SUMMARY
Brown and beige/brite fats generate heat via uncoupled respiration to defend against cold. The total mass and activity of thermogenic adipose tissues are also tightly linked to systemic energy and nutrient homeostasis. Despite originating from distinct progenitors, brown and beige adipocytes acquire remarkably similar molecular and metabolic characteristics during differentiation through the action of a network of transcription factors and cofactors. How this regulatory network interfaces with long noncoding RNAs (lncRNAs), an emerging class of developmental regulators, remains largely unexplored. Here we globally profiled lncRNA gene expression during thermogenic adipocyte formation and identified Brown fat lncRNA 1 (Blnc1) as a novel nuclear lncRNA that promotes brown and beige adipocyte differentiation and function. Blnc1 forms a ribonucleoprotein complex with transcription factor EBF2 to stimulate the thermogenic gene program. Further, Blnc1 itself is a target of EBF2, thereby forming a feedforward regulatory loop to drive adipogenesis toward thermogenic phenotype.
INTRODUCTION
Metabolic syndrome has become a global epidemic that raises the risk for type 2 diabetes, cardiovascular disease, and non-alcoholic fatty liver disease. White adipose tissue (WAT) is important for energy storage, endocrine signaling, and metabolic-immune crosstalk (Gesta et al., 2007; Rosen and Spiegelman, 2014), whereas brown adipose tissue (BAT) contains abundant mitochondria and expresses high levels of uncoupling protein 1 (UCP1), an inner mitochondrial membrane protein that dissipates proton gradient for heat generation (Kozak and Harper, 2000; Lowell and Spiegelman, 2000; Nedergaard and Cannon, 2010). Brown fat thermogenesis defends against cold and contributes to energy expenditure. Genetic ablation of brown fat or deletion of Ucp1 renders mice cold-sensitive and prone to high-fat diet-induced obesity (Enerback et al., 1997; Feldmann et al., 2009; Lowell et al., 1993), whereas activation of brown fat thermogenesis by cold exposure has been linked to increased energy expenditure, reduced adiposity, and lower plasma lipids (Bartelt et al., 2011; van der Lans et al., 2013; Yoneshiro et al., 2013). Recent work has demonstrated that brown-like fat is present in adult humans (Cypess et al., 2009; Nedergaard et al., 2007; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009), and responds to physiological and environmental stimuli (Orava et al., 2011; Ouellet et al., 2012; van der Lans et al., 2013). As such, augmenting brown fat abundance and/or function may provide a potentially effective treatment for obesity and its associated metabolic disorders.
A hallmark of brown adipocyte differentiation is transcriptional activation of gene programs underlying mitochondrial fuel oxidation and thermogenesis. A number of transcription factors and cofactors, including peroxisome proliferator-activated receptor γ (Pparγ), Pparα, PR domain containing 16 (Prdm16), early B cell factor 2 (Ebf2), CCAAT/enhancer binding protein β (C/EBPβ), and Pparγ coactivator 1α and 1β (PGC-1α and PGC-1β), have been identified to regulate different aspects of brown fat development (Farmer, 2008; Kajimura et al., 2010; Wu et al., 2013). Further, several microRNAs have been identified to regulate brown adipocyte determination, differentiation, and uncoupling (Trajkovski and Lodish, 2013). While sharing key molecular and metabolic characteristics with the classical rodent BAT, brown fat in adult humans appears to contain both classical and brown-like adipocytes (Cypess et al., 2013; Jespersen et al., 2013; Lidell et al., 2013; Sharp et al., 2012). UCP1-positive adipocytes also emerge in WAT in response to certain stimuli, such as rosiglitazone and CL-316,243, a selective β3-adrenergic agonist; these potential thermogenic adipocytes have been termed beige, brite, or inducible brown adipocytes (referred hereafter as beige), but their exact identity remains uncertain (Petrovic et al., 2010; Schulz et al., 2011; Wu et al., 2012). Like brown adipocytes, beige adipocytes also have high mitochondrial content and express Ucp1 in an inducible manner. In rodents, brown and beige fats appear to have distinct developmental origins (Berry and Rodeheffer, 2013; Seale et al., 2008; Timmons et al., 2007; Wu et al., 2012), and are differentially influenced by genetic factors (Kozak and Koza, 2010; Xue et al., 2007).
Long non-coding RNAs (lncRNAs) are a unique class of transcripts that share similarities with mRNA with regard to their transcriptional regulation and biogenesis (Atkinson et al., 2012; Rinn and Chang, 2012). Like mRNA, lncRNAs are transcribed by RNA polymerase II and undergo splicing and polyadenylation. However, these RNA transcripts lack functional open reading frames and are not predicted to encode proteins. The expression of lncRNAs exhibits remarkable tissue specificity and is responsive to developmental and physiological signals (Gupta et al., 2010; Huarte et al., 2010; Pauli et al., 2012; Sun et al., 2013; Vollmers et al., 2012). Through interaction with DNA/RNA or proteins, lncRNAs regulate epigenetic states of chromatin, gene expression, tissue development, and tumorigenesis (Guttman and Rinn, 2012; Hu et al., 2012; Lee, 2012; Wang and Chang, 2011). Recent studies demonstrated that lncRNAs, including steroid receptor RNA activator, play a role in white adipocyte differentiation (Sun et al., 2013; Xu et al., 2010). However, the extent to which and the significance of lncRNAs in thermogenic adipocyte differentiation have not been explored. In this study, we defined a set of highly regulated lncRNAs in brown fat and identified Blnc1 as an inducible lncRNA that regulates brown and beige adipocyte differentiation. Mechanistically, Blnc1 physically interacts with EBF2 and promotes the induction of the thermogenic gene program via a feedforward regulatory loop.
RESULTS
Identification of Blnc1 as an inducible lncRNA during brown adipogenesis
To identify inducible lncRNAs involved in brown fat development and thermogenesis, we performed transcriptional profiling in adipose tissues and during brown adipocyte (BAC) differentiation using chips that cover 25,376 protein-coding transcripts and 31,423 annotated lncRNAs. These analyses revealed a cluster of 30 coding genes and 21 lncRNAs whose expression were enriched in BAT, induced in epididymal WAT (eWAT) by CL-316,243, a selective β3-adrenergic agonist, and increased during BAC differentiation (Figure 1A). As expected, the protein-coding gene set included those participating in mitochondrial fuel oxidation and uncoupling, such as long-chain acyl-CoA dehydrogenase (Acadl), cytochrome c oxidase subunit 7a polypeptide 1 (Cox7a1), cell death-inducing DFFA-like effector a (Cidea) and Ucp1, and was enriched for Gene Ontology (GO) terms associated with mitochondrial organelle and function (Supplementary Table S1 and Figure S1A). The noncoding gene set included 10 intergenic lncRNAs and 11 lncRNAs located near mRNA transcripts (Supplementary Table S1). We focused on three highly conserved intergenic lncRNAs (AK080070, 3930402G23Rik, AK038898) and examined their role in brown adipogenesis by retroviral expression of short hairpin RNA (shRNA) targeting individual lncRNA (data not shown). RNAi knockdown of AK038898 significantly impaired brown adipogenesis, and as such, we renamed this lncRNA Brown fat lncRNA 1 (Blnc1).
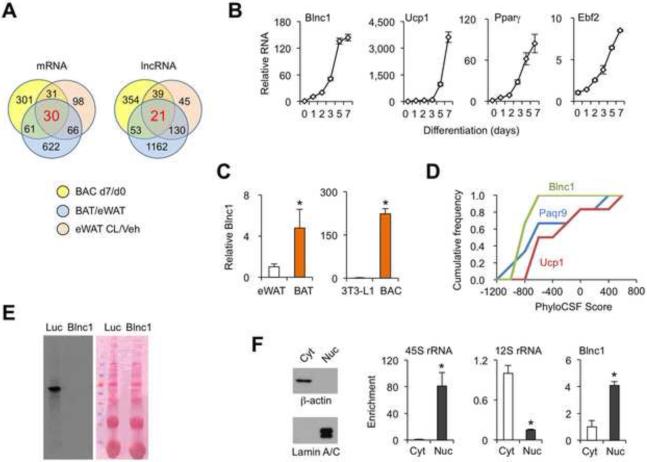
Figure 1. Identification of Blnc1 as an inducible lncRNA in brown fat.
(A) Venn diagrams of protein-coding (left) and lncRNA (right) genes induced during brown adipocyte differentiation (BAC d7/d0), enriched in BAT vs. eWAT, and induced in eWAT by CL-316,243 (CL/veh).
(B) qPCR analyses of gene expression during BAC differentiation.
(C) qPCR analyses of Blnc1 expression in adipose tissues and differentiated adipocytes.
(D) PhyloCSF analysis of Blnc1, Paqr9 and Ucp1.
(E) In vitro transcription/translation assay using luciferase (Luc) and Blnc1 constructs. Shown are 35S autoradiograph (left) and Ponceau S stained blot (right).
(F) Immunoblotting and qPCR analyses of cytosolic (Cyt) and nuclear (Nuc) fractions. Data represent mean ± sd. *p<0.05 eWAT vs. BAT; 3T3-L1 vs. BAC (C); and Cyt vs. Nuc (F). (See also Figure S1 and S2)
Blnc1 expression was highly induced during brown adipocyte differentiation along with known adipogenic markers, such as Ucp1, Pparγ, and Ebf2, a transcriptional regulator of brown fat development (Figure 1B). Blnc1 RNA was also enriched in brown fat compared to white fat (Figure 1C), consistent with recent RNA sequencing (RNA-seq) studies (Sun et al., 2013), and reached higher levels in differentiated brown adipocytes compared to 3T3-L1 adipocytes. Expression of AK080070 and 3930402G23Rik exhibited similar patterns (Supplementary Figure S1B). Rapid Amplification of cDNA Ends (RACE) revealed that Blnc1 transcript was polyadenylated and transcribed from a single exon of approximately 965 bp in length (Supplementary Figure S2A-B). We performed phylogenetic information-based codon substitution frequency (PhyloCSF) analyses, a comparative genomic tool that distinguishes protein-coding from non-coding transcripts (Lin et al., 2011). While Ucp1 and Paqr9, the latter being a protein-coding gene near the Blnc1 locus, were predicted to encode proteins, Blnc1 RNA had a low probability of containing a protein-coding open reading frame (Figure 1D). Further, a Blnc1 expression vector failed to produce a protein(s) using in vitro transcription/translation assay (Figure 1E). Analysis of a global RNA sequencing and ribosome footprinting dataset (Ingolia et al., 2011) indicated that Blnc1 RNA was largely free of ribosome association (Supplementary Figure S2C).
LncRNAs are targeted to discrete subcellular locations to carry out their biological functions. QPCR analyses of fractionated nuclear and cytoplasmic RNA indicated that Blnc1 was primarily localized in the nuclear compartment (Figure 1F). As control, β-actin and Lamin A/C proteins were detected exclusively in the cytoplasmic and nuclear fractions, respectively. In addition, 45S ribosomal RNA (rRNA) precursor was primarily localized in the nucleus, whereas 12S rRNA, a mitochondrial rRNA, was found in the cytoplasmic fraction.
Blnc1 stimulates the thermogenic gene program in brown adipocytes and in vivo
To investigate the role of Blnc1 in brown adipocyte differentiation, we transduced brown preadipocytes with control or a retroviral vector (pMSCV) expressing Blnc1 cDNA and monitored adipocyte gene expression at different time points following adipogenic induction. While Pparγ was similarly induced, retroviral-mediated overexpression of Blnc1 significantly increased mRNA expression of Ucp1, Cox7a1, Prdm16, Pparα, and Ebf2 (Figure 2A). Microarray analyses of differentiated adipocytes indicated that Blnc1 increased the expression of a cluster of mitochondrial genes involved in fatty acid β-oxidation, including mitochondrial trans-2-enoyl-CoA reductase (Mecr), acyl-CoA thioesterase 2 (Acot2), and acetyl-CoA acyltransferase 2 (Acaa2), and glucose metabolism, such as pyruvate dehydrogenase kinase 1 (Pdk1) and Pdk4 (Figure 2B). Gene ontology analysis revealed that mitochondrion was among the top GO terms associated with Blnc1-inducible genes (Supplementary Figure S3A). The induction of thermogenic gene expression correlated with increased histone H3 acetylation (Supplementary Figure S3B). Further, mRNA expression of key thermogenic markers, including Ucp1, Deiodinase 2 (Dio2), and Pparα, was higher in Blnc1-expressing adipocytes after CL-316,243 treatments (Figure 2C). UCP1 protein levels were also increased in Blnc1 overexpressing adipocytes at basal state and following treatments with CL-316,243, norepinephrine, or forskolin, which increased intracellular cAMP levels (Figure 2D), whereas FABP4 and PPARγ protein levels remained similar.
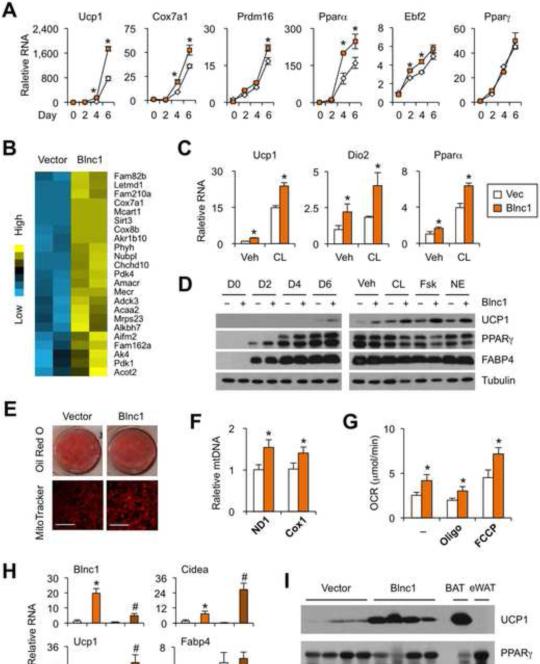
Figure 2. Blnc1 promotes brown adipocyte differentiation.
(A) qPCR analyses of gene expression during differentiation of brown preadipocytes transduced with vector (open) or Blnc1 (orange) retrovirus.
(B) Heat map of mitochondrial genes induced by Blnc1.
(C) qPCR analyses of adipocyte genes following treatments with vehicle (veh) or CL-316,243 (CL).
(D) Immunoblots of adipocyte lysates during differentiation (left) and following treatments with veh, CL, forskolin (Fsk), or norepinephrine (NE) for 4 hrs (right).
(E) Oil Red O and MitoTracker staining of differentiated brown adipocytes. Scale bar=50 μM.
(F) qPCR analyses of mitochondrial DNA content in vector (open) and Blnc1 (orange) expressing adipocytes.
(G) Oxygen consumption rate (OCR) in the absence or presence of oligomycin (Oligo) or FCCP.
(H) qPCR analyses of gene expression in fat pads formed from transplanted preadipocytes transduced with vector or Blnc1 retrovirus. BAT and eWAT from transplanted mice were used as control.
(I) Immunoblotting analyses of transplanted fats. Data represent mean ± sd. *p<0.05 Blnc1 vs. vector (A, C, F, G, H); #p<0.05 BAT vs. eWAT. (See also Figure S3)
Oil Red O staining revealed that Blnc1 overexpression had modest effects on lipid accumulation in differentiated adipocytes (Figure 2E). In contrast, mitochondrial mass and DNA content were increased in Blnc1-expressing adipocytes (Figure 2E-F). Measurements of oxygen consumption rate (OCR) in differentiated adipocytes indicated that Blnc1 increased total respiratory capacity when measured in the presence of carbonyl cyanide-ptrifluoromethoxyphenylhydrazone (FCCP), a chemical uncoupler that dissipates the proton gradient across mitochondrial inner membrane (Figure 2G). In addition, basal OCR and uncoupled respiration, the latter measured in presence of oligomycin, an inhibitor of mitochondrial ATP synthase, were also higher in adipocytes overexpressing Blnc1. As such, retroviral-mediated Blnc1 expression is sufficient to stimulate mitochondrial gene expression and increase oxidative capacity and uncoupled respiration in brown adipocytes.
To further assess the effects of Blnc1 on brown adipocyte differentiation in vivo, we performed transplantation experiments in immunodeficient nude mice. Brown preadipocytes transduced with control or Blnc1 retroviral vectors were injected subcutaneously on two sides at the base of sternum, where endogenous fat is nearly absent (Guo et al., 2009). We performed analyses two weeks after transplantation. While fat pads formed from transplanted preadipocytes expressed comparable levels of adipogenic marker Fabp4, Blnc1-transduced preadipocytes gave rise to fat pads with significantly higher Ucp1 and Cidea mRNA expression, reaching approximately 30% of the levels observed in endogenous brown fat (Figure 2H). UCP1 protein levels were also markedly higher in fat pads expressing Blnc1 (Figure 2I). Together, these gain-of-function studies strongly implicated Blnc1 as a lncRNA activator of brown adipogenesis and brown fat formation.
Blnc1 is required for brown adipocyte differentiation
We next performed RNAi knockdown studies to assess the significance of Blnc1 in BAC differentiation using two independent shRNA vectors targeting Blnc1. We stably transduced brown preadipocytes with control or Blnc1 shRNA retroviruses and subjected transduced cells to adipogenic induction. As expected, Blnc1 shRNA expression significantly decreased of Blnc1 levels in both nuclear and cytoplasmic fractions (Supplementary Figure S4A). RNAi knockdown of Blnc1 in brown preadipocytes severely impaired adipogenesis, as revealed by reduced lipid accumulation and brown fat marker gene expression, including Ucp1, Cox7a1, and Ebf2 (Figure 3A). Gene ontology analysis indicated that gene sets downregulated by Blnc1 knockdown were enriched for mitochondrial metabolism (Supplementary Figure S4B). Consistently, Ucp1 mRNA and protein levels were also greatly reduced by RNAi knockdown of Blnc1 under basal and stimulated conditions (Figure 3B-D).
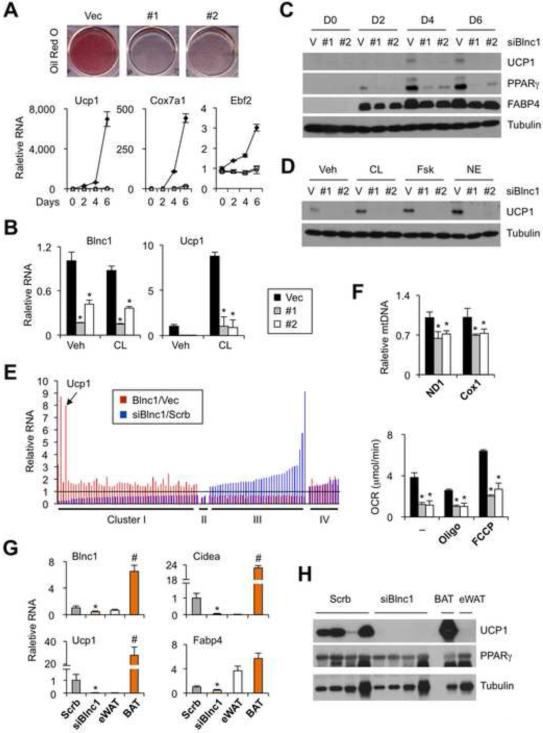
Figure 3. Blnc1 is required for brown adipogenesis.
(A) Oil Red O staining and gene expression. Brown preadipocytes transduced with vector (Vec, filled) or two independent shRNAs targeting Blnc1 (#1 and #2, open) were differentiated for 6 days.
(B) qPCR analyses in adipocytes treated with vehicle (veh) or CL-316,243 (CL).
(C) Immunoblots of total lysates during BAC differentiation.
(D) Immunoblots of UCP1 in differentiated BAC treated with vehicle (veh), CL-316,243 (CL), forskolin (Fsk), or norepinephrine (NE) for 4 hrs.
(E) Fold change of genes regulated by Blnc1 overexpression (red) or RNAi knockdown (blue). Four clusters of genes were differentially regulated by Blnc1.
(F) Mitochondrial DNA content (top) and oxygen consumption rate (OCR, bottom) in differentiated adipocytes.
(G) qPCR analyses of gene expression in fat pads formed from transplanted preadipocytes transduced with control (Scrb) or siBlnc1 retrovirus. BAT and eWAT from transplanted mice were used as control.
(H) Immunoblotting analyses of transplanted fats. Data represent mean ± sd. *p<0.05 Vec vs. #1 or #2 (B, F, G); #p<0.05 BAT vs. eWAT. (See also Figure S4)
We analyzed gene expression profile in BAC and identified a set of 110 genes that are differentially regulated by Blnc1 overexpression and knockdown (Figure 3E and Supplementary Table S2). Interestingly, the expression of Blnc1-inducible genes (cluster I) was generally lower in knockdown adipocytes; this cluster includes many genes involved in mitochondrial function and thermogenesis. In contrast, those suppressed by Blnc1 (cluster III) were increased in response to Blnc1 knockdown. Consistently, mitochondrial DNA content was significantly lower in knockdown adipocytes (Figure 3F). Total respiratory capacity and uncoupled respiration were also lower in adipocytes with depleted Blnc1 expression. Further, brown preadipocytes expressing shRNA that targeted Blnc1 failed to form fat pads characteristic of brown fat, as indicated by lower Ucp1 and Cidea expression (Figure 3G-H). Similar decrease in UCP1, but not PPARγ, protein levels was also observed in the knockdown group. These loss-of-function studies illustrate that Blnc1 is required for the activation of adipogenic and thermogenic gene programs in culture and in vivo.
Regulation of beige adipocyte differentiation by Blnc1
Beige fat is emerging as a new type of thermogenic fat that is inducible in response to cold acclimation, chronic adrenergic activation, or PPARγ agonists (Lee et al., 2012; Ohno et al., 2012; Wu et al., 2013). To determine whether Blnc1 expression is regulated during browning, we treated C57BL/6 mice with CL-316,243 daily for one week and analyzed adipose tissue expression. As expected, Ucp1 mRNA levels were markedly elevated in interscapular BAT, inguinal WAT (ingWAT), eWAT, and perirenal WAT (prWAT) (Figure 4A). In parallel to Ucp1, Blnc1 RNA expression was also significantly increased by CL-316,243 treatments for 7 days. Recent studies have begun to unravel the transcriptional signatures of brown and beige adipocytes. We noted that a probeset for Blnc1 was present on Affymetrix Mouse 430 2.0 chips and examined its levels in a microarray dataset on adipocytes differentiated from mouse white, brown, and beige preadipocyte clones (Wu et al., 2012). Compared to white adipocytes, Blnc1 RNA expression was enriched in brown and beige adipocytes, similar to that of Pgc-1α and Acox1 (Figure 4B). Similar to previous studies, Slc27a1 and Klhl13 expression was higher in beige adipocytes. We next examined Blnc1 expression during adipocyte differentiation of X9E cells, an established beige progenitor clone (Wu et al., 2012). Gene expression analyses indicated that, similar to Cidea, the expression of Blnc1 and Ebf2 was markedly induced during beige adipocyte differentiation (Figure 4C). These findings raised the possibility that Blnc1 may play a role in beige adipocyte development.
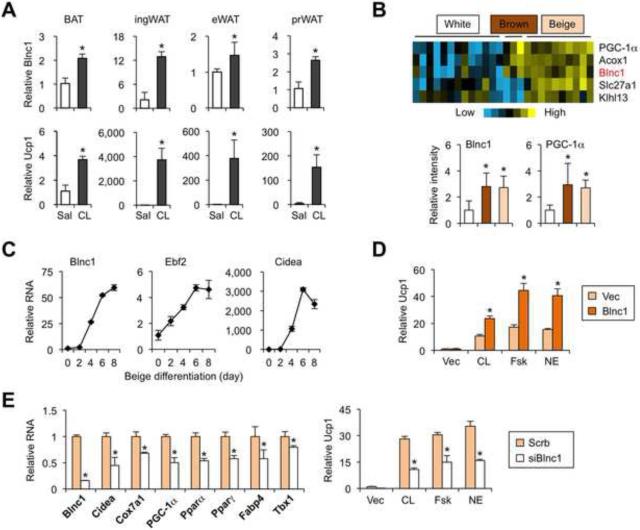
Figure 4. Blnc1 induces thermogenic gene program in beige adipocytes.
(A) qPCR analyses of Blnc1 and Ucp1 expression in BAT, inguinal WAT (ingWAT), eWAT, and perirenal WAT (prWAT) from mice treated with saline (Sal) or CL-316,243 (CL) for 7 days.
(B) Heat map of gene expression in adipocytes differentiated from preadipocyte clones of white (open), brown (brown), and beige lineages (beige). Quantitation of microarray values was shown below.
(C) qPCR analyses of gene expression during beige adipogenesis.
(D) Ucp1 expression in transduced beige adipocytes following vehicle (veh), CL, forskolin (Fsk), or norepinephrine (NE) treatments for 6 hrs.
(E) qPCR analyses of gene expression (left) and Ucp1 (right) in transduced beige adipocytes after 7 days of differentiation before (left) and after veh, CL, Fsk and NE treatment (right) for 6 hrs . Data represent mean ± sd. *p<0.05 Sal vs. CL (A); brown/beige vs. white (B); Vec vs. Blnc1 (D); Scrb vs. siBlnc1 (E, F).
We next performed Blnc1 gain- and loss-of-function studies to assess its role in beige adipocyte differentiation. Retroviral-mediated expression of Blnc1 had a modest effect on lipid accumulation in X9E cells following adipogenic induction (data not shown). As previously reported (Wu et al., 2012), basal Ucp1 expression was relatively low yet highly inducible by adrenergic stimuli. In response to CL-316,243, forskolin, or norepinephrine, Ucp1 mRNA expression reached significantly higher levels in beige adipocytes overexpressing Blnc1 (Figure 4D). RNAi knockdown of Blnc1 impaired the expression of several genes enriched in brown and beige adipocytes, including Cidea, Cox7a1, Pgc-1α, and Tbx1, a recently identified beige fat marker (Jespersen et al., 2013) (Figure 4E). The induction of Ucp1 mRNA expression was also diminished by RNAi knockdown of Blnc1. These results demonstrate that Blnc1 is sufficient and required for beige adipocytes to acquire thermogenic phenotype.
EBF2 and Blnc1 cooperatively stimulate the thermogenic adipocyte gene program
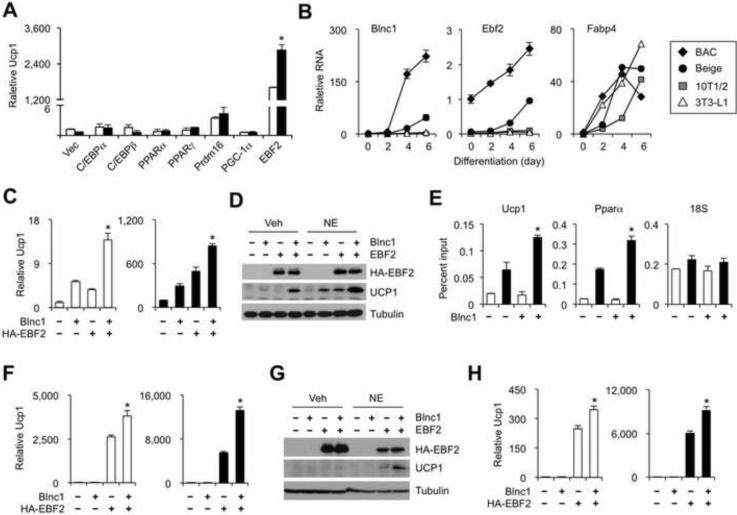
As shown above, retroviral-mediated expression of Blnc1 in brown and beige preadipocytes promoted the induction of thermogenic gene program and function, whereas Blnc1 depletion by RNAi knockdown had the opposite effects. Despite this, Blnc1 failed to increase the expression of thermogenic adipocyte markers when overexpressed in C3H10T1/2 mesenchymal stem cells and 3T3-L1 preadipocytes (Figure 5A and data not shown). These observations suggest that Blnc1 may selectively act on committed brown and beige adipocyte progenitors and/or may require the presence of one or more factors enriched in these adipocytes. To test this, we used recombinant retroviral vectors to drive Blnc1 expression in 10T1/2 fibroblasts in combination with individual factors known to regulate brown fat gene expression. As expected, overexpression of Prdm16 and Ebf2 augmented Ucp1 mRNA expression following adipogenic induction. Blnc1 further increased Ucp1 mRNA levels in the presence of EBF2, but not other factors, including C/EBPα, C/EBFβ, PPARα, PPARγ, Prdm16, and PGC-1α (Figure 5A). Gene expression analyses indicated that both Blnc1 and Ebf2 mRNA expression was increased during brown and beige adipogenesis, but not during adipocyte differentiation of 10T1/2 and 3T3-L1 cells (Figure 5B).
Figure 5. Blnc1 and EBF2 cooperatively stimulate brown adipogenesis.
(A) Ucp1 mRNA expression in adipocytes differentiated from 10T1/2 fibroblasts transduced with individual factors alone (open) or in combination with Blnc1 (filled).
(B) qPCR analyses of gene expression during adipogenesis from different preadipocytes.
(C) Ucp1 mRNA expression in adipocytes differentiated from transduced brown preadipocytes treated with vehicle (open) or NE (filled) for 4 hrs.
(D) Immunoblots of total lysates from differentiated adipocytes in C.
(E) ChIP assay of EBF2 occupancy in BAC differentiated from transduced preadipocytes using IgG (open) or EBF2 antibody (filled).
(F) Ucp1 mRNA expression in adipocytes differentiated from transduced 10T1/2 cells treated with vehicle (open) or NE (filled) for 4 hrs.
(G) Immunoblots of total lysates from differentiated adipocytes in F.
(H) Ucp1 mRNA expression in adipocytes differentiated from transduced 3T3-L1 cells treated with vehicle (open) or NE (filled) for 4 hrs. Data represent mean ± sd. *p<0.05 Vec vs. Blnc1 (A); EBF2/Blnc1 vs. EBF2 alone (C, E, F, H). (See also Figure S5)
To elucidate the cooperative action of Blnc1 and Ebf2 in adipogenesis, we generated stable preadipocyte cell lines that expressed vector, Ebf2 and Blnc1 alone, or a combination of these two factors in brown preadipocytes, 10T1/2 and 3T3-L1 fibroblasts. We analyzed gene expression following standard adipogenic induction without or with norepinephrine stimulation. Gene expression analyses indicated that co-expression of Blnc1 and EBF2 significantly increased Ucp1 gene expression when compared to adipocytes expressing these factors alone (Figure 5C-D). This stimulatory effect was also observed following adrenergic stimulation. EBF2 stimulates brown adipocyte gene expression through direct chromatin association (Rajakumari et al., 2013). As expected, we found that EBF2 was enriched on its binding sites on Ucp1 and Pparα promoters and its occupancy was enhanced by Blnc1 overexpression (Figure 5E). Similarly, while Blnc1 alone had a modest effect on Ucp1 expression in adipocytes differentiated from 10T1/2 fibroblasts, it significantly enhanced Ucp1 expression in the presence of EBF2 (Figure 5F-G). This stimulatory effect of Blnc1 on EBF2 in the induction of Ucp1 expression was also observed in 3T3-L1 adipocytes (Figure 5H), and for other genes characteristic of thermogenic adipocytes, such as Cidea and Pparα (Supplementary Figure S5). Together, these results demonstrate that Blnc1 acts in concert with EBF2 in the induction of thermogenic adipocyte gene program.
EBF2 and Blnc1 form a feedforward regulatory loop to direct adipogenesis toward thermogenic phenotype
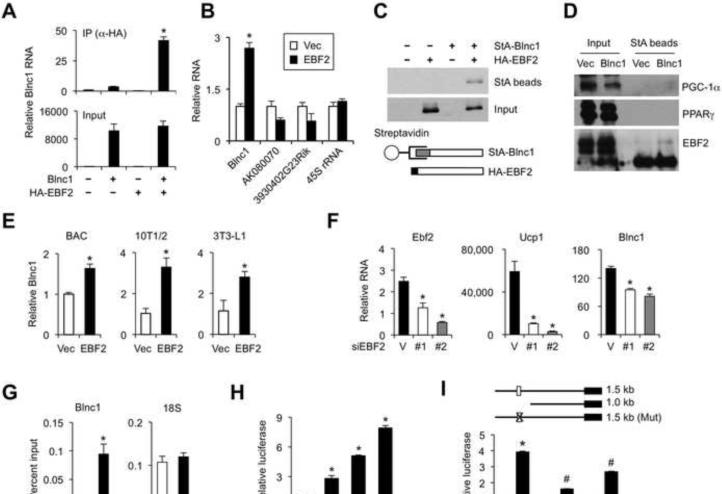
LncRNAs control gene expression through cis-regulation to modulate their neighboring genes or trans-regulation by physical interaction with other factors. To determine whether Blnc1 and EBF2 form a ribonucleoprotein transcriptional complex, we transiently transfected HEK293T cells with plasmids expressing Blnc1 RNA and HA-tagged EBF2 and performed immunoprecipitation (IP) using α-HA agarose beads. QPCR analyses indicated that Blnc1 was present in the EBF2 immunocomplex (Figure 6A). We performed similar IP studies in differentiated brown adipocytes stably expressing HA-EBF2. IP/qPCR assays indicated that EBF2 physically associated with Blnc1, but not 45S rRNA and other brown fat-enriched lncRNAs (Figure 6B). To confirm this physical interaction, we constructed a plasmid expressing Blnc1 fused to Streptavidin-binding Aptamer (StA-Blnc1), a 44-nucleotide RNA tag that mimics biotin and binds to streptavidin with high affinity (Srisawat and Engelke, 2001; Walker et al., 2011). Streptavidin precipitation of lysates from HEK293T cells transiently transfected with StA-Blnc1 and HA-EBF2 alone or in combination revealed that EBF2 was detected in the Blnc1 RNA complexes (Figure 6C). Similar experiment was performed in differentiated brown adipocytes stably expressing StA-Blnc1 (Figure 6D). We found that endogenous EBF2, but not PPARγ and PGC-1α, was present in Blnc1 RNA affinity complexes. These protein-RNA interaction studies demonstrate that Blnc1 and EBF2 form a ribonucleoprotein transcriptional complex in the cell.
Figure 6. EBF2 physically interacts Blnc1 and induces Blnc1 expression.
(A) qPCR analyses of Blnc1 in RNA isolated from α-HA immunocomplex (top) or input (bottom) from transiently transfected HEK293T cells.
(B) qPCR analyses of endogenous RNA in the EBF2 immunocomplex from differentiated BAC.
(C) Immunoblots of HA-EBF2 after strepavidin-Blnc1 (StA-Blnc1) precipitation from transfected HEK293T cell lysates. Schematic diagrams of StA-Blnc1 and HA-EBF2 were shown at the bottom.
(D) Immunoblots using total lysates (left) or StA-Blnc1 precipitation from brown adipocytes expressing StA-Blnc1.
(E) qPCR analyses of Blnc1 expression in adipocytes transduced with vector (Vec) or EBF2 retrovirus.
(F) qPCR analyses of genes in differentiated brown adipocytes transduced with vector control (V) or EBF2 RNAi knockdown (#1 and #2) retroviruses.
(G) ChIP assay in differentiated BAC using control IgG or EBF2 antibody. (H) Blnc1 reporter gene assay (1.5 kb) without (open) or with different amounts of EBF2 expression plasmid (filled).
(I) Blnc1 reporter gene assay using truncation and EBF2 binding site (rectangular box) mutant constructs without (open) or with (filled) EBF2 expression plasmid. Data represent mean ± sd. *p<0.05 EBF2/Blnc1 vs. EBF2 alone (A); Vec vs. EBF2 (B, E, G, H, I); Vector vs. siEBF2 (F); #p<0.05 Mutants vs. 1.5 kb WT construct.
We noted that the expression of Blnc1 closely correlated with EBF2 during brown and beige adipocyte differentiation (Figure 5B). Interestingly, retroviral-mediated EBF2 expression resulted in increased Blnc1 RNA levels in several cell types, including brown preadipocytes, and 10T1/2 and 3T3-L1 fibroblasts (Figure 6E), suggesting that Blnc1 may be a direct target of EBF2. Consistently, RNAi knockdown of EBF2 significantly reduced Blnc1 and Ucp1 gene expression in BAC (Figure 6F). ChIP studies revealed that EBF2 was recruited to a predicted EBF2 binding site (-1.2 kb) on the proximal Blnc1 promoter (Figure 6G). In reporter gene assays, EBF2 dose-dependently stimulated a Blnc1 promoter reporter construct containing this binding site (Figure 6H). Truncation or mutant reporter constructs lacking the putative EBF2 site had significantly diminished response to EBF2 (Figure 6I).
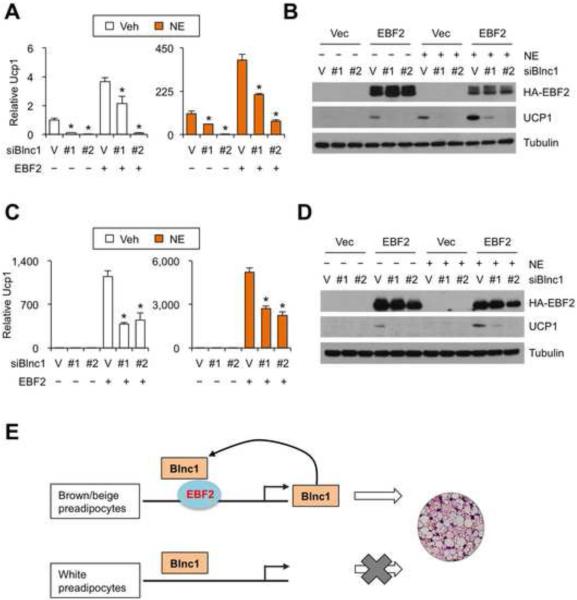
EBF2 was recently demonstrated to regulate brown fat gene expression and is required for brown fat development in mice (Rajakumari et al., 2013). Our studies identified Blnc1 as an RNA component of the EBF2 transcriptional complex that stimulates the expression of thermogenic genes. To assess the significance of Blnc1, we performed Blnc1 RNAi knockdown in the absence or presence of EBF2 overexpression in brown preadipocytes. We analyzed Ucp1 mRNA and protein expression on day 6 before and after norepinephrine induction. As expected, retroviral-mediated EBF2 expression robustly stimulated Ucp1 mRNA and protein expression at baseline as well as following adrenergic activation (Figure 7A-B). RNAi knockdown of Blnc1 severely impaired the induction of Ucp1 during differentiation. mRNA expression of Cidea and Pparα was also lower by knockdown of Blnc1 in adipocytes expressing control or EBF2 (Supplementary Figure S6). Similarly, depletion of Blnc1 also significantly reduced the induction of Ucp1 expression by EBF2 in 10T1/2 cells (Figure 7C-D). These studies demonstrate that Blnc1 is a required component of the EBF2 transcriptional complex in adipocyte gene expression.
Figure 7. Blnc1 is required for full transcriptional activity of EBF2.
(A) Ucp1 mRNA expression. Differentiated BAC expressing Vector (V) or shRNAs targeting Blnc1 (#1 and #2) were treated with vehicle (Veh) and norepinephrine (NE) for 4 hrs.
(B) Immunoblots of adipocytes treated in A.
(C) qPCR analyses of Ucp1 expression in adipocytes differentiated from 10T1/2 cells expressing Vector (V) or shRNAs targeting Blnc1 (#1 and #2).
(D) Immunoblots of adipocytes treated in C. (E) Model depicting the EBF2/Blnc1 feedforward loop in the induction of genes involved thermogenesis. Data represent mean ± sd. *p<0.05 vector vs. siBlnc1 (A, C). (See also Figure S6)
DISCUSSION
The transcriptional network that controls the commitment, differentiation, and function of thermogenic adipocytes is coupled to the physiological demand for thermogenesis. In rodents, cold exposure stimulates adaptive thermogenesis in brown fat and also recruits adipogenic progenitors, leading to the expansion of brown and beige adipose tissues during persistent cold stress. In this study, we identified Blnc1 as the RNA component of the EBF2 ribonucleoprotein complex that promotes the induction of thermogenic gene program during brown and beige adipogenesis. Blnc1 is a direct target gene of EBF2 and is required for its full transcriptional activity, thus forming a feedforward regulatory loop that directs adipogenesis toward thermogenesis (Figure 7E).
LncRNAs are emerging as a novel class of regulatory molecules that impinge on diverse biological processes. Like protein regulators, many lncRNAs exhibit remarkable tissue specificity and are regulated by developmental and physiological signals (Gupta et al., 2010; Huarte et al., 2010; Pauli et al., 2012; Sun et al., 2013; Vollmers et al., 2012). In the context of adipogenesis, a cluster of regulated lncRNAs was recently identified using RNA-seq. To our surprise, the overlap between RNA-seq and microarray analyses was relatively low. Among 21 lncRNAs that passed our selection filters, only AK080070 was present in the dataset obtained in RNA-seq study. As noncoding RNA transcripts remain poorly annotated at present, this lack of significant overlap between microarray and RNA-seq studies likely reflects the complexity and diversity of potential RNA regulators.
The expression of Blnc1 is highly induced during brown and beige adipogenesis, but not during differentiation of 3T3-L1 adipocytes, which more closely resembles white adipocytes. In response to β3-adrenergic stimulation, Blnc1 RNA levels were significantly increased in mouse adipose tissues, including BAT, inguinal WAT, epididymal WAT, and perirenal WAT. This induction of Blnc1 expression correlates with the stimulation of the thermogenic gene program in these adipose tissues. Several lines of evidence established Blnc1 as a highly regulated lncRNA in thermogenic adipocyte development. First, Blnc1 is sufficient to promote the expression of key thermogenic markers, including Ucp1 and mitochondrial genes, and uncoupled respiration during brown and beige adipocyte differentiation. Moreover, in transplanted fats derived from Blnc1 overexpressing brown preadipocytes, Ucp1 mRNA and protein levels reached approximately 30-40% that of endogenous brown fat. Finally, RNAi knockdown of Blnc1 greatly impaired adipogenesis in cultured cells and in vivo. Recent studies have revealed an array of circulating factors that regulate brown and/or beige adipocyte development (Harms and Seale, 2013); whether Blnc1 serves as a downstream target of these factor remains to be investigated.
Blnc1 is primarily localized in the nucleus and forms a ribonucleoprotein complex with EBF2, a member of the EBF family of transcription factor that has been implicated in the control of adipogenesis (Akerblad et al., 2002; Jimenez et al., 2007; Rajakumari et al., 2013). Using reciprocal protein-RNA interaction assays, we found that Blnc1 physically interacts with EBF2 in cultured brown adipocytes. The association of Blnc1 with EBF2 resulted in augmented transcriptional activity of EBF2, particularly in the induction of Ucp1 and mitochondrial gene expression. Other EBF members have also been demonstrated to regulate adipogenesis (Akerblad et al., 2002; Jimenez et al., 2007), it remains possible that Blnc1 may also physically interacts with other EBF factors to regulate adipocyte gene expression. Interestingly, Blnc1 alone was unable to activate the thermogenic gene program in 3T3-L1 and 10T1/2 cells where endogenous EBF2 expression was low. In support of this, retroviral-mediated expression of EBF2 in these progenitors rendered them responsive to the stimulatory effects of Blnc1, whereas RNAi knockdown of Blnc1 significantly impaired EBF2-induced brown adipogenesis, suggesting that Blnc1 is an important transcriptional partner for EBF2. ChIP assay and reporter gene studies indicate that Blnc1 is a direct target of EBF2; EBF2 is recruited to the proximal promoter of Blnc1 and stimulates its expression. As such, Blnc1 appears to function as a critical RNA component of the EBF2 transcriptional complex adipocyte gene regulation. Together, these findings support a novel mechanism in which Blnc1 and EBF2 form a feedforward regulatory loop that potentially functions as a switch that directs brown and beige adipocyte differentiation.
EXPERIMENTAL PROCEDURES
Microarray
Total RNA was isolated from brown and white adipose tissues, during brown adipocyte differentiation (0, 2, 4, 6 days) and from WAT following 7 days of CL treatment. LncRNA profiling was performed using a chip containing 31,423 lncRNAs and 25,376 coding transcripts (mouse lncRNA v2.0, Arraystar). We used a cutoff of normalized array values (log2 transformed values > 7.0) to enrich for transcripts present in brown fat and brown adipocytes. We performed Student's t-test to identify transcripts exhibiting significant differences of over 2.5-fold for BAT/WAT and 2-fold for CL/saline groups. For BAC differentiation, we calculated fold induction by comparing day 6 vs. day 0 undifferentiated samples (> 4-fold). For Mouse Genome 430 2.0 Array, we used total RNA isolated from differentiated brown adipocytes with Blnc1 overexpression and RNAi knockdown. Gene Ontology analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, available at http://david.abcc.ncifcrf.gov).
Animal studies
All animal studies were performed according to procedures approved by the University Committee on Use and Care of Animals. NU/J Foxn1nu (nude) and C57BL/6 wild type mice were obtained from the Jackson Laboratory. Mice were maintained under 12/12hr light/dark cycle and fed with standard rodent chow. Mice were injected intraperitoneally daily with saline or CL316,243 (1 mg/kg) for 7 days. For transplantation, 3×107 brown preadipocytes were subcutaneously injected at the base of sternum on two sides in 8-week old male nude mice. Fat pads were carefully dissected after 2 weeks for gene expression analyses.
Adipocyte differentiation
Brown preadipocytes were isolated and immortalized as previously described (Klein et al., 2002). C3H10T1/2 cells and brown preadipocytes were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). 3T3-L1 fibroblasts were cultured in DMEM containing 10% bovine growth serum. To initiate differentiation, a cocktail containing 0.5mM IBMX, 125μM indomethacin and 1μM dexamethasone was added to confluent preadipocytes maintained in DMEM supplemented with 10% FBS, 20nM insulin and 1nM T3. Cells were switched to differentiation medium (DMEM, 10% FBS, 20nM insulin and 1nM T3) after two days. Mitochondrial abundance was observed following MitoTracker staining under a Leica DM IRB microscope.
Mitochondrial DNA content and respiration
Mitochondrial DNA content was measured using qPCR analysis of total DNA isolated from differentiated brown adipocytes. Primers for mitochondrial DNA (mtND1 and mtCox1) and nuclear DNA (PECAM) were listed in Supplementary Table S3. Oxygen consumption rate in differentiated brown adipocytes was measured using Oxygen Meter (Strathkelvin Instruments) with a Mitocell (MT200) mixing chamber. Cells were suspended in 400 μL differentiation medium and oxygen concentration was recorded for 5 min followed by injections of FCCP (25 μM) or oligomycin (5 μg/mL) to the chamber. Oxygen consumption rate was calculated using software (782 Oxygen System version 4.0) and normalized to protein content.
Gene expression analyses
Gene expression was analyzed by qPCR using SYBR Green method. Ribosomal protein 36B4 was used as an internal control. For immunoblotting, total lysates were prepared in a lysis buffer containing 50mM Tris (pH=7.5), 150mM NaCl, 5mM NaF, 25mM β-glycerolphosphate, 1mM sodium orthovanadate, 10% glycerol, 1% Triton X-100, 1 mM dithiothreitol (DTT), and freshly added protease inhibitors. Protein lysates were separated by 10% SDS-polyacrylamide gel, transferred to PVDF membrane, and immunoblotted with antibodies against UCP1 (Alpha Diagnosis), PPARγ, HA-tag, and Lamin A/C (Santa Cruz Biotechnology), FABP4 (Cell Signaling Technology), and β-actin and α-tubulin (Sigma-Aldrich).
RNA-protein Interaction Assays
HEK293T cells transiently transfected with HA-EBF2 and Blnc1 alone or in combination and differentiated brown adipocytes expressing HA-EBF2 were crosslinked with 1% formaldehyde followed by sonication. Immunoprecipitation was performed using anti-HA agarose beads. After washing, RNA was extracted with Trizol, treated with RNase-free DNase, and analyzed by RT-qPCR. For reverse precipitation, Blnc1 was precipitated from total lysates prepared from transfected HEK293T cells and brown adipocytes using streptavidin agarose beads. Proteins associated with beads were analyzed by immunoblotting.
Reporter gene and ChIP assays
Reporter gene assays were performed in transiently transfected HEK293 cells. For ChIP assay, chromatin extracts were prepared from differentiated brown adipocytes, precleared, and immunoprecipitated using control IgG or antibodies against EBF2 (R&D Systems) or acetylated histone H3 (Millipore). Chromatin association was measured using qPCR with primers listed in Supplementary Table S3.
Statistics
Data were analysed using two-tailed Student's t-test for independent groups. A p-value of less than 0.05 was considered statistically significant.
Supplementary Material
HIGHLIGHTS.
Blnc1 is a highly regulated lncRNA during thermogenic adipocyte development
Blnc1 drives the thermogenic gene program in brown and beige adipocytes
Blnc1 forms a ribonucleoprotein transcriptional complex with EBF2
Blnc1 and EBF2 are assembled into a feedforward regulatory loop
ACKNOWLEDGEMENTS
We thank Drs. J. Wu and B.M. Spiegelman for sharing beige adipocyte progenitor clones and lab members for discussion. This work was supported by NIH (DK095151 and DK077086, J.D.L.). X.Y.Z. and S. L. are supported by Postdoctoral Fellowship and Scientist Development Grant from American Heart Association, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Molecular and cellular biology. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SR, Marguerat S, Bahler J. Exploring long non-coding RNAs through sequencing. Seminars in cell & developmental biology. 2012;23:200–205. doi: 10.1016/j.semcdb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nature medicine. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nature cell biology. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature medicine. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes & development. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Guo K, Mogen J, Struzzi S, Zhang Y. Preadipocyte transplantation: an in vivo study of direct leptin signaling on adipocyte morphogenesis and cell size. American journal of physiology Regulatory, integrative and comparative physiology. 2009;296:R1339–1347. doi: 10.1152/ajpregu.90691.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nature medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO reports. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homoe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell metabolism. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Molecular and cellular biology. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell metabolism. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Fasshauer M, Klein HH, Benito M, Kahn CR. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- Kozak LP, Harper ME. Mitochondrial uncoupling proteins in energy expenditure. Annual review of nutrition. 2000;20:339–363. doi: 10.1146/annurev.nutr.20.1.339. [DOI] [PubMed] [Google Scholar]
- Kozak LP, Koza RA. The genetics of brown adipose tissue. Progress in molecular biology and translational science. 2010;94:75–123. doi: 10.1016/B978-0-12-375003-7.00004-2. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell metabolism. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nature medicine. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American journal of physiology Endocrinology and metabolism. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell metabolism. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, Scheinin M, Taittonen M, Niemi T, Enerback S, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell metabolism. 2011;14:272–279. doi: 10.1016/j.cmet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Ouellet V, Labbe SM, Blondin DP, Phoenix S, Guerin B, Haman F, Turcotte EE, Richard D, Carpentier AC. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. The Journal of clinical investigation. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. The Journal of biological chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell metabolism. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PloS one. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends in endocrinology and metabolism: TEM. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lans AA, Hoeks J, Brans B, Vijgen GH, Visser MG, Vosselman MJ, Hansen J, Jorgensen JA, Wu J, Mottaghy FM, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of clinical investigation. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell metabolism. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Good PD, Gipson TA, Engelke DR. The dual use of RNA aptamer sequences for affinity purification and localization studies of RNAs and RNA-protein complexes. Methods in molecular biology. 2011;714:423–444. doi: 10.1007/978-1-61779-005-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes & development. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, Chen XW, Cawthorn WP, MacDougald OA, Koenig RJ. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PloS one. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of lipid research. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, Saito M. Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of clinical investigation. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.