Summary
Human pluripotent stem cells (hPSCs) offer a unique platform for elucidating the genes and molecular pathways that underlie complex traits and diseases. To realize this promise, methods for rapid and controllable genetic manipulations are urgently needed. By combining two newly developed gene-editing tools, the TALEN and CRISPR/Cas systems, we have developed a novel genome-engineering platform in hPSCs, which we named iCRISPR. iCRISPR enabled rapid and highly efficient generation of biallelic knockout hPSCs for loss-of-function studies, as well as homozygous knockin hPSCs with specific nucleotide alterations for precise modeling of disease conditions. We further demonstrate, for the first time, efficient one-step generation of double- and triple-gene knockout hPSC lines, as well as stage-specific inducible gene knockout during hPSC differentiation. Thus the iCRISPR platform is uniquely suited for dissection of complex genetic interactions and pleiotropic gene functions in human disease studies, and has the potential to support high-throughput genetic analysis in hPSCs.
Introduction
The identification and functional validation of sequence variants affecting diverse human traits, including disease susceptibility, is key to understanding human biology and disease mechanisms. Advances in next-generation sequencing and genome-wide association studies have led to the rapid discovery of numerous disease-associated sequence variants in recent years. To functionally validate this increasing number of disease-associated mutations, an ideal platform should not only closely recapitulate their genomic, cellular and human-specific contexts (Cooper and Shendure, 2011), but also offer superior speed and capacity to meet the growing demand.
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and the closely related human induced pluripotent stem cells (hiPSCs), offer a promising solution to meet these challenges (Zhu and Huangfu, 2013). First, hPSCs have unlimited self-renewal capacity, providing a renewable source of experimental cells suitable for rapid, large-scale analyses. Second, they have the potential to generate all adult cell types, including rare or inaccessible human cell populations, providing a unique platform to recapitulate the cellular and human-specific contexts required for disease studies. Yet, to fulfill this potential, it is of paramount importance to develop methods for rapid, efficient and controllable genetic manipulation in hPSCs. Unfortunately, while classic gene-targeting technology via homologous recombination in mouse ESCs (mESCs) has proven a powerful tool to dissect gene function (Capecchi, 2005; Thomas and Capecchi, 1986; Thomas et al., 1986), this approach has been extremely inefficient when applied to hPSCs (Hockemeyer and Jaenisch, 2010).
Recently, with the advent of programmable site-specific nucleases, genome engineering has become a much easier task in a wide range of organisms and cultured cell types including hPSCs (Joung and Sander, 2013; Ran et al., 2013b; Urnov et al., 2010). Acting as “DNA scissors”, they induce double strand breaks (DSBs) at desired genomic loci, triggering the endogenous DNA repair machinery. Processing of DSBs by the error-prone non-homologous end-joining (NHEJ) pathway leads to small insertions and deletions (Indels) useful for generating loss-of-function mutations; whereas error-free homology directed repair (HDR) enables targeted integration of exogenously provided DNA sequences for introducing precise nucleotide alterations or knockin reporters.
Transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have emerged as powerful and versatile site-specific nucleases for genome modification in a variety of model systems. TALENs are typically designed as pairs to bind the genomic sequences flanking the target site. Each TALEN arm consists of a programmable, sequence-specific TALE DNA-binding domain linked to a nonspecific DNA cleavage domain derived from the bacterial restriction endonuclease FokI (Cermak et al., 2011; Miller et al., 2011). Recent studies have also successfully adapted the prokaryotic type II CRISPR/Cas system for genome editing in eukaryotic systems (Cong et al., 2013; DiCarlo et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Mali et al., 2013b). The type II CRISPR/Cas system requires two components: the DNA endonuclease Cas9 protein for DNA cleavage, and a variable CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) duplex for DNA target recognition (Jinek et al., 2012). Binding of crRNA/tracrRNA to the target sequence via Watson-Crick base pairing directs Cas9 to any genomic locus of interest for site-specific DNA cleavage. The CRISPR/Cas system has now been further improved for use in mammalian systems through Cas9 codon optimization and replacement of the crRNA/tracrRNA duplex with a single chimeric guide RNA (gRNA) (Cho et al., 2013a; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013b).
While programmable site-specific nucleases have significantly improved our capacity to genetically modify hPSCs (Ding et al., 2013a; Ding et al., 2013b; Hockemeyer et al., 2009; Hockemeyer et al., 2011; Hsu et al., 2013; Mali et al., 2013b; Soldner et al., 2011), none of the available methods has achieved multiplexed gene targeting or inducible gene knockout in hPSCs. These two features are crucial for interrogating complex genetic interactions and pleiotropic gene functions, which are often difficult to study using animal models due to the involvement of multiple alleles and random segregation of these alleles through breeding.
To achieve rapid, multiplexable and inducible genome editing in hPSCs, we have developed a novel genome-engineering platform. Through TALEN-mediated gene targeting, we created a panel of hPSC lines for robust, doxycycline-inducible expression of Cas9 (referred to as iCas9 hPSCs). By transfecting iCas9 hPSCs with gRNAs targeting different genes, we have generated biallelic knockout hPSC lines for 6 individual genes with high efficiency (~20% to ~60%). This highly efficient platform enabled us to generate, for the first time, double- and triple-gene knockout hPSC lines in a single step with up to ~10% efficiency. Moreover, co-transfection of gRNAs with a single-stranded DNA (ssDNA) HDR template yielded homozygous knockin clones at a rate of up to ~10%, allowing efficient and scarless introduction of defined nucleotide modifications in different hPSC lines for disease modeling. Finally, we achieved stage-specific inducible gene knockout during hPSC differentiation. This versatile new platform, which we called “iCRISPR”, allows rapid generation of mutant hPSCs for analysis of complex disease phenotypes in isogenic background and could be easily scalable for high-throughput genetic analysis.
Results
An iCRISPR platform for rapid and versatile genome editing
A recent study by Jaenisch and colleagues successfully applied CRISPR/Cas for highly efficient genome modifications in mESCs (Wang et al., 2013), suggesting similar approaches may work in hPSCs. Encouraged by these findings, we investigated the efficiency of CRISPR/Cas-mediated genome editing in hESCs through plasmid electroporation to transiently express Cas9 and a specific gRNA, a method that is currently used in hESCs (Figure 1A, upper panel). Although we successfully generated heterozygous GATA6 mutant lines at 2-6% efficiencies, no homozygous mutants were identified out of 384 total hESC clones analyzed (Table 1). This finding is consistent with the variable and generally low gene editing efficiencies observed by others in hPSCs (Ding et al., 2013b; Hsu et al., 2013; Mali et al., 2013b) and highlights the need for a more efficient method to model human traits caused by recessive or multiple-gene mutations.
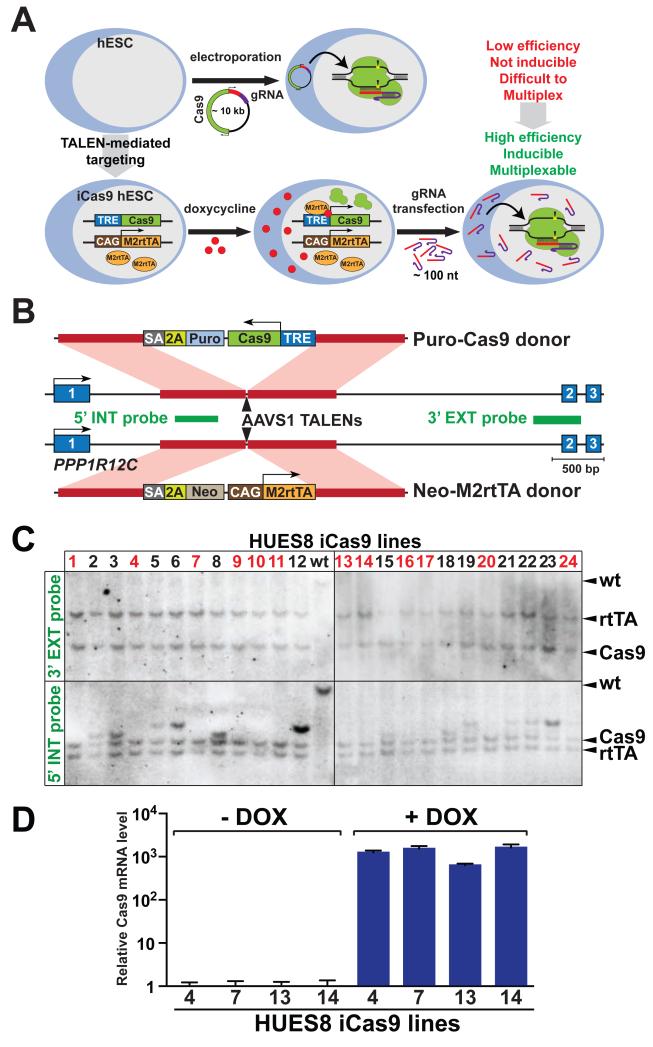
Figure 1. Engineering an iCRISPR platform through generating iCas9 hPSCs.
(A) Schematic comparison of the current genome editing approach in hPSCs by electroporation of a ~10 kilobases (kb) CRISPR/Cas9 vector (upper panel) with the iCRISPR platform (lower panel) for genome editing in hPSCs. Cas9 protein (green) binds a chimeric gRNA composed of a constant transactivating structural region (purple) and a variable DNA recognition site (red). The Cas9/gRNA complex binds to DNA and induces a DSB (yellow). TRE, tetracycline response element; CAG, constitutive synthetic promoter; M2rtTA, reverse tetracycline transactivator sequence and protein; doxycycline, red dots. (B) Generation of iCas9 hPSCs through TALEN-mediated gene targeting at the AAVS1 locus. Red lines indicate homology to PPP1R12C intron 1; SA, splice acceptor; 2A, self-cleaving 2A peptide; Puro, Puromycin resistance gene; Neo, Neomycin resistance gene. (C) Southern blot analysis of HUES8 iCas9 lines using 3′ external and 5′ internal probes. Lines carrying desired targeted insertions of the Puro-Cas9 and Neo-M2rtTA donor sequences without random integrations are indicated in red. (D) Quantitative real-time RT-PCR (qRT-PCR) analysis of Cas9 transcript levels with or without doxycycline treatment in HUES8 iCas9 lines.
See also Figure S1 and Table S1.
Table 1. CRISPR/Cas-mediated single-gene targeting in hESC.
| Gene | CRISPR | Delivery method | Mutant alleles per clone |
|||
|---|---|---|---|---|---|---|
| 1 | 2 |
|||||
| Compound heterozygotes |
Homozygotes | Total biallelic mutant clones |
||||
| NGN3 | Cr5 | gRNA transfection | 2/48 (4.2%) | 2/48 | 10/48 | 12/48 (25%) |
| Cr6 | gRNA transfection | 1/36 (2.8%) | 1/36 | 5/36 | 6/36 (16.7%) | |
| GATA4 | Cr2 | gRNA transfection | 2/96 (2.1%) | 19/96 | 5/96 | 24/96 (25%) |
| GATA6 | Cr1 | gRNA transfection | 14/40 (35%) | 7/40 | 0/40 | 7/40 (17.5%) |
| Cr8 | gRNA transfection | 6/48 (12.5%) | 13/48 | 1/48 | 14/48 (29.2%) | |
| Cr8 | Plasmid electroporation | 2/96 (2.1%) | 0/96 | 0/96 | 0/96 (0%) | |
| Cr8 | Plasmid electroporation* | 17/288 (5.9%) | 0/288 | 0/288 | 0/288 (0%) | |
| TET1 | Cr2 | gRNA transfection | 4/48 (8.3%) | 10/48 | 3/48 | 13/48 (27.1%) |
| TET2 | Cr4 | gRNA transfection | 6/48 (12.5%) | 14/48 | 14/48 | 28/48 (58.3%) |
| TET3 | Cr4 | gRNA transfection | 4/48 (8.3%) | 24/48 | 8/48 | 32/48 (66.6%) |
In vitro transcribed gRNAs targeting NGN3, GATA4, GATA6, NGN3, TET1, TET2, or TET3 were transfected in doxycycline-induced iCas9 hESC, or electroporated as plasmids (GATA6 Cr8) in wild type HUES8 or HUES9 (*) hESCs. The number of lines containing each specific number of mutated alleles (1 or 2) is shown in relation to the total number of lines screened in each experiment.
We reasoned that one could develop a more efficient and versatile genome-editing platform by first generating hPSCs that express Cas9, the invariable component of the CRISPR/Cas system. We anticipated that Cas9-expressing hPSCs would be easily transfected with gRNAs due to their small size (~100 nucleotides), which in turn could lead to reproducible and highly efficient genome editing (Figure 1A, lower panel). We first determined the efficiency of lipid-mediated transfection of small RNAs. Using a control fluorescence-labeled, double-stranded RNA (dsRNA) probe, we estimated that ~60% hESCs would be transfected with gRNAs (Figure S1A). In contrast, only ~10% GFP+ hESCs were detected by flow cytometry after electroporation of a GFP-expressing plasmid of comparable size with the Cas9/gRNA vectors (Figure S1A-D). Importantly and in contrast to plasmid electroporation, lipid-mediated transfection is associated with very low cytotoxicity. This opens up the possibility of repeated transfections and cotransfection of multiple gRNAs for multiplexed genome editing, as well as gRNA transfection during a specific stage of hPSC differentiation for inducible gene knockout.
We next engineered iCas9 hPSC lines for doxycycline-inducible expression of Cas9 through TALEN-mediated gene targeting. We chose to target the transgenes into the AAVS1 (also known as PPP1R12C) locus, as it has been shown to support robust and sustained transgene expression similar to the Rosa26 locus in mice (Smith et al., 2008). Based on a gene trap approach used by Jaenisch and colleagues (Hockemeyer et al., 2011), we co-electroporated the AAVS1 TALEN constructs with two donor plasmids targeting the first intron of the PPP1R12C gene in three hESC lines (HUES8, HUES9 and MEL-1) and one hiPSC line (BJ iPSC) (Cowan et al., 2004; Huangfu et al., 2008). One donor plasmid contains a doxycycline-inducible Cas9 expression cassette (Puro-Cas9 donor), and the other carries a constitutive reverse tetracycline transactivator (M2rtTA) expression cassette (Neo-M2rtTA donor) (Figure 1B and S1E, F). Southern blot analysis revealed a high biallelic targeting efficiency: >50% of the clonal lines have both transgenes correctly inserted without additional random integrations (Figure 1C and S1G). qRT-PCR analysis confirmed the induction of Cas9 expression upon doxycycline treatment in all clonal iCas9 lines examined (Figure 1D and S1H). Further analysis showed that iCas9 hPSCs display uniform expression of the pluripotency markers OCT4, NANOG and SOX2 (Figure S1I), and maintain the capacity to differentiate into tissue derivatives of the three embryonic germ layers in teratoma assays (Figure S1J). Furthermore, the AAVS1 targeting strategy does not introduce apparent chromosomal aberrations, as confirmed by karyotyping analysis (two of the iCas9 lines inherited a duplication present in the parental line before the targeting experiment) (Table S1).
One-step creation of single-, double- and triple-gene knockout hPSCs
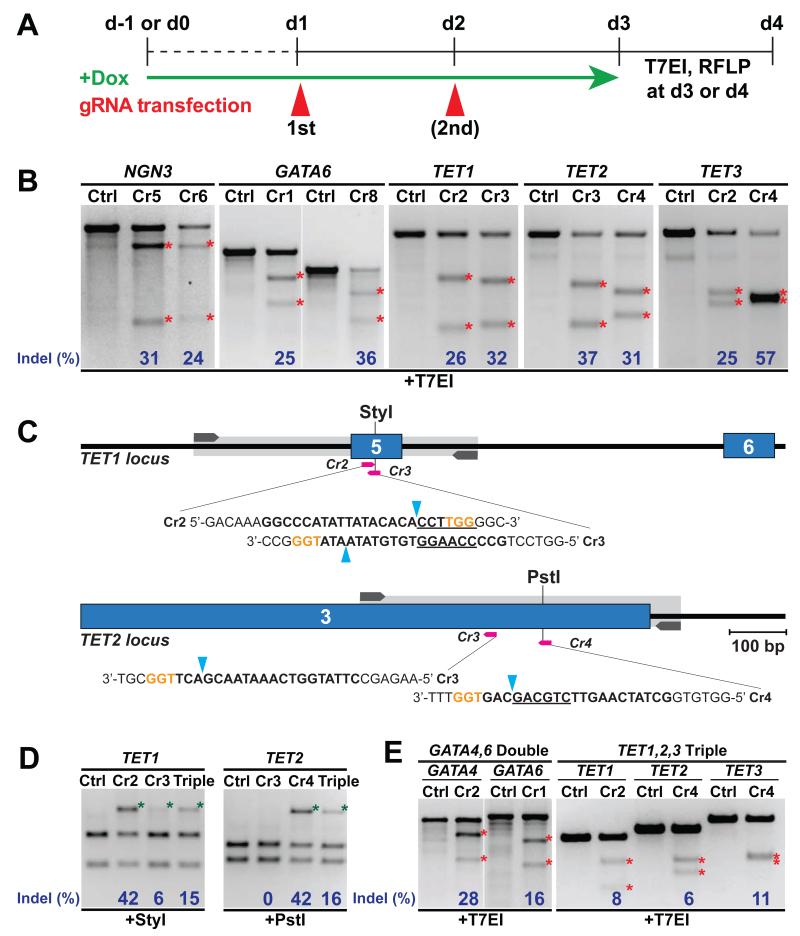
To determine the capacity of iCas9 hPSCs for gene editing, we designed a panel of gRNAs targeting six genes (NGN3, GATA4, GATA6, TET1, TET2 and TET3) located on five different chromosomes (Figure S2A). We first selected gRNAs effectively targeting each gene by performing experiments in 293T cells and assessing Indel rates using T7 Endonuclease I (T7EI), which specifically cleaves heteroduplexes formed by the hybridization of wild-type and mutant DNA sequences (Mashal et al., 1995) (Figure S2B). Next, iCas9 hPSCs were treated with doxycycline and transfected with selected gRNAs generated through in vitro transcription (Figure 2A). We observed ~30% Indel rates based on T7EI assays performed on genomic DNA extracted two or three days after gRNA transfection (Figure 2B). For TET1 and TET2, we designed Restriction Fragment Length Polymorphism (RFLP) assays to directly assess the loss of a restriction site in proximity of the predicted Cas9 cleavage site (Figure 2C). RFLP assays revealed a higher mutation rate compared to T7EI (42% versus 26% for TET1-Cr2, 42% versus 31% for TET2-Cr4, Figure 2D), suggesting T7EI assays may underestimate the rate of mutations as suggested for similar Indel assays (Guschin et al., 2010). Encouraged by these high Indel rates, we then performed multiplexed genome editing through co-transfection of gRNAs targeting 2 or 3 genomic loci. Multiplexed targeting of GATA4 and GATA6 induced 28% and 16% of Indels in the respective loci (Figure 2E). Similar efficiencies were observed for multiplexed targeting of TET1, 2 and 3 (to 17%, 11% and 18% respectively) through optimizing the transfection conditions (Figure 2E and Figure S3A-D). These results suggest that our system supports efficient genome editing including multiplexing.
Figure 2. Single and multiplexed gRNA transfection efficiently induces Indels in iCas9 hPSCs.
(A) Schematic representation of the experimental procedure. (B) T7EI assay for Cas9-mediated cleavage in HUES8 iCas9 cells using single gRNAs targeting NGN3 (Cr5, 6), GATA6 (Cr1, 8), TET1 (Cr2, 3), TET2 (Cr3, 4), and TET3 (Cr2, 4). All gRNAs were transfected twice, except for GATA6 (Cr1, 8), which were transfected once. In all figures in this study red asterisks indicate the expected T7EI-specific fragments used to quantify Indel frequency (blue). (C) Schematic of Cas9/gRNA-targeting sites (pink arrows in all figures in this study) in TET1 and TET2 loci showing exon structure (blue boxes in all figures in this study), PCR amplicons (light grey boxes in all figures in this study) and StyI or PstI restriction sites used for RFLP analysis. gRNA-targeting sequences, bold; protospacer-adjacent motif (PAM) sequence, orange; Cas9 cleavage site (blue arrow heads); restriction sites, underlined, as in all figures in this study. (D) RFLP analysis upon TET1 (Cr2, 3), TET2 (Cr3, 4) or multiplexed TET1, 2, 3 (Triple) gRNA transfection. In all figures in this study green asterisks indicate uncut PCR fragment used to quantify Indel frequency by RFLP (blue). (E) T7EI assay in HUES8 iCas9 cells transfected with multiplexed GATA4,6 (Double) or TET1, 2, 3 (Triple) gRNAs.
See also Figure S2.
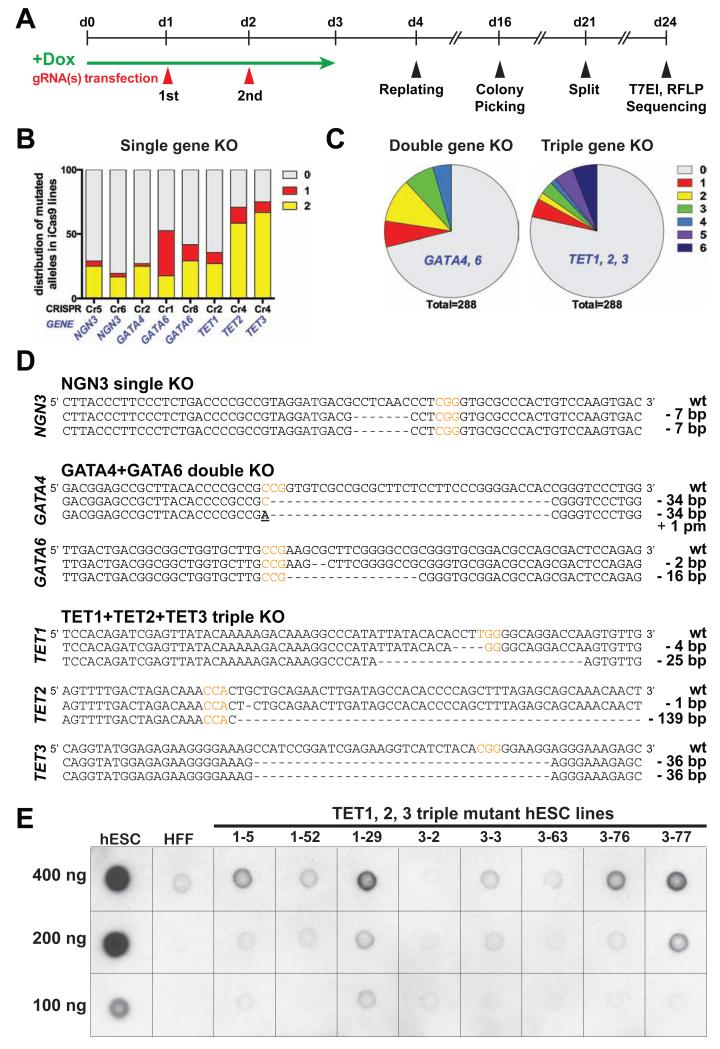
We next established clonal lines after gRNA transfection (Figure 3A). For single-gene targeting, we generated mutant hESC lines affecting 6 individual genes after typically sequencing 36 to 96 clones for each targeting experiment. ~42% (ranging from 20% to 75%) clones carried mutations in at least one allele. Notably, the majority of mutant clones carried mutations in both alleles, including both homozygous and compound heterozygous mutations (Figure 3B, D and Table 1). Some gRNAs induced non-random deletions on target sequences in a significant number of clones, a phenomenon which has been reported by others in the murine system (Wang et al., 2013). For instance, 10 out of 12 biallelic mutant lines generated with an NGN3-targeting gRNA (NGN3-Cr6) contained an identical homozygous 7-bp deletion, and 9 out of 14 biallelic mutant clones generated with a TET2-targeting gRNA (TET2-Cr2) carried the same 3-bp deletion (Figure S3E). Preferential generation of these alleles is likely caused by microhomology-mediated repair that utilizes short stretches of repeated sequences flanking the DSB site.
Figure 3. Single and multiplexed gene targeting.
(A) Strategy of gene targeting. (B and C) Allelic sequence distribution in HUES8 clonal lines generated with single gRNAs targeting NGN3 (Cr5, 6), GATA4 (Cr2), GATA6 (Cr1, 8), TET1 (Cr2), TET2 (Cr4), TET3 (Cr4) (B) or multiplexed gRNAs targeting GATA4 and 6 (Cr2, Cr1 respectively) or TET1, 2 and 3 (Cr2, Cr4, Cr4, respectively) (C). (D) Representative sequences of various knockout (KO) mutant clones with PAM sequences labeled in red. pm: point mutation. (E) Analysis of 5hmC levels in DNA isolated from TET1, 2, 3 triple-targeted hESC clones by dot blot assay using an anti-5hmC antibody. Wild-type HUES8 iCas9 cells and human foreskin fibroblasts (HFF) are used as controls. See also Figure S3, Table S2, S3.
The use of iCas9 hPSCs also enabled efficient one-step multiplexed gene editing. We were able to identify mutant clones affecting 2 genes (i.e. all 4 alleles of GATA4 and GATA6) and 3 genes (i.e. all 6 alleles of TET1, TET2 and TET3) with ~5% efficiencies (Figure 3C, D and Table S2). Further optimization of gRNA transfection conditions (Figure S3A-D) improved the efficiency of biallelic triple-gene targeting to close to 10% (Table S2). Since TET proteins are responsible for catalyzing the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (Ito et al., 2010; Tahiliani et al., 2009), we analyzed the 5hmC levels in triple-targeted hESC lines. As expected from loss-of-function alleles, we detected a significant reduction of 5hmC levels in all triple-gene mutant lines compared to wild-type control hESCs (Figure 3E). These findings demonstrate rapid, single-step generation of multiple-gene knockout lines for loss-of-function studies. Importantly, despite the high gene-editing efficiencies, we did not detect off-target mutations in multiple single- and triple-gene mutant hESC lines analyzed (Table S3).
Precise HDR-mediated genome editing for disease modeling
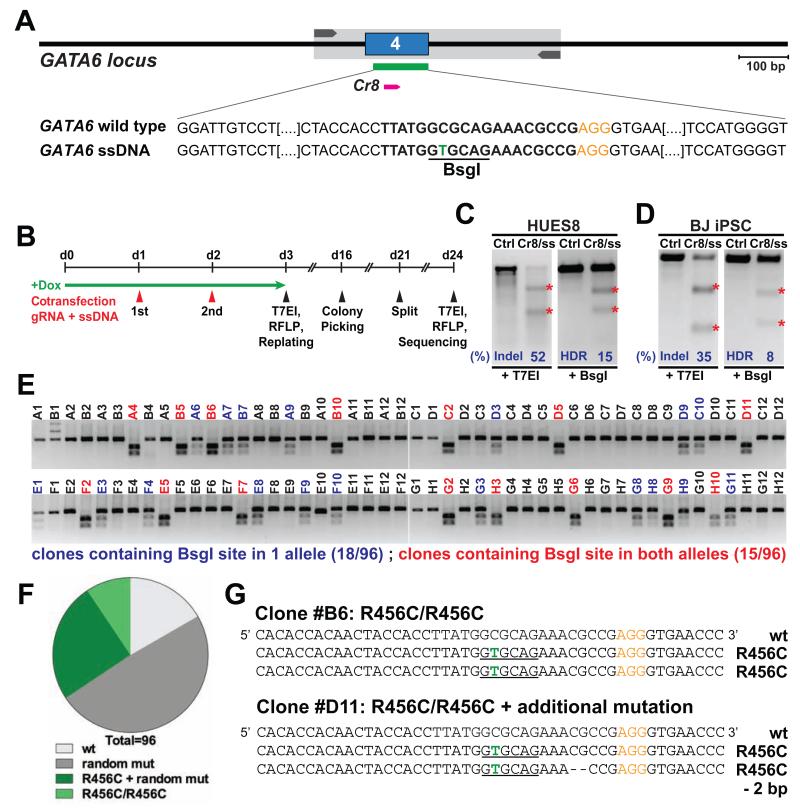
To accurately dissect gene function in human development and disease, precise nucleotide alterations are required to either create disease-specific variants in wild-type cells or correct disease-associated mutations in patient cells. To test this approach, we co-transfected doxycycline-treated iCas9 hPSCs with a GATA6-targeting gRNA (GATA6-Cr8) and a 110-nt ssDNA repair template introducing a single nucleotide mutation (Figure 4A, B). T7EI analysis revealed ~50% and ~35% mutation rates in HUES8 and BJ iPSCs respectively. As the HDR ssDNA template creates a silent BsgI restriction site in GATA6, we next performed RFLP analysis to discriminate successful HDR-mediated gene editing from NHEJ-mediated Indels, and determined that ~15% of HUES8 and ~8% of BJ iPSC genomic DNA sequences integrated the desired modification (Figure 4C, D). Next, we replated transfected cells to establish mutant clones (Figure 4B). RFLP analysis on 96 clones identified 33 (34%) clones with at least one BsgI site, and 15 (16%) clones containing one BsgI site in each GATA6 allele (Figure 4E). Sequence analysis verified that 9 clones carried the desired homozygous mutations without additional sequence alterations (Figure 4F, G). Similar results were obtained with the NGN3 locus as determined by RFLP analysis (Figure S4A, B). Since our platform allows rapid generation of precise homozygous knockin mutations, we tested the feasibility of generating an allelic series for modeling disease susceptibility.
Figure 4. HDR-mediated genome editing.
(A) Schematic of Cas9/gRNA and ssDNA oligo targeting sites at the GATA6 locus. A C>T substitution (green) was introduced in the ssDNA HDR template, generating a new BsgI restriction site (underlined), resulting in an R456C amino acid substitution in GATA6. (B) Strategy of HDR-mediated genome editing. (C and D) T7EI and RFLP assay in HUES8 (C) and BJ iPSCs (D) cotransfected with GATA6 gRNA (Cr8) and ssDNA HDR repair template. (E and F) RFLP analysis (E) and allelic sequence distribution (F) in clones generated with GATA6 gRNA/ssDNA. wt, wild-type; mut, mutation. “R456C + random mut” includes clones with undesired mutations in addition to the R456C modification in one or both alleles. (G) Representative sequences of one homozygous (R456C/ R456C) and one compound heterozygous (R456C/R456C + additional mutations) GATA6 mutant clone.
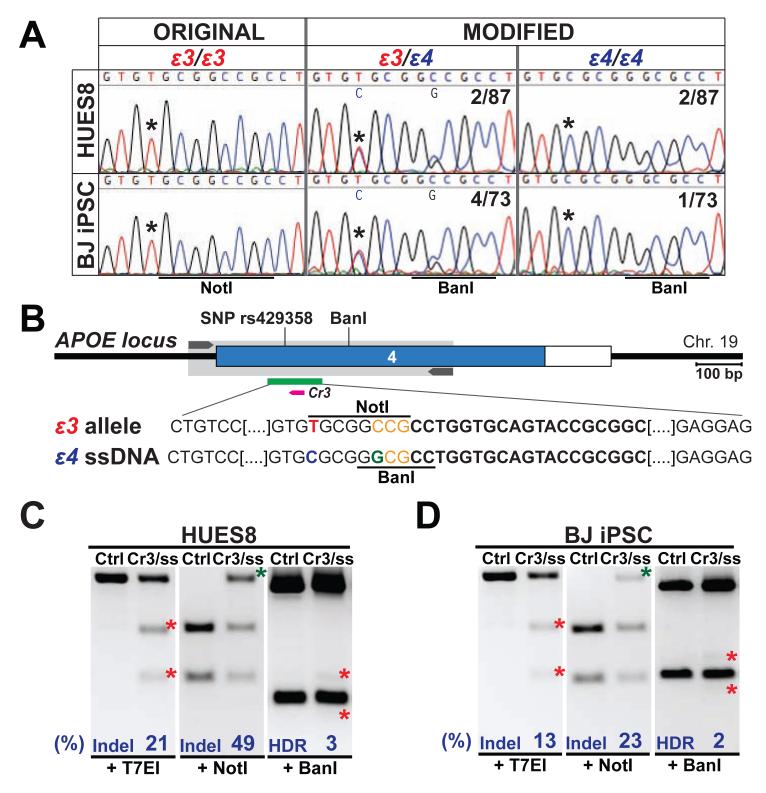
APOE is the most common risk locus associated with late-onset Alzheimer’s disease (LOAD). There are three common allelic variants of APOE, known as ε2, ε3, and ε4. The most common ε3 variant is considered “neutral”, whereas the ε4 variant is the major known risk factor with a dose dependent effect: increased number of the ε4 alleles (i.e. from 0 to 1 and to 2) is associated with increased risk and decreased onset age (Corder et al., 1993). Sequence analysis showed that both HUES8 and BJ iPSCs carry the common ε3/ε3 genotype (Figure 5A). We therefore sought to create hPSCs carrying the ε3/ε4 and ε4/ε4 genotypes, which would be useful for modeling disease susceptibility.
Figure 5. Generation of an allelic series at the APOE locus.
(A) Sequence analysis of SNP rs429358 in parental HUES8 hESCs and BJ hiPSCs lines (ε3/ε3) and derived HDR-mediated edited clones (ε3/ε4 or ε4/ε4). Ratios indicate the number of colonies with the specified genotype out of the total number of colonies analyzed. (B) Schematic of Cas9/gRNA and ssDNA oligo targeting sites at the APOE locus. A T(red)>C(blue) substitution was introduced in the ε4 ssDNA to convert the ε3 allele into ε4, in addition a C>G substitution (green) was introduced generating a novel BanI, and disrupting the endogenous NotI restriction site (underlined). SNP rs429358 and BanI sites are indicated in APOE exon 4. (C and D) T7EI and RFLP assay in HUES8 hESCs (C) and BJ iPSCs (D) cotransfected with APOE gRNA (Cr3) and ε4 ssDNA HDR repair template. Ratios indicate the number colonies with the specified genotype out of the total number of colonies analyzed.
See also Figure S4.
We targeted the single-nucleotide polymorphism (SNP) rs429358 in APOE exon 4 to introduce a T-to-C transition for conversion from ε3 to ε4 (Figure 5B). We designed gRNAs targeting APOE (Figure S4C) and selected the most effective one (APOE Cr3) through the T7EI assay (Figure S4D). Next, we co-transfected doxycycline-treated iCas9 hPSCs with APOE Cr3 gRNA and a 120-nt ssDNA repair template carrying the ε4 polymorphism and a BanI silent restriction site (Figure 5B). T7EI analysis revealed ~21% and ~13% mutation rates in the HUES8 and BJ iPSC APOE loci respectively. As observed previously, RFLP assay using NotI revealed a higher mutation rate (49% in HUES8 and 23% in BJ iPSCs, Figure 5C, D). Using BanI RFLP we further discriminated HDR-mediated gene editing from NHEJ-mediated Indels, and estimated that ~2-3% of the target sequence had the desired modification (Figure 5C, D). Indeed sequence analysis confirmed successful generation of clonal lines carrying an APOE allelic series in both HUES8 and BJ iPSCs: the original ε3/ε3, and the modified ε3/ε4 and ε4/ε4 genotypes (Figure 5A). This strategy could be easily modified to convert the risk-associated ε4 variant to the standard ε3 variant to create isogenic control cells for studies of hPSCs carrying the ε4 variant or for therapeutic purposes.
Inducible gene knockout
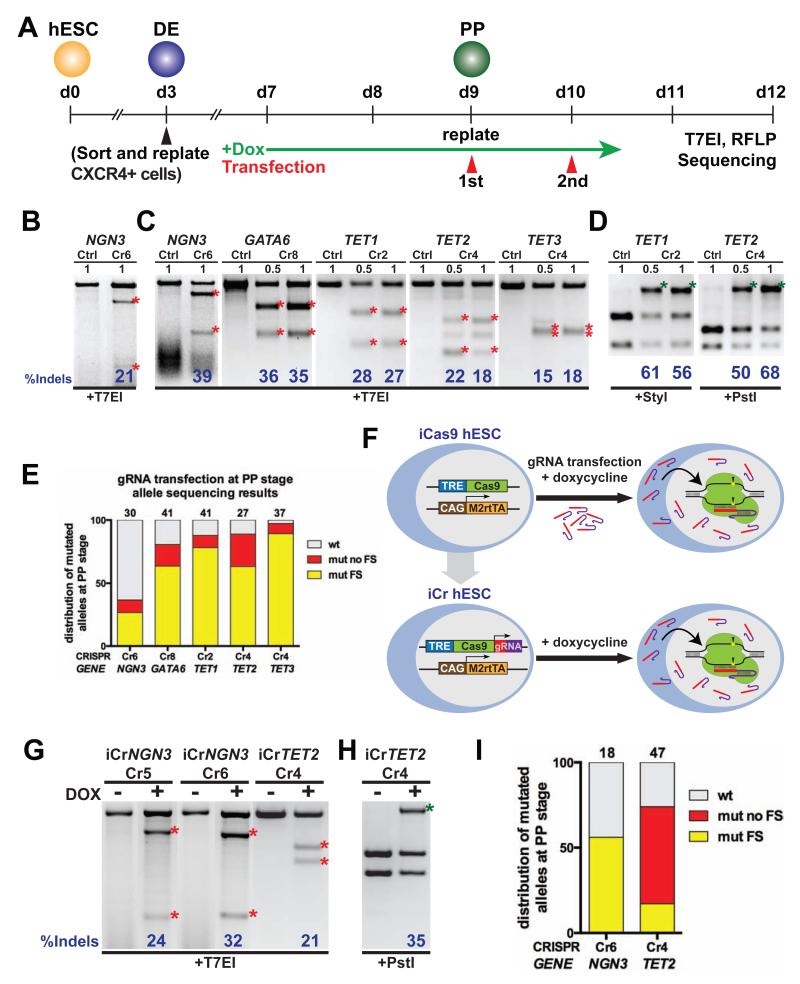
Inactivating genes in a temporal or tissue-specific manner has greatly facilitated the study of genes with pleiotropic effects. Since the iCRISPR platform allows inducible Cas9 expression and tightly regulated delivery of gRNA with minimal toxicity, we investigated the feasibility of conducting inducible gene knockout. We were able to differentiate iCas9 hESCs first into definitive endoderm (DE), and subsequently pancreatic progenitor (PP) and insulin-expressing β-like cells based on an established protocol (Kroon et al., 2008) (Figure S5A, S5B). We proceeded to determine the efficiencies of inducible knockout by inducing Cas9 expression and performing gRNA transfection at the PP stage (Figure 6A). T7EI analysis revealed a ~20% Indel rate in PP cells transfected with an NGN3-targeting gRNA (NGN3-Cr6) (Figure 6B). To further investigate the feasibility of using iCas9 cells for knocking out genes in a tissue-specific manner, we enriched CXCR4-expressing DE cells through fluorescence-activated cell sorting (FACS) (D’Amour et al., 2005), and differentiated the cells further to the PP stage for gRNA transfection (Figure 6A). T7EI analysis showed that PP cells transfected with single gRNAs targeting different genes carried on average ~30% Indels in optimal transfection conditions (Figure 6C). As observed in other experiments, RFLP assays detected higher Indel rates, around 60% with gRNAs targeting TET1 and TET2 (Figure 6D). Sequencing analysis of the PCR amplicon clones, a more direct measurement than T7EI or RFLP, revealed an average of ~78% mutation rate for all targeting experiments and reaching up to ~97% with TET3 (Figure 6E). Notably, the majority of mutations were frameshift, supporting the usage of this inducible gene-editing approach for examination of loss-of-function phenotypes in a temporal and cell-type-specific manner.
Figure 6. Inducible gene knockout.
(A) Strategy of inducible gene knockout. DE, definitive endoderm stage; PP pancreatic progenitor stage. (B and C) T7EI assay in HUES8 iCas9 cells transfected with gRNAs targeting NGN3 (Cr6) at the PP stage (B), or sorted CXCR4+ DE cells differentiated to the PP stage, and transfected with gRNAs targeting NGN3 (Cr6), GATA6 (Cr8), TET1 (Cr2), TET2 (Cr4) and TET3 (Cr4) (C). (D) RFLP analysis of TET1 Cr2 and TET2 Cr4 transfected samples from (C). (E) Allelic sequence distribution in differentiated HUES8 iCas9 cells transfected with different gRNAs at PP stage. wt, wild-type; mut, mutation; FS, frameshift. (F) Schematic illustrating the TALEN-mediated establishment of iCr hESCs for inducible gene knockout. A U6 Pol III driving constitutive expression of a specific gRNA is included 3′ of the inducible Cas9 expression cassette at the AAVS1 locus, allowing gene knockout in all doxycycline treated iCr hESCs. (G) T7EI assay in differentiated iCr hESC lines expressing NGN3-Cr5 (#7), Cr6 (#12) or TET2-Cr4 (#2) gRNAs, treated with doxycycline at the PP stage. (H) RFLP analysis of iCrTET2 (Cr4, #2) hESCs from (G). (I) Allelic sequence distribution in differentiated iCr hESCs treated with doxycycline at PP stage. The number of independent clones analyzed is indicated above each column in panel E and I.
See also Figure S5.
The gRNA transfection method described above may interfere with some differentiation procedures. To overcome this limitation, we generated a Puro-Cr donor by modifying the Puro-Cas9 AAVS1 donor vector to include a constitutive gRNA expression module in addition to the doxycycline-inducible Cas9 expression cassette (Figure S5C). Co-electroporation of this Puro-Cr donor with the Neo-M2rtTA donor and the AAVS1-TALEN constructs allows the generation of hPSC lines for convenient, inducible knockout studies: all cells would express the gRNA of interest, which, upon doxycycline treatment, would target the induced Cas9 protein to the desired genomic locus (Figure 6F). Following this strategy, we generated hESC lines for inducible knockout of NGN3 and TET2, named iCrNGN3 and iCrTET2 respectively (Figure S5D). Doxycycline treatment of differentiated iCrNGN3 and iCrTET2 hESCs at the PP stage resulted in consistent induction of Indels as revealed by T7EI and RFLP assays (Figure 5G, H). Sequencing analysis of the PCR amplicons, revealed ~55% and ~75% mutation rate in induced iCrNGN3 and iCrTET2 hESCs (Figure 6I). The ratio of frameshift vs non-frameshift mutations differed significantly between induced iCrNGN3 and iCrTET2 hESCs, suggesting that the knockout efficiencies also depend on the types of mutations generated. Importantly, to determine the tightness of our inducible system, we analyzed PCR amplicons after prolonged in vitro culture of iCrNGN3 hESCs (15 passages after establishment of the line) in the absence of doxycycline treatment, and identified no mutations associated with potential “leaky” targeting at the NGN3 locus (Figure S5E). These findings demonstrate the versatile use of the iCRISPR platform as a novel approach for inducible gene knockout in hPSCs and their differentiated progeny.
Discussion
Efficient genetic engineering in hPSCs is crucial for elucidating the genes and molecular pathways that underlie complex human traits. While recent approaches based on TALEN and CRISPR/Cas systems have led to encouraging results, a more efficient and universal platform would be highly desirable for large-scale analysis of gene function in the post-genomic era. The iCRISPR platform offers a rapid and efficient approach to introduce mutations in any gene of interest. Based on the high targeting efficiencies observed in our study, the analysis of ~24 colonies should be sufficient to establish multiple mono- and biallelic mutant lines for loss-of-function studies of a single gene. We expect a trained individual could readily perform gRNA transfection, and analyze ~300 colonies at a time, enabling the generation of mutant lines affecting 12 genes in just one month, or 144 genes in a year. Further optimization would enable the use of the iCRISPR platform for high-throughput genetic analysis of disease phenotypes in hPSCs. For instance, one may use the iCRISPR platform to screen an array of human disease-associated genes for their functional relevance.
Using hESC lines in the current NIH hESC registry (HUES8, HUES9 and MEL-1) as well as BJ iPSCs, we have generated multiple iCas9 lines, which will be available to the academic community upon request. Establishing the iCRISPR platform in different hPSC backgrounds requires minimal efforts due to the highly efficient TALEN-mediated AAVS1 targeting approach: only a handful of clonal lines need to be analyzed and the entire procedure takes around a month. Once the system is established, it can be used repeatedly and reliably for rapid and versatile genetic studies in hPSCs, which is highly desirable for laboratories interested in systematically interrogating biological and disease mechanisms in isogenic background. Despite the stable integration of Cas9 in hPSCs, Cas9 activity is tightly regulated by doxycycline treatment, and no adverse effects were observed. Two observations support this conclusion: first, the karyotype of wild-type iCas9 lines remains normal even after 16 additional passages following line establishment, and karyotypically normal mutant lines have been established using the iCRISPR system (Table S1); second, even though iCrNGN3 hESCs constitutively express the gRNA targeting the NGN3 locus, sequencing analysis revealed no mutations in the targeted site even after 15 additional passages in the absence of doxycycline treatment (Figure S5E). Future studies can also include the option to remove the Cas9 gene when desired, by flanking the Cas9 gene with loxP sites for example.
It is worth noting that iCRISPR may create complete loss-of-function (null), partial loss-of-function (hypomorphic) or less frequently, gain-of-function (e.g. dominant negative) alleles. Studying such allelic series may provide valuable insights into the molecular basis of distinct phenotypes caused by different mutations in a single gene, though careful analysis is needed to determine the exact nature of individual mutant alleles. Recent studies suggest that the CRISPR/Cas system may have off-target effects (Cho et al., 2013b; Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013a; Pattanayak et al., 2013), potentially confounding genetic studies in hPSCs. Although our analysis so far has not identified off-target mutations, CRISPR design using recently developed algorithms can reduce potential off-target effects (Hsu et al., 2013; Mali et al., 2013b). Characterizing the full extent of off-target activities in future studies would likely allow more rational design of gRNAs with higher specificity. To increase target specificity, a double nicking approach has been developed recently using a nickase version of Cas9 (nCas9), which requires the cooperation between two gRNAs to create a DNA DSB (Mali et al., 2013a; Ran et al., 2013a). This strategy significantly ameliorates but does not completely eliminate off-target activities. We propose two practical solutions to overcome this limitation for genetic studies in hPSCs. In a first approach, one may generate independent mutant lines using gRNAs targeting different sequences in the same gene: observing the same phenotype from multiple independent mutant lines would strongly suggest that the phenotype is caused by the disruption of the gene of interest. In a complementary approach, one may perform rescue experiments using the iCRISPR platform, which supports highly efficient HDR-mediated scarless correction of mutant sequences.
The iCRISPR platform is also highly versatile. Our platform allows the generation of hPSCs carrying biallelic mutations in multiple genes in just a month, which will greatly accelerate the study of complex multigenic interactions that are often challenging to model in vivo due to the random segregation of alleles. This is highly valuable not only for studying multigenic disorders, but also for genetic epistasis analysis involving multiple genes. Secondly, iCRISPR can also be used to introduce specific nucleotide modifications with high efficiency. This is critical for dissecting protein functional domains, precisely modeling human diseases, and potentially correcting disease-associated mutations for therapeutic intervention. A challenge for modeling complex diseases lies in the large number of susceptibility loci each associated with multiple sequence variants. For instance, APOE is associated with 3 common polymorphisms, and it is among the close to 20 LOAD-susceptibility loci identified so far (Alperovitch et al., 2013). The iCRISPR platform offers an ideal solution: it enables the rapid generation of allelic series, as demonstrated for the APOE locus; and it is expected to greatly facilitate the perturbation of multiple disease-associated loci either individually or in combination in isogenic background.
Finally, we demonstrate, for the first time, that the CRISPR/Cas system can be used for inducible gene knockout, which has not yet been shown in any in vivo or cell culture systems. While the ubiquitous rtTA expression system used in the present study only allowed temporal regulation of Cas9 expression, we were able to achieve tissue-specific regulation by enriching defined cell populations through FACS. Future studies may also achieve tissue specificity by expressing rtTA from tissue-specific promoters. This approach may be further extended to in vivo systems as a simpler alternative to conventional conditional knockout strategies, which generally require complex genetic configurations involving tissue-specific expression of a site-specific recombinase (e.g. Cre) combined with a conditional allele (e.g. a “floxed” allele).
In addition to generating mutant alleles with small Indels or precise nucleotide alterations through NHEJ- or HDR-mediated repair mechanisms as shown in the present study, the iCRISPR platform is likely to also greatly facilitate other types of genome engineering in hPSCs. For instance, iCRISPR may be used to create larger deletions for study of non-coding RNAs or gene regulatory regions such as promoters and enhancers. It may also facilitate the generation of reporter alleles through HDR-mediated gene targeting using long donor DNA templates, or defined chromosomal rearrangements such as translocations. Also, by swapping Cas9 with other newly developed Cas9 variants (e.g. nCas9, dCas9 or dCas9-KRAB) (Gilbert et al., 2013; Mali et al., 2013a; Qi et al., 2013; Ran et al., 2013b), one may repurpose iCRISPR for additional tasks such as gene regulation. Because of the highly efficient TALEN-mediated gene targeting at the AAVS1 locus and the superior speed, one can easily apply iCRISPR to any human cell line of interest, including patient-specific hiPSCs or cancer cell lines. This versatility allows studying gene function in practically any hPSC line or genetic background of choice.
Experimental Procedures
Construction of TALENs, AAVS1 targeting vectors, and Cas9/gRNA expressing vectors
A pair of TALENs (AAVS1-TALEN-L targeting CCCCTCCACCCCACAGT and AAVS1-TALEN-R targeting TTTCTGTCACCAATCCT) was generated to target the first intron of the constitutively expressed gene PPP1R12C at the AAVS1 locus (Hockemeyer et al., 2011). The TALEN constructs were constructed following a published protocol (Sanjana et al., 2012). In brief, a library of TALE monomers with complementary overhangs was built by PCR using vector templates from Addgene (32180, 32181, 32182 or 32183). Monomers were joined into hexamers according to the target DNA sequence. Next the hexamers were linked together and incorporated into the full-length TALEN expression backbone (Addgene 32190).
Neo-M2rtTA donor (Figure S1F) was kindly provided by D. Hockemeyer. Puro-Cas9 donor (Figure S1E) was constructed by replacing EGFP in the TRE-TIGHT-EGFP-BW plasmid (Addgene plasmid 22077) (Hockemeyer et al., 2011) with the human codon-optimized Streptococcus pyogenes Cas9 cDNA amplified by PCR from pX260 (Addgene plasmid 42229) (Cong et al., 2013). Puro-Cr donor was generated by introducing a chimeric gRNA expression cassette, PCR amplified from pX330 (Addgene plasmid 42230) (Cong et al., 2013), 3′ of Cas9 in Puro-Cas9 donor (Figure S5C). piCRct Entry (Figure S1C), a human codon-optimized S. pyogenes Cas9 expression entry vector carrying a bicistronic crRNA/tracrRNA duplex expression cassette was generated by modifying pX260. piCRg Entry (Figure S1D), was built by deleting the bicistronic crRNA/tracrRNA duplex expression cassette from piCRct Entry and replacing it with the chimeric gRNA expression cassette PCR amplified from pX330. To generate CRISPR/Cas9 expression vectors targeting specific genomic loci, 30 or 20 base pairs (bp) of sequence located 5’ of the PAM sequence was cloned in piCRct Entry or piCRg Entry, respectively, following an established protocol (Cong et al., 2013). Briefly, piCRct Entry or piCRg Entry were digested with BbsI, dephosphorylated and gel purified. A pair of oligos including either 30 or 20 bp homology (Table S4) was annealed and phosphorylated, generating BbsI overhangs that can be cloned into the BbsI-digested, dephosphorylated vector. Vectors described in this manuscript will be available to academic researchers through Addgene.
Production of gRNAs through in vitro transcription (IVT)
For production of gRNA, we first generated a T7-gRNA IVT template by adding the T7 promoter to the gRNA sequence in the piCRg Entry vector through PCR amplification using CRISPR-specific forward primers and a universal reverse primer (Table S4). Alternatively, for APOE gRNAs we designed a 120-nt oligo including the T7 promoter and the full-length gRNA sequence. This oligo was used as a template for PCR amplification using T7 and gRNA universal primers (Table S4).
T7-gRNA PCR products were used as templates for IVT using the MEGAshortscript T7 kit (Life Technologies). The resulting gRNAs were purified using the MEGAclear kit (Life Technologies), eluted in RNase-free water and stored at −80°C until use.
gRNA or gRNA+ssDNA transfection
iCas9 hPSCs were treated with doxycycline (2 μg/ml) for one or two days before and during transfection. For transfection, cells were dissociated using Accutase (Stem Cell Technologies) or TrypLE (Life Technologies), replated onto iMEF-coated plates and transfected in suspension with gRNAs or gRNA+ssDNA using Lipofectamine RNAiMAX (Life Technologies) following manufacturer’s instructions. Briefly, gRNA and ssDNA were added at a 10 nM and 20 nM final concentrations respectively, unless otherwise indicated. gRNAs (or gRNA+ssDNA) and Lipofectamine RNAiMAX were diluted separately in Opti-MEM (Life Technologies), mixed together, incubated for 5 min at RT, and added dropwise to cultured hPSCs. A second transfection was performed 24 hours later in some experiments.
T7EI, RFLP analysis for assessment of genome modification and off-target analysis
Genomic DNA was extracted two or three days after last gRNA transfection. Genomic regions flanking the CRISPR target sites were PCR amplified (Table S4). For T7EI assays, 12 μL of PCR products were denatured and re-annealed in NEB Buffer 2 (New England Biolabs) in a total volume of 25 μL using the following protocol: 95°C, 5 min; 95-85°C at −2°C/s; 85-25°C at −0.1°C/s; hold at 4°C. 12.5 μL of hybridized PCR products were treated with 5 U of T7EI at 37°C for 15 min in 13 μL final reaction volume. Products were then analyzed on 2.5% agarose gels and imaged with a Gel Doc gel imaging system (Bio-Rad). Quantification was based on relative band intensities using ImageJ. Indel percentage was determined by the formula: 100 × (1 − (1 − (b + c) / (a + b + c))1/2), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product (Hsu et al., 2013). For RFLP analysis, 10 μL PCR products were digested with enzymes and analyzed on 2.5% agarose gel. Indel percentage was determined by the formula: 100 × a / (a+b+c).
For off-target analysis, ectopic gRNAs targets were identified using the rules outlined in a previous study (Mali et al., 2013b). The most likely off targets falling in gene coding sequences (4-5 sites per gRNA-mediated targeting experiment) were analyzed through sequencing. Primers for PCR amplification and sequencing of each off-target site are summarized in Table S4.
Establishment of knockout or knockin lines through NHEJ- or HDR-mediated repair
Two days after the last gRNA or gRNA+ssDNA (Table S4) transfection, hPSCs were dissociated into single cells and replated at ~2,000 cells per 10-cm dish. Cells were allowed to grow until colonies from single cells became visible (~10 days). Single colonies were randomly picked based on hPSC morphology, mechanically disaggregated and replated into individual wells of 96-well plates. Colonies were amplified, replated as described above and analyzed by Sanger sequencing to identify mutant clones. Clonal cell lines carrying desired mutations were amplified and frozen down. Alternatively, for APOE experiments, colonies were picked and directly analyzed by Sanger sequencing to identify mutant clones.
Supplementary Material
Highlights.
Rapid and efficient generation of biallelic gene knockouts
Rapid one-step creation of biallelic mutations in multiple genes
Efficient introduction of biallelic, precise nucleotide alterations
Inducible gene knockout during specific stages of hPSC differentiation
Acknowledgments
We thank Feng Zhang and Rudolf Jaenisch for providing vectors through Addgene; Dirk Hockemeyer for the Neo-M2rtTA donor and the AAVS1 3′ external probe vector; Qiurong Ding from Chad Cowan’s laboratory for providing technical advice on picking hPSC colonies for identification and establishment of mutant clones; Drs. Ed Stanley and Andrew Elefanty for kindly providing MEL-1 cells; Gouri Nanjangud at the MSKCC Molecular Cytogenetics Core for performing karyotype analysis; Daniela Georgieva for technical assistance; and Lorenz Studer, Wenjun Guo and members of the Huangfu laboratory for insightful discussions and critical reading of the manuscript. This study was funded in part by NIH (R01DK096239), Basil O’Connor Starter Scholar Award from March of Dimes Birth Defects Foundation, Tri-Institutional Stem Cell Initiative, Louis V. Gerstner Jr. Young Investigators Award, and MSKCC Society Special Projects Committee. F.G. and Z.Z. were supported by the New York State Stem Cell Science fellowship from the Center for Stem Cell Biology of the Sloan-Kettering Institute. N.V. was supported by the Howard Hughes Medical Institute (HHMI) Medical Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.H., F.G., Z.Z., Z.S. and K.L. conceived the experiments; F.G., Z.Z., Z.S., K.L., N.V. and Q.L. performed the experiments; and D.H., F.G. and Z.Z. wrote the manuscript.
References
- Alperovitch A, Boland A, Delepoine M, Dubois B, Duron E, Epelbaum J, Van Cauwenberghe C, Engelborghs S, Vandenberghe R, De Deyn PP, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nature reviews Genetics. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic acids research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013a;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013b;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature reviews Genetics. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. Derivation of embryonic stem-cell lines from human blastocysts. The New England journal of medicine. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic acids research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell. 2013a;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013b;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods in molecular biology. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. Gene targeting in human pluripotent cells. Cold Spring Harbor symposia on quantitative biology. 2010;75:201–209. doi: 10.1101/sqb.2010.75.021. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nature reviews Molecular cell biology. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal RD, Koontz J, Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature genetics. 1995;9:177–183. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013a;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013b;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, ffrench-Constant C, Pedersen RA. Robust, Persistent Transgene Expression in Human Embryonic Stem Cells Is Achieved with AAVS1-Targeted Integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Targeting of genes to specific sites in the mammalian genome. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 2):1101–1113. doi: 10.1101/sqb.1986.051.01.128. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Huangfu D. Human pluripotent stem cells: an emerging model in developmental biology. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.