Abstract
Background
The age at onset of alcohol dependence (AD) is a critical moderator of genetic associations for alcohol dependence. The present study evaluated whether single nucleotide polymorphisms (SNPs) can influence the age at onset of AD in large high-risk families from the Collaborative Study on the Genetics of Alcoholism (COGA).
Methods
Genomewide SNP genotyping was performed in 1788 regular drinkers from 118 large European American families densely affected with alcoholism. We used a genome-wide Cox proportional hazards regression model to test for association between age at onset of AD and SNPs.
Results
This family-based analysis identified an intergenic SNP, rs2168784 on chromosome 3 that showed strong evidence of association (p= 5 × 10−9) with age at onset of AD among regular drinkers. Carriers of the minor allele of rs2168784 had 1.5 times the hazard of AD onset as compared with those homozygous for the major allele. By the age of 20 years, nearly 30% of subjects homozygous for the minor allele were alcohol dependent while only 19% of those homozygous for the major allele were. We also identified intronic SNPs in the ADP-ribosylation factor like 15 (ARL15) gene on chromosome 5 (P = 1.11 × 10−8) and the UTP20 small subunit (UTP20) gene on chromosome 12 (P = 4.32 × 10−8) that were associated with age at onset of AD.
Conclusions
This extended family based genome-wide cox-proportional hazards analysis identified several loci that might be associated with age at onset of AD.
Keywords: GWAS, alcohol dependence, age at onset, survival analysis, SNP
1. INTRODUCTION
The hazardous and harmful use of alcohol is a major contributing factor to death, disease and injury globally. Almost 4% of all deaths worldwide are due to alcohol, more than the percent of deaths caused by HIV/AIDS, violence or tuberculosis (World Health Organization, 2011). Both genetic and environmental factors influence the initiation of alcohol use and subsequent alcohol dependence (AD; Bierut, 2011; Kendler et al., 2008; Wang et al., 2012). AD is a complex disorder with high heritability (50-60%; Heath et al., 1997; Kendler et al., 2010, 1994). However, the relationship between genetic risk factors and AD is likely complex, due to the interplay between genes and the environment (Blomeyer et al., 2013; Perry et al., 2013; van der Zwaluw and Engels, 2009).
There is strong evidence of familial aggregation of AD and related traits. Numerous studies have shown that first degree relatives of alcoholics are two to seven times more likely to develop problems with alcohol at some time in their lives, than individuals without a family history of AD (Grant, 1998; Nurnberger et al., 2004; Reich et al., 1998). Family history of alcoholism might be an indicator of shared or common environmental factors, genetic influences, or a combination of both (Grant, 1998). Familiality of alcoholism is also associated with age at onset and severity of AD (Limosin et al., 2001).
A large proportion of variation (~49%) in age at onset of AD can be attributed to genetic factors (Le Strat et al., 2010). Indeed candidate gene based studies and genome-wide linkage studies have reported variants and genomic regions, respectively that were associated with early age at onset of AD (Edenberg et al., 2008; Tayo et al., 2005; Zhong and Zhang, 2005).
The emergence of genome-wide association studies (GWAS) has provided an opportunity to employ an unbiased approach to identify additional genes related to AD. Prior GWAS have focused on the detection of susceptibility genes to AD (Bierut et al., 2010; Edenberg et al., 2010; Gelernter et al., 2014; Treutlein et al., 2009) and related traits (Baik et al., 2011; Heath et al., 2011; Kapoor et al., 2013; Kendler et al., 2011; Schumann et al., 2011; Wang et al., 2013; Wetherill et al., 2014) but none have investigated genes influencing age at onset. In this study we took advantage of a family-based study design in a well-characterized European American (EA) cohort and performed genome-wide Cox proportional hazards analysis to identify genes associated with age at onset of AD. The extended AD families might be enriched for common factors increasing risk for AD, hence providing additional power to identify the genes associated with age at onset of AD. The analysis demonstrated genome-wide evidence of association of age at onset of AD with several novel loci.
2. MATERIAL AND METHODS
2.1. Subjects
Six sites participating in the Collaborative Study on the Genetics of Alcoholism (COGA; Begleiter et al., 1995; Foroud et al., 2000a) recruited alcohol dependent probands from inpatient and outpatient facilities. The probands and their family members were administered a poly-diagnostic interview, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA; Bucholz et al., 1994; Hesselbrock et al., 1999). Individuals 18 years of age or younger were administered an adolescent version of the SSAGA. Institutional review boards at all sites approved the study. The present COGA sample consisted of 1788 regular drinkers subjects of European descent from 118 extended families. Further details about the complete cohort are described elsewhere (Kang et al., 2012; Wang et al., 2013).
2.2. Phenotype
Only individuals who reported drinking once per month for 6 months or more (regular drinking) at any evaluation were included in the analysis. If an individual was interviewed more than once, data from the SSAGA interview with the maximum total number of endorsed DSMIV alcohol dependence criteria was utilized. Age at onset of DSM-IV alcohol dependence (defined as age when full criteria were met), age at last interview, year of birth, gender and principal component 1 (PC1) for ancestry was included in the analysis. Table 1 shows the distribution of age at interview and age at onset of AD for the controls and cases respectively.
Table 1.
Distribution of age at last interview and age at onset of AD in COGA cohort
| Age at last Interview (Mean + S.D.) yrs |
Age at onset of AD (Mean ± S.D.) yrs |
|
|---|---|---|
| Alcohol Dependent (N = 685) | 38.61 + 12.94A | 22.50 + 7.65 |
| Non Dependent (N = 1103) | 35.81 + 15.49A | - |
t = 3.96; P < 0.0001
2.3. Genotyping
Genotyping was performed at the Genome Technology Access Center at Washington University School of Medicine in St. Louis (http://gtac.wustl.edu/) using the Illumina Human OmniExpress array 12.VI (Illumina, San Diego, CA, USA). A total of 707,557 autosomal SNPs passed quality control (Wang et al., 2013). SNPs with a minor allele frequency (MAF) below 5% (n=115,872) were excluded from further analysis. EIGENSTRAT was utilized in the HapMap European reference samples to identify ethnic stratification within the sample.
2.4. Imputation
We used BEAGLE version 3.3.1 (Browning and Browning, 2007) to impute SNPs that were not genotyped on the Illumina Omni Express array. Since our sample was European American, we used as a reference set the genotypic data from the EUR in the August 2010 release of the 1000 Genomes Project, provided with the Beagle 3.3.1 release. To account for uncertainty, we used the mean of the distribution of imputed genotypes, which corresponds to an expected allelic or genotypic count (dosage) for each individual. SNPs with a correlation between the best-guess genotype and allele dosage greater than 0.3 (r2>0.3), were used in the analyses. For individual-level genotype data, we retained genotypes having a probability ≥80% (from the gprob metric in Beagle); all other genotypes were set to missing. We converted genotypic probability data into most-likely genotypes. This allowed us to detect genotypic errors in families. The same rigorous quality control process used for genotyped SNPs was also applied to imputed SNPs. A total of 4,058,415 SNPs (MAF > 5%) that passed quality control and Mendelian inheritance checks were used for association analysis.
2.5. Data Analysis
The density plot of age at interview for DSM-IV AD and non-dependent subjects shows that a large proportion of non-dependent subjects are younger than the AD subjects at their last interview (Figure 1). We included these high-risk non-AD subjects in the analysis after censoring them at age at latest interview and performed the age at onset analysis of AD using survival models. We used a genome-wide Cox proportional hazards (coxph) analysis, a survival analysis package in R (www.r-project.org), to estimate the hazard ratio for AD occurrence based on genotype, controlling for sex, birth cohort and first principal component (PC1) from EIGENSTRAT (Price et al, 2006). This analysis incorporates a clustered sandwich estimator to account for the familial correlation among observations. Violation of the proportional hazards assumption was tested with non-zero slope of Schoenfeld residuals versus time, using the survival analysis package in R. The analyses of AD for the strongest signals were conducted using the GWAF, an R package for genome-wide association analyses with family data (Chen and Yang, 2010). A logistic regression model was employed with gender, age and cohort included as covariates, and a log additive model for each SNP was tested for association. The generalized estimating equation (GEE) framework was used to control for relatedness. The association of individual SNPs with SC was performed using PROC GLIMMIX from SAS (http://support.sas.com/rnd/app/da/glimmix.html). Birth cohorts and gender were included as covariates for the association analysis.
Fig. 1.
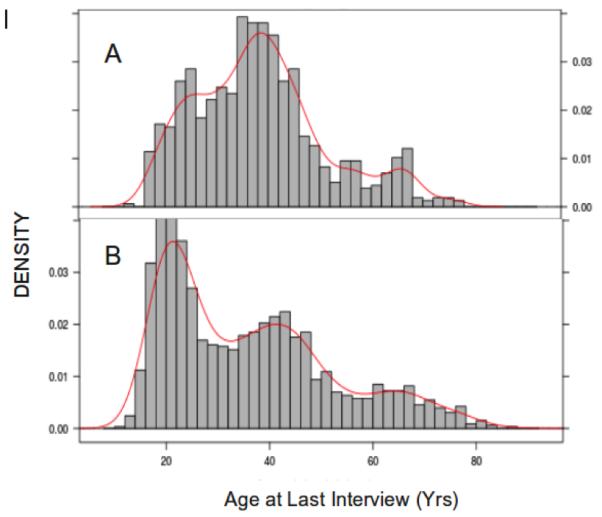
Parts A-B: Distribution of age at last interview among DSM-IV alcohol dependent subjects (A) and non- alcohol dependent subjects (B).
X axis represents the age at last interview and Y axis represents the density of subjects. The observed values of age at last interview were used to construct the relative likelihoods (density) at given age.
3. RESULTS
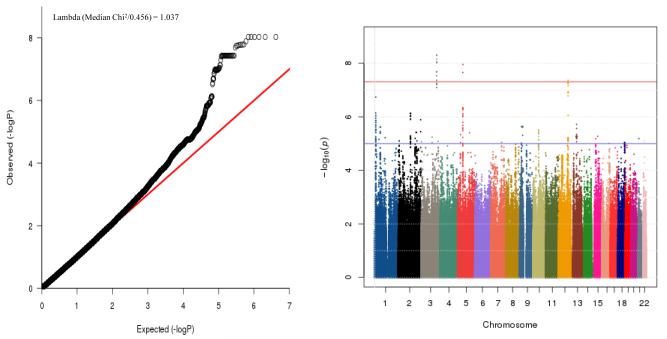
Genome-wide cox-proportional hazards analysis with 4,058,415 SNPs identified genome wide significant SNPs (p < 5.0 × 10−8) in 3 loci: 35 SNPs clustered on chromosome 3, 2 on chromosome 5 and one on chromosome 12 (Table 2, Fig. 2). A total of 225 SNPs showed suggestive association at p < 1.0 × 10−5 (please see supplemental e Table 1 for SNPs with genome-wide suggestive association1). Among these 225 SNPs, 95 SNPs were in strong LD (D’=1; r2 > 0.8) with the most significant SNPs on chromosomes 3, 5 and 12.
Table 2.
Summary of results for the SNPs showing genome-wide significant evidence of association (p < 5 × 10−8) in COGA dataset
| COGA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | BP | MAF | A1 | A2 | N | HR | SE | P |
| rs57350471 | 3 | 164520763 | 0.10 | A | T | 1761 | 1.45 | 0.07 | 2.11E-08 |
| rs9871275 | 3 | 164520783 | 0.10 | T | C | 1761 | 1.45 | 0.07 | 2.11E-08 |
| rs9875364 | 3 | 164520853 | 0.10 | C | G | 1762 | 1.44 | 0.07 | 2.06E-08 |
| rs9862987 | 3 | 164531227 | 0.10 | C | A | 1772 | 1.43 | 0.07 | 4.31E-08 |
| rs9817317 | 3 | 164554801 | 0.11 | A | G | 1783 | 1.44 | 0.06 | 9.29E-09 |
| rs7630142 | 3 | 164555706 | 0.11 | G | A | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9874932 | 3 | 164556648 | 0.11 | A | G | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9824625 | 3 | 164557575 | 0.11 | C | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9880338 | 3 | 164557652 | 0.11 | A | G | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs4541411 | 3 | 164557917 | 0.11 | C | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9847462 | 3 | 164558595 | 0.11 | G | A | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs28821642 | 3 | 164559430 | 0.11 | T | C | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs7645282 | 3 | 164560182 | 0.11 | G | A | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs7647704 | 3 | 164560194 | 0.11 | A | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs7640074 | 3 | 164562164 | 0.11 | T | C | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9855885 | 3 | 164563201 | 0.11 | C | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9873722 | 3 | 164563390 | 0.11 | G | A | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9819217 | 3 | 164565763 | 0.11 | T | C | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9830200 | 3 | 164567759 | 0.11 | G | A | 1755 | 1.44 | 0.07 | 2.91E-08 |
| rs9290244 | 3 | 164568563 | 0.11 | G | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs7628882 | 3 | 164571138 | 0.11 | A | G | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9881091 | 3 | 164573311 | 0.11 | G | T | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs4602392 | 3 | 164578266 | 0.11 | T | C | 1783 | 1.44 | 0.06 | 2.07E-08 |
| rs9865078 | 3 | 164579310 | 0.11 | A | G | 1786 | 1.45 | 0.06 | 9.71E-09 |
| rs7619524 | 3 | 164580672 | 0.11 | T | C | 1786 | 1.45 | 0.06 | 9.71E-09 |
| rs6791103 | 3 | 164585575 | 0.11 | C | A | 1783 | 1.45 | 0.06 | 8.85E-09 |
| rs5023277 | 3 | 164586715 | 0.11 | G | C | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs9879548 | 3 | 164587302 | 0.11 | T | G | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs7641732 | 3 | 164587350 | 0.11 | T | A | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs6808582 | 3 | 164587559 | 0.11 | A | G | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs6763281 | 3 | 164588205 | 0.11 | T | G | 1783 | 1.45 | 0.06 | 9.02E-09 |
| rs6788557 | 3 | 164588388 | 0.11 | G | C | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs9810313 | 3 | 164588702 | 0.11 | T | G | 1786 | 1.45 | 0.06 | 6.95E-09 |
| rs56303997 | 3 | 164589150 | 0.11 | A | G | 1783 | 1.45 | 0.06 | 9.02E-09 |
| rs2168784 | 3 | 164589939 | 0.11 | T | C | 1784 | 1.46 | 0.06 | 4.99E-09 |
| rs35952 | 5 | 53592163 | 0.14 | C | T | 1780 | 1.42 | 0.06 | 2.21E-08 |
| rs35951 | 5 | 53592361 | 0.14 | G | T | 1776 | 1.43 | 0.06 | 1.11E-08 |
| rs57083693 | 12 | 101742180 | 0.22 | C | T | 1744 | 1.35 | 0.06 | 4.32E-08 |
CHR = Chromosome, BP = Base pairs; MAF = minor allele frequency; A1 = minor allele; A2 = Major allele; N= number of regular drinkers; HR = Hazard ratio; SE = standard error
Fig. 2.
Parts A-B: (A) Quantile-Quantile (QQ) plot and (B) Manhattan plot for the genome-wide association analysis of the age at onset of DSM-IV alcohol dependence in COGA (a) Observed p values for the 4,058,415 SNPs (black dots) were plotted against the expected p value (x--axis). The genomic inflation factor value (lambda) was 1.037.
(b) Observed log p values for the 4,058,415 SNPs were plotted according to chromosomal position.
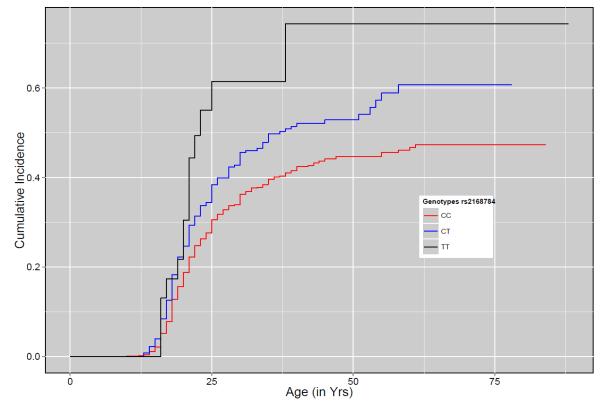
An intergenic variant on chromosome 3, rs2168784, emerged as the SNP most significantly associated with age at onset of AD (MAF = 0.11, HR = 1.46, P = 4.99 × 10−9). The results of Cox hazard analysis indicated that the incidence of AD in carriers of the rs2168784 minor allele was 1.5 times greater than the subjects homozygous for the major allele. Twenty one percent of subjects were carriers of the minor allele of rs2168784, 47% of whom were diagnosed with AD. In this high-risk sample, the cumulative incidence plots indicate that by the age of 20 years, nearly 30% of the subjects homozygous for the minor allele of rs2168784 met criteria of AD. In comparison, only 19% of subjects homozygous for the major allele were diagnosed with AD by age 20 (Fig. 3(a). This SNP was also associated with AD symptom count (p = 4.1 × 10−6) and DSM-IV AD (p = 2.6 × 10−7) in the same sample of COGA families. Although rs2168784 has a variable MAF across populations the frequency in unrelated individuals from the COGA families (0.111) was similar to the MAF for Utah Residents with Northern and Western European ancestry (CEU) population estimated in 1000 genome project (0.118).
Fig. 3.
Part A: Cumulative Incidence plot for rs2168784 on chromosome 3
X-axis represents the age at onset for alcohol dependence. Y-axis represents the cumulative incidence of AD. The red, blue and black lines show the cumulative incidence of AD for subjects with CC, CT and TT genotypes respectively. Steps on each line represent the occurrence of event (AD).
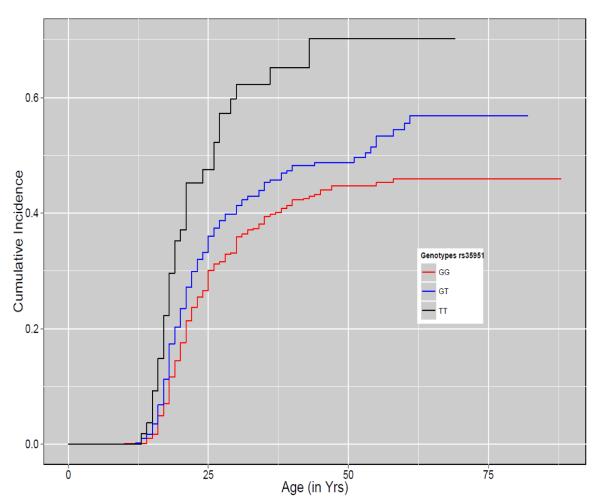
Part B: Cumulative Incidence plot for rs35951 on chromosome 5
X-axis represents the age at onset for alcohol dependence. Y-axis represents the cumulative incidence of AD. The red, blue and black lines show the cumulative incidence of AD for subjects with GG, GT and TT genotypes respectively. Steps on each line represent the occurrence of event (AD).
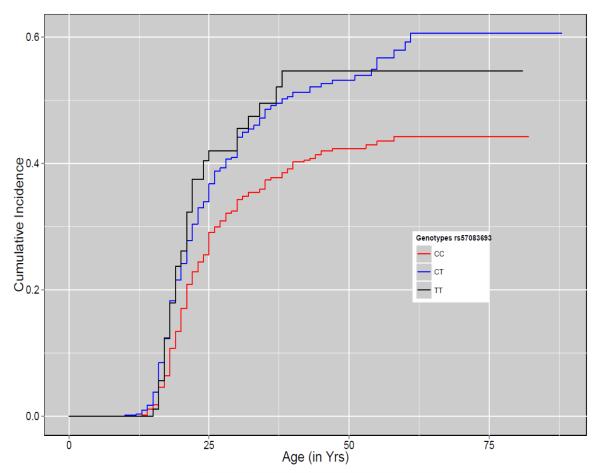
Part C: Cumulative Incidence plot for rs57083693 on chromosome 12
X-axis represents the age at onset for alcohol dependence. Y-axis represents the cumulative incidence of AD. The red, blue and black lines show the cumulative incidence of AD for subjects with CC, CT and TT genotypes respectively. Steps on each line represent the occurrence of event (AD).
Two intronic SNPs, rs35951 and rs35952 (MAF = 0.14, HR = 1.43, P = 1.11 × 10−8) in ADP-ribosylation factor like 15 (ARL15) gene on chromosome 5 and one intronic SNP, rs57083693 (MAF = 0.22, HR = 1.35, P = 4.32 × 10−8) in UTP20 small subunit (UTP20) gene on chromosome 12, were also associated with incidence of AD at genome-wide significance. The cumulative incidence plot showed that by the age of 20 years nearly 37% of subjects homozygous for the minor allele of rs35951 were diagnosed with AD, while only 17% of subjects homozygous for major allele developed AD (Fig. 3B). The CI plot for rs57083693 showed a dominant effect of the minor allele on the age at onset of AD (Fig. 3C). By age of 20 years nearly 25% carriers of minor allele of rs57083693 were diagnosed with AD, while only 19% subjects homozygous for major allele were diagnosed with AD at age 20.
4. DISCUSSION
Here we present the results from the first GWAS of age at onset of AD. Survival analysis identified 38 SNPs showing genome-wide significance (p < 5.0 × 10−8) and 909 SNPs showing suggestive association (p < 5.0 × 10−5) with age at onset of AD. The genome-wide significant SNP rs2168784 was in strong LD with 33 imputed/genotyped SNPs spanning a 60 kb region on chromosome 3, from a long intergenic non-coding RNA (lincRNA) to 106 kb 3′ of the Sucrase Isomaltase (SI) gene (please see supplemental e Fig. 1 for regional plot for variants flanking rs2168784). The same SNPs also showed association with AD but with less significance than the age at onset analyses, this is most likely due to the fact that there are many people in this dataset who are young and have therefore not yet passed through the peak period of risk for developing AD. The chromosomal region identified in the current association analysis partially overlaps with the alcohol dependence linkage signal on chromosome 3 previously reported in a COGA dataset (Foroud et al., 2000b) from which these subjects were drawn. In contrast to the GWAS dataset used in this study the linkage sample consisted of both non-Hispanic white families and African American (AA) families. Indeed. The linkage signal is apparent in each population when analyzed separately suggesting that there is an AD risk locus in this region in both populations. However, the imputed and genotyped SNPs showing the strongest association in EAs exhibit much lower LD (D’ = 0.84, r2 = 0.12, 1000 genome pilot dataset) with each other in subjects of African ancestry. As a result, although we have linkage data and exome chip data in the AA families we cannot accurate impute the associated SNPs. Furthermore, assuming these SNPs tag the functional variation but are not the functional alleles the differences in LD may mean that these SNPs are not in LD with the functional alleles in AAs. Further genotyping and sequencing in the AA families will be needed to determine whether the linkage signals observed on chromosome 3 in the two populations reflect the association observed in this study in EA families.
LincRNAs are usually associated with open chromatin signatures such as histone modification sites. There is emerging evidence suggesting that lincRNAs regulate gene expression both during normal development and under pathological conditions, including neuropsychiatric disorders (Dudley et al., 2011; Mattick, 2009). We utilized the HaploReg tool (Ward and Kellis, 2012) to explore the annotations of the non-coding genome for the significant SNPs and found no significant enrichment of regulatory markers at this locus. The strongest signals on chromosome 3 were also in moderate LD (D’=1, r2 = 0.42) with SNPs in the neighboring SI gene. The SI gene encodes a sucrase-isomaltase enzyme, which is essential for the digestion of dietary carbohydrates including starch, sucrose and isomaltase.
Other genome-wide significant SNPs were in the ARL15 gene on chromosome 5 (rs35951, rs35952) and the UTP20 gene on chromosome 12 (rs57083795). These SNPs also showed suggestive evidence of association (P < 5 × 10−5) with the AD and AD symptom counts in this family based dataset. ARL15 was previously shown to be associated with adiponectin levels, but the exact function of this gene is still unknown (Richards et al., 2009). Several studies have found that adiponectin levels are elevated in patients with chronic alcohol abuse (Buechler et al., 2009) and might also be associated with alcohol craving (Hillemacher et al., 2009). The enrichment analysis using HaploReg showed that SNPs identified at this locus in the COGA dataset predicted significant (p < 5.2 × 10−4) enrichment of enhancers in H1 and leukemia cell lines. There was also evidence (p = 3.1 × 10−2) of enrichment of DNase hypersensitive sites in undifferentiated embryonic stem cells. The UTP20 gene on chromosome 12 is a component of the U3 small nucleolar RNA protein complex and is involved in 18S rRNA processing (Wang et al., 2007). One of the most strongly associated SNPs, rs2270861 (p = 6.79 × 10−8) in UTP20 is predicted to be a trans eQTL for the olfactory receptor family 5, subfamily T, member 2 (OR5T2) gene on chromosome 11, as observed in data of Dixon et al (2007).
One assumption of our model is that all subjects in each family are at risk of developing AD, because they belong to high-risk families that were selected for high prevalence of alcohol use disorders. Many people in these COGA families are aged between 12-25yrs. These individuals may be unaffected but are still at substantial risk of developing AD because they have not yet passed through the age of peak risk for developing AD. In contrast, there is little or no increase in power to be gained by using the age at onset of AD as a phenotype in population based cohorts or case-control datasets because the unaffected subjects in these datasets are generally older than the mean age at onset of AD, are not necessarily ascertained from high-risk alcoholic families and might not be exposed to predisposing genetic or environmental factors. We are not aware of any other dataset with a similar study design to the densely affected COGA families. This lack of a well-matched study design was the main reason we did not attempt to replicate the strongest signals. The lack of direct replication is one of the biggest limitations of the study, but the linkage signal on chromosome 3 in EAs and AAs families provides some additional support for an AD risk locus in this region. Nonetheless the current study provides a well-characterized dataset that can be used in genome-wide meta-analysis of AD or age at onset of AD.
In summary, the present study used high-risk extended COGA families and identified 3 novel genome-wide significant loci associated with age at onset of AD. The identified genetic variants predisposed subjects to higher risk of AD and increased incidence of AD in these families. Further large family based studies will be needed to validate these GWAs loci and the role of genetic variants on the age at onset of AD.
Supplementary Material
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA): COGA, Principal Investigators B Porjesz, V Hesselbrock, H Edenberg, L Bierut includes ten different centers: University of Connecticut (V Hesselbrock); Indiana University (HJ Edenberg, J Nurnberger Jr, T Foroud); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (B Porjesz); Washington University in Saint Louis (L Bierut, A Goate, J Rice, K Bucholz); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield); Southwest Foundation (L Almasy), Howard University (R Taylor) and Virginia Commonwealth University (D Dick). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Role of funding source The Collaborative Study on the Genetics of Alcoholism (COGA): This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Dr. Kapoor is supported through COGA and R21AA021235.
Footnotes
Contributors: MK and AG conceived the analysis. MK performed all the analysis. LW, TF and AG designed the family based dataset. SB, TH, NL, JB, MK and JCW were involved in QC of genotyped and imputed SNPs. Phenotype expertise was provided by LB. VH, JK, JN, JR, MS, JT, BP, HE, LB, KB are involved in data collection, data management and overall designing of project. Support and suggestions for analysis were also provided by LA, DD, HE, AG, TF, AA, XL and OH. All authors read and critically reviewed the manuscript.
Conflict of interest: We disclose that Drs. LJ Bierut, AM Goate and JC Wang are listed as inventors on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:…
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:…
REFERENCES
- Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am. J. Clin. Nutr. 2011;93:809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, Edenberg H, Rice JP. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res. World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ. Genetic vulnerability and susceptibility to substance dependence. Neuron. 2011;69:618–627. doi: 10.1016/j.neuron.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, et al. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomeyer D, Buchmann AF, Lascorz J, Zimmermann US, Esser G, Desrivieres S, Schmidt MH, Banaschewski T, Schumann G, Laucht M. Association of PER2 genotype and stressful life events with alcohol drinking in young adults. PLoS One. 2013;8:e59136. doi: 10.1371/journal.pone.0059136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr., Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J. Stud. Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Buechler C, Schaffler A, Johann M, Neumeier M, Kohl P, Weiss T, Wodarz N, Kiefer P, Hellerbrand C. Elevated adiponectin serum levels in patients with chronic alcohol abuse rapidly decline during alcohol withdrawal. J. Gastroenterol. Hepatol. 2009;24:558–563. doi: 10.1111/j.1440-1746.2008.05693.x. [DOI] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW. Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neurosci. Biobehav. Rev. 2011;35:1544–1551. doi: 10.1016/j.neubiorev.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Almasy LA, Nurnberger JI, Jr., Foroud T. Association of NFKB1, which encodes a subunit of the transcription factor NF-kappaB, with alcohol dependence. Hum. Mol. Genet. 2008;17:963–970. doi: 10.1093/hmg/ddm368. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol. Clin. Exp. Res. 2000a;24:933–945. [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol. Clin. Exp. Res. 2000b;24:933–945. [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, et al. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF. The impact of a family history of alcoholism on the relationship between age at onset of alcohol use and DSM-IV alcohol dependence: results from the National Longitudinal Alcohol Epidemiologic Survey. Alcohol Health Res. World. 1998;22:144–147. [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol. Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol. Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Weinland C, Heberlein A, Groschl M, Schanze A, Frieling H, Wilhelm J, Kornhuber J, Bleich S. Increased levels of adiponectin and resistin in alcohol dependence--possible link to craving. Drug Alcohol Depend. 2009;99:333–337. doi: 10.1016/j.drugalcdep.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, et al. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum. Genet. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin. Exp. Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol. Clin. Exp. Res. 2010;34:1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am. J. Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Schmitt E, Aggen SH, Prescott CA. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch. Gen. Psychiatry. 2008;65:674–682. doi: 10.1001/archpsyc.65.6.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Strat Y, Grant BF, Ramoz N, Gorwood P. A new definition of early age at onset in alcohol dependence. Drug Alcohol Depend. 2010;108:43–48. doi: 10.1016/j.drugalcdep.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limosin F, Gorwood P, Ades J. Clinical characteristics of familial versus sporadic alcoholism in a sample of male and female patients. Eur. Psychiatry. 2001;16:151–156. doi: 10.1016/s0924-9338(01)00556-9. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Wiegand R, Bucholz K, O’Connor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, Bierut L, Hinrichs AL, et al. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch. Gen. Psychiatry. 2004;61:1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- Perry BL, Pescosolido BA, Bucholz K, Edenberg H, Kramer J, Kuperman S, Schuckit MA, Nurnberger JI., Jr. Gender-specific gene-environment interaction in alcohol dependence: the impact of daily life events and GABRA2. Behav. Genet. 2013;43:402–414. doi: 10.1007/s10519-013-9607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Richards JB, Waterworth D, O’Rahilly S, Hivert MF, Loos RJ, Perry JR, Tanaka T, Timpson NJ, Semple RK, Soranzo N, Song K, Rocha N, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5:e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayo BO, Liang Y, Stranges S, Trevisan M. Genome-wide linkage analysis of age at onset of alcohol dependence: a comparison between microsatellites and single-nucleotide polymorphisms. BMC Genet. 2005;6(Suppl. 1):S12. doi: 10.1186/1471-2156-6-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, et al. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC. Gene-environment interactions and alcohol use and dependence: current status and future challenges. Addiction. 2009;104:907–914. doi: 10.1111/j.1360-0443.2009.02563.x. [DOI] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, Almasy L, Brooks AI, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol. Psychiatry. 2013;18:1218–1224. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kapoor M, Goate AM. The genetics of substance dependence. Annu. Rev. Genomics Hum. Genet. 2012;13:241–261. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu J, Zhao H, Lu W, Zhao J, Yang L, Li N, Du X, Ke Y. Human 1A6/DRIM, the homolog of yeast Utp20, functions in the 18S rRNA processing. Biochim. Biophys. Acta. 2007;1773:863–868. doi: 10.1016/j.bbamcr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill L, Kapoor M, Agrawal A, Bucholz K, Koller D, Bertelsen SE, Le N, Wang JC, Almasy L, Hesselbrock V, Kramer J, Nurnberger JI, Jr., et al. Family-based association analysis of alcohol dependence criteria and severity. Alcohol. Clin. Exp. Res. 2014;38:354–366. doi: 10.1111/acer.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Management of Substance Abuse Team . Global Status Report on Alcohol and Health. World Health Organization; Geneva, Switzerland: 2011. [Google Scholar]
- Zhong X, Zhang H. Linkage analysis and association analysis in the presence of linkage using age at onset of COGA alcoholism data. BMC Genet. 2005;6(Suppl. 1):S31. doi: 10.1186/1471-2156-6-S1-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.