderson PJ, Iwata S, Kunji ER, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nat Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 6.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 7.Tate CG. Overexpression of mammalian integral membrane proteins for structural studies. FEBS Lett. 2001;504:94–98. doi: 10.1016/s0014-5793(01)02711-9. [DOI] [PubMed] [Google Scholar]
- 8.Quilici LS, Silva-Pereira I, Andrade AC, Albuquerque FC, Brigido MM, Maranhao AQ. A minimal cytomegalovirus intron A variant can improve transgene expression in different mammalian cell lines. Biotechnol Lett. 2013;35:21–27. doi: 10.1007/s10529-012-1043-z. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, Liu Y, Li Q, Yang J. High activity expression of D-amino acid oxidase in Escherichia coli by the protein expression rate optimization. Protein Expr Purif. 2013;88:120–126. doi: 10.1016/j.pep.2012.11.020. • Expression rate of D-amino acid oxidase(DAAO) was optimized by rational promoter recombination and delicate RBS substitution in E.coli, contributing to a 20-fold increase of DAAO activity.
- 10.Lin CH, Jarvis DL. Utility of temporally distinct baculovirus promoters for constitutive and baculovirus-inducible transgene expression in transformed insect cells. J Biotechnol. 2013;165:11–17. doi: 10.1016/j.jbiotec.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nannenga BL, Baneyx F. Enhanced expression of membrane proteins in E. coli with a P(BAD) promoter mutant: synergies with chaperone pathway engineering strategies. Microb Cell Fact. 2011;10:105. doi: 10.1186/1475-2859-10-105. • A downregulated PBAD promoter mutant was used in conjunction with an E.coli strain lacking the molecular chaperone Trigger Factor for membrane protein expression. 4-fold increase in productivity was achieved with both GPCRs tested.
- 12.Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Hogbom M, van Wijk KJ, Slotboom DJ, Persson JO, et al. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandjean M, Girod PA, Calabrese D, Kostyrko K, Wicht M, Yerly F, Mazza C, Beckmann JS, Martinet D, Mermod N. High-level transgene expression by homologous recombination-mediated gene transfer. Nucleic Acids Res. 2011;39:e104. doi: 10.1093/nar/gkr436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura H, Sekine S, Adachi H, Uematsu Y, Mitani A, Futaki N, Shimizu N. High levels of human recombinant cyclooxygenase-1 expression in mammalian cells using a novel gene amplification method. Protein Expr Purif. 2011;80:41–46. doi: 10.1016/j.pep.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Araki Y, Hamafuji T, Noguchi C, Shimizu N. Efficient recombinant production in mammalian cells using a novel IR/MAR gene amplification method. PLoS One. 2012;7:e41787. doi: 10.1371/journal.pone.0041787. •• A novel and efficient IR/MAR gene amplification method was introduced in mammalian cell lines, leading to a dramatic improvement in antibody production with no significant loss of stability over the passages.
- 16.Gantke T, Boussouf S, Janzen J, Morrice NA, Howell S, Muhlberger E, Ley SC. Ebola virus VP35 induces high-level production of recombinant TPL-2-ABIN-2-NF-kappaB1 p105 complex in co transfected HEK-293 cells. Biochem J. 2013;452:359–365. doi: 10.1042/BJ20121873. • Plasmid activiation of PKR(double-stranded-RNA-dependent protein kinase) during transient transfection was blocked by co-expression of Ebola virus VP35, thus substantially increased production of recombinant protein complex in HEK-293 cells.
- 17.Teng CY, Chang SL, van Oers MM, Wu TY. Enhanced protein secretion from insect cells by co-expression of the chaperone calreticulin and translation initiation factor eIF4E. Mol Biotechnol. 2012;54:68–78. doi: 10.1007/s12033-012-9545-4. [DOI] [PubMed] [Google Scholar]
- 18.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 19.Folwarczna J, Moravec T, Plchova H, Hoffmeisterova H, Cerovska N. Efficient expression of Human papillomavirus 16 E7 oncoprotein fused to C-terminus of Tobacco mosaic virus (TMV) coat protein using molecular chaperones in Escherichia coli. Protein Expr Purif. 2012;85:152–157. doi: 10.1016/j.pep.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Voulgaridou GP, Mantso T, Chlichlia K, Panayiotidis MI, Pappa A. Efficient E. coli expression strategies for production of soluble human crystallin ALDH3A1. PLoS One. 2013;8:e56582. doi: 10.1371/journal.pone.0056582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate CG, Whiteley E, Betenbaugh MJ. Molecular chaperones stimulate the functional expression of the cocaine-sensitive serotonin transporter. J Biol Chem. 1999;274:17551–17558. doi: 10.1074/jbc.274.25.17551. [DOI] [PubMed] [Google Scholar]
- 22.Qian W, Aguilar F, Wang T, Qiu B. Secretion of truncated recombinant rabies virus glycoprotein with preserved antigenic properties using a co-expression system in Hansenula polymorpha. J Microbiol. 2013;51:234–240. doi: 10.1007/s12275-013-2337-0. [DOI] [PubMed] [Google Scholar]
- 23.Shen Q, Wu M, Wang HB, Naranmandura H, Chen SQ. The effect of gene copy number and co-expression of chaperone on production of albumin fusion proteins in Pichia pastoris. Appl Microbiol Biotechnol. 2012;96:763–772. doi: 10.1007/s00253-012-4337-0. • Increasing gene copy number and co-expression of protein disulfide isomerase improved expression of two albumin fusion proteins in P. pastoris.
- 24.Le Fourn V, Girod PA, Buceta M, Regamey A, Mermod N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab Eng. 2012 doi: 10.1016/j.ymben.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nannenga BL, Baneyx F. Reprogramming chaperone pathways to improve membrane protein expression in Escherichia coli. Protein Sci. 2011;20:1411–1420. doi: 10.1002/pro.669. •• The chaperone pathway was reprogrammed in E. coli through overexpression of desired chaperones (SRP and YidC) together with deletion of competing chaperone (TF). This strategy was used to improve production of 3 model membrane proteins.
- 27.Peng RW, Abellan E, Fussenegger M. Differential effect of exocytic SNAREs on the production of recombinant proteins in mammalian cells. Biotechnol Bioeng. 2010;108:611–620. doi: 10.1002/bit.22986. [DOI] [PubMed] [Google Scholar]
- 28.Hou J, Tyo K, Liu Z, Petranovic D, Nielsen J. Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab Eng. 2012;14:120–127. doi: 10.1016/j.ymben.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Rahimpour A, Vaziri B, Moazzami R, Nematollahi L, Barkhordari F, Kokabee L, Adeli A, Mahboudi F. Engineering the Cellular Protein Secretory Pathway for Enhancement of Recombinant Tissue Plasminogen Activator Expression in Chinese Hamster Ovary Cells: Effects of CERT and XBP1s Genes. J Microbiol Biotechnol. 2013;23:1116–1122. doi: 10.4014/jmb.1302.02035. [DOI] [PubMed] [Google Scholar]
- 30.Florin L, Pegel A, Becker E, Hausser A, Olayioye MA, Kaufmann H. Heterologous expression of the lipid transfer protein CERT increases therapeutic protein productivity of mammalian cells. J Biotechnol. 2009;141:84–90. doi: 10.1016/j.jbiotec.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Nasoohi N, Khajeh K, Mohammadian M, Ranjbar B. Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement. Int J Biol Macromol. 2013;60:56–61. doi: 10.1016/j.ijbiomac.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Selvaraj SR, Scheller AN, Miao HZ, Kaufman RJ, Pipe SW. Bioengineering of coagulation factor VIII for efficient expression through elimination of a dispensable disulfide loop. J Thromb Haemost. 2012;10:107–115. doi: 10.1111/j.1538-7836.2011.04545.x. • A dispensable disulfide loop of coagulation factor VIII was identified and mutated to improve secretion. It was combined with other expression-improving point mutations and the additive effect resulted in 35-45 fold higher production of FVIII in CHO cells.
- 33.Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, Pluckthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci U S A. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heggeset TM, Kucharova V, Naerdal I, Valla S, Sletta H, Ellingsen TE, Brautaset T. Combinatorial mutagenesis and selection of improved signal sequences and their application for high-level production of translocated heterologous proteins in Escherichia coli. Appl Environ Microbiol. 2012;79:559–568. doi: 10.1128/AEM.02407-12. •• The previously designed consensus signal peptide was subject to combinatorial mutagenesis and selection based on ampicillin tolerance in E.coli. The selected signal sequences significantly improved expression of β-lactamase and alkaline phosphatase.
- 35.Majors BS, Chiang GG, Pederson NE, Betenbaugh MJ. Directed evolution of mammalian anti-apoptosis proteins by somatic hypermutation. Protein Eng Des Sel. 2011;25:27–38. doi: 10.1093/protein/gzr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majors BS, Betenbaugh MJ, Chiang GG. Links between metabolism and apoptosis in mammalian cells: applications for anti-apoptosis engineering. Metab Eng. 2007;9:317–326. doi: 10.1016/j.ymben.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Ohsfeldt E, Huang SH, Baycin-Hizal D, Kristoffersen L, Le TM, Li E, Hristova K, Betenbaugh MJ. Increased expression of the integral membrane proteins EGFR and FGFR3 in anti-apoptotic Chinese hamster ovary cell lines. Biotechnol Appl Biochem. 2012;59:155–162. doi: 10.1002/bab.1000. [DOI] [PubMed] [Google Scholar]
- 38.Macaraeg NF, Reilly DE, Wong AW. Use of an anti-apoptotic CHO cell line for transient gene expression. Biotechnol Prog. 2013;29:1050–1058. doi: 10.1002/btpr.1763. • Two pro-apoptotic factors were deleted for construction of an anti-apoptotic CHO-K1 cell line. A polyethylenimine transient transfection system was developed using this cell line and produced 3- to 4- fold higher antibody titers.
- 39.Druz A, Son YJ, Betenbaugh M, Shiloach J. Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab Eng. 2013;16:87–94. doi: 10.1016/j.ymben.2012.12.004. • Stable inhibition of a pro-apoptotic microRNA in CHO cells led to delayed apoptosis onset, higher maximum viable cell density, increased integral viable cells and improved SEAP production.
- 40.Ferndahl C, Bonander N, Logez C, Wagner R, Gustafsson L, Larsson C, Hedfalk K, Darby RA, Bill RM. Increasing cell biomass in Saccharomyces cerevisiae increases recombinant protein yield: the use of a respiratory strain as a microbial cell factory. Microb Cell Fact. 2010;9:47. doi: 10.1186/1475-2859-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreesen IA, Fussenegger M. Ectopic expression of human mTOR increases viability, robustness, cell size, proliferation, and antibody production of chinese hamster ovary cells. Biotechnol Bioeng. 2010;108:853–866. doi: 10.1002/bit.22990. [DOI] [PubMed] [Google Scholar]
- 42.Jadhav V, Hackl M, Bort JA, Wieser M, Harreither E, Kunert R, Borth N, Grillari J. A screening method to assess biological effects of microRNA overexpression in Chinese hamster ovary cells. Biotechnol Bioeng. 2012;109:1376–1385. doi: 10.1002/bit.24490. [DOI] [PubMed] [Google Scholar]
- 43.Jardon MA, Sattha B, Braasch K, Leung AO, Cote HC, Butler M, Gorski SM, Piret JM. Inhibition of glutamine-dependent autophagy increases t-PA production in CHO cell fed-batch processes. Biotechnol Bioeng. 2012;109:1228–1238. doi: 10.1002/bit.24393. • In fed-batch processes, chemical inhibition of autophagy with 3-methyl adenine increased the t-PA yield by 2.8-fold, without compromising the glycosylation capacity of cells.
- 44.Lamppa JW, Tanyos SA, Griswold KE. Engineering Escherichia coli for soluble expression and single step purification of active human lysozyme. J Biotechnol. 2012;164:1–8. doi: 10.1016/j.jbiotec.2012.11.007. •• In order to overexpress soluble functional lysozyme in E.coli, its inhibitor Ivy was co-expressed to repress lysozyme lytic activity in cytoplasm. Multiple chaperones were co-expressed, contributing to a 3000- fold improvement in soluble lysozyme production.
- 45.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dostalova Z, Liu A, Zhou X, Farmer SL, Krenzel ES, Arevalo E, Desai R, Feinberg-Zadek PL, Davies PA, Yamodo IH, et al. High-level expression and purification of Cys-loop ligand-gated ion channels in a tetracycline-inducible stable mammalian cell line: GABAA and serotonin receptors. Protein Sci. 2010;19:1728–1738. doi: 10.1002/pro.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao S, White JF, Betenbaugh MJ, Grisshammer R, Shiloach J. Transient and stable expression of the neurotensin receptor NTS1: a comparison of the baculovirus-insect cell and the T-REx-293 expression systems. PLoS One. 2013;8:e63679. doi: 10.1371/journal.pone.0063679. • A tetracycline-inducible HEK-293 cell line was shown to substantially improve functional expression level of neurotensin receptor. This mammalian expression system was quantitatively compared with baculovirus- insect cell system throughout expression and purification process.
- 48.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruswitz F, Chaudhary S, Ho JD, Schlessinger A, Pezeshki B, Ho CM, Sali A, Westhoff CM, Stroud RM. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A. 2010;107:9638–9643. doi: 10.1073/pnas.1003587107. [DOI] [PMC free article] [PubMed] [Google Scholar]