Abstract
With the recent dawn of synthetic biology, the old idea of man-made artificial life has gained renewed interest. In the context of a bottom-up approach, this entails the de novo construction of synthetic cells that can autonomously sustain themselves and proliferate. Reproduction of a synthetic cell involves the synthesis of its inner content, replication of its information module, and growth and division of its shell. Theoretical and experimental analysis of natural cells shows that, whereas the core synthesis machinery of the information module is highly conserved, a wide range of solutions have been realized in order to accomplish division. It is therefore to be expected that there are multiple ways to engineer division of synthetic cells. Here we survey the field and review potential routes that can be explored to accomplish the division of bottom-up designed synthetic cells. We cover a range of complexities from simple abiotic mechanisms involving splitting of lipid-membrane-encapsulated vesicles due to physical or chemical principles, to potential division mechanisms of synthetic cells that are based on prokaryotic division machineries.
Keywords: Synthetic cells, Cell division, Minimal cells, Vesicle splitting
Introduction
The advent of synthetic biology signals an increasing emphasis on the engineering of biology. One of the main goals of the field is to create biological systems that will serve as a ‘chassis’ for the engineering of higher-level functionalities (Drubin et al. 2007; Purnick and Weiss 2009; Porcar et al. 2011). The attempt to construct a man-made cell is arguably the most challenging goal of synthetic biology. In efforts to achieve this, one can distinguish three main approaches that are mutually connected at the level of mechanistic understanding. In the top-down approach, existing biological cells are analyzed, manipulated and minimized in order to identify the smallest necessary number of genes that can sustain life (Hutchison et al. 1999; Koonin 2000; Glass et al. 2006; Forster and Church 2006). Within the framework of the origin-of-life one attempts to build model protocells, i.e., possible models for the ancestors of natural cells, from materials that may have existed during the prebiotic era (Schrum et al. 2010; Dzieciol and Mann 2012; Murtas 2013). Finally, bottom-up synthetic biologists start from basic components to add and assemble these one by one according to established engineering principles, with an end goal to build a de novo functional cell (Luisi et al. 2006; Schwille 2011).
In essence, all attempts to construct man-made cells are connected to the definition of life (Rasmussen et al. 2004). Although the question of what life is lies beyond the scope of this review, it is intriguing to note that according to the accepted cellular theory, every cell must form from a pre-existing one (Turner 1890; Mazzarello 1999; Noireaux et al. 2011). Thus, a prerequisite for any minimal or synthetic cell is that it will be able to reproduce itself, that is, to grow, split and distribute its content and its shell among its progenies. This relates to the notion that encapsulation (and thus also splitting of the cells) is needed for any differentiation of self and non-self, which forms the bases for subsequent evolution. Therefore, splitting the shell of synthetic cells, which is the main focus of this review, is as essential for cellular life as the accepted necessity of replication of the genetic material (Szostak et al. 2001).
Though this review concentrates on the bottom-up approach for synthetic cell division, it is of interest to first ask what lessons regarding cell division can be learned from the top-down approach. Although different top-down studies identified somewhat different sets of essential genes, a conservative estimate suggests that the minimal set consists of about 200 genes that are involved in functions such as DNA replication, transcription, translation and basic metabolism. For building robust artificial cells, it is essential to go beyond the level of merely continued sustainance and analyze the operation of different functional modules that provide the mechanism for their shell growth and division. Surprisingly, however, inspection of the minimal-genome lists shows that only few genes (or even none) were associated with cell division, suggesting a neglect of this essential process (Forster and Church 2006; Moya et al. 2009; Henry et al. 2010; Jewett and Forster 2010). Assuming that one would like to build synthetic cells, it is thus essential to ask how will they be able to grow their shells and divide.
In fact, this point becomes even more pronounced by noticing that the vast majority of existing organisms devote numerous genes to this task. For example, in yeast and higher eukaryotes at least 130 genes are involved in the process of cytokinesis (Pollard 2010; Balasubramanian et al. 2012), while in Escherichia coli (E. coli) and other rod-shaped bacteria, the division machinery is composed of more than 20 different proteins, with another 10 proteins participating in the regulation of the process (Kirkpatrick and Viollier 2011; Egan and Vollmer 2013), see also a list of divisome proteins in the reference list (Division-Proteins). Two reasons may contribute to the discrepancy between the large number of genes that are known to participate in cell division and their general absence from the minimal genome lists. First, the vast majority of prokaryotes are encapsulated by a stiff cell wall (Typas et al. 2012; Albers and Meyer 2011), and many of these genes are involved in the processes of cell-wall synthesis and degradation. However, for minimal cells or first-generation synthetic cells, these functions might be superfluous, as cells may still survive with partly defective cell walls, or without a cell wall at all. Second, in spite of the high conservation of many division proteins, and in contrast to some other core cellular machineries like translation or transcription, the division system exhibits a high level of redundancy and plasticity. For example, a comparison the genomes of three different Mollicutes bacteria species, known for their reduced genomes, has identified only two cell-division proteins that are common to all three species (Fisunov et al. 2011). Hence, multiple realizations can probably be found to address the question of how to divide simple synthetic cells.
Here, we review different mechanisms that may be used for synthetic cell division. In doing so, we adopt a bottom-up perspective and we cover a range of complexities from simple abiotic mechanisms of vesicles division to those that are based on current prokaryote machineries. The structure of this review is as follows: First, we discuss physical mechanisms that can lead to vesicle splitting. Next, we present mechanisms for vesicle splitting that depend on the chemical synthesis of the shell material. Subsequently, we describe simple protein machineries that can induce cell or vesicle separation. Finally, we dwell upon the question of the localization of the division machinery within a cell/vesicle.
In contrast to top-down-defined minimal cells that, by definition, contain only the minimal set of natural proteins and nucleotides that sustain life, bottom-up-defined synthetic cells can be realized from a variety of materials. Thus, they can contain elements from existing cells but also non-natural components, and concurrently, they can be more complex or less complex than top-down minimal cells. However, since the most direct way to envision synthetic cells is through materials and machineries that at least resemble those of modern cells, we concentrate on the division of unilamellar vesicles of amphipathic molecules, particularly on lipid vesicles (liposomes). As an important remark, we note that, synthetic cells and model protocells are not the same, though they share common themes. Similar to the distinction between the top-down minimal cell and the bottom-up synthetic cell approaches, the main difference between the origin-of-life approach and the bottom-up synthetic biology one is that the first is solely based on the use of possible pure prebiotic chemicals, while in the later approach both highly evolved proteins, simple pure chemicals and/or other highly evolved cellular components can be used side-by-side in order to achieve the final aim (Stano and Luisi 2013). Thus, although some division mechanisms that contribute to the proliferation of model protocells can inspire the field of synthetic cells, and vice versa, the proliferation of synthetic cell can involve many other factors.
Division processes based on physical mechanisms
Physically induced shape transformation of vesicles
All cells are enveloped by a phospholipid membrane that can take different closed forms. In spite of the fact that cellular shapes mostly depend on a stiff exoskeleton (cell-wall) or a macromolecular cytoskeleton, it is interesting to ask what physical parameters determine the shape of a bare lipid vesicle. Similarly, though intracellular transport of vesicles in eukaryote cells is usually regulated by protein machineries, physical processes alone can already drive fission or fusion of lipid membranes. Since, as mentioned above, we think of synthetic cells as artificial cells that are enveloped by a lipid membrane, physical processes that induce shape changes of vesicles or promote their fission can form the basis for physical division of synthetic cells (Svetina 2009). Here, we discuss such physical processes.
Originating from the important development by Helfrich (1973) of the bending energy functional, theoretical and experimental work has shown that the shape of a liposome with a homogeneous composition is determined by the minimum of its elastic energy (Seifert et al. 1991). This energy is constrained by the volume (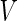 ), area (
), area (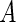 ), area-difference between the two leaflets of the lipid double layer (
), area-difference between the two leaflets of the lipid double layer (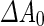 ), and the spontaneous curvature of the lipids (
), and the spontaneous curvature of the lipids (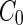 ) (see Box 1). Without growth of the shell of a vesicle, the surface area is fixed and the volume is determined by the osmotic pressure difference between the inside and the outside of the liposome. However,
) (see Box 1). Without growth of the shell of a vesicle, the surface area is fixed and the volume is determined by the osmotic pressure difference between the inside and the outside of the liposome. However, 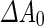 or
or  are adjustable parameters that can drive shape transformation of the vesicle. Such a transformation of the shape of a vesicle under the control of the adjustable parameters is called a shape trajectory. Of particular interest in the context of this paper, is the fact that the adjustable parameters can drive the transformation a vesicle into two (or more) spheres that are connected by a very narrow neck(s). It is clear that such a shape transformation brings the division of synthetic cells at arm’s length.
are adjustable parameters that can drive shape transformation of the vesicle. Such a transformation of the shape of a vesicle under the control of the adjustable parameters is called a shape trajectory. Of particular interest in the context of this paper, is the fact that the adjustable parameters can drive the transformation a vesicle into two (or more) spheres that are connected by a very narrow neck(s). It is clear that such a shape transformation brings the division of synthetic cells at arm’s length.
Box 1.
Vesicle-shape phase diagram
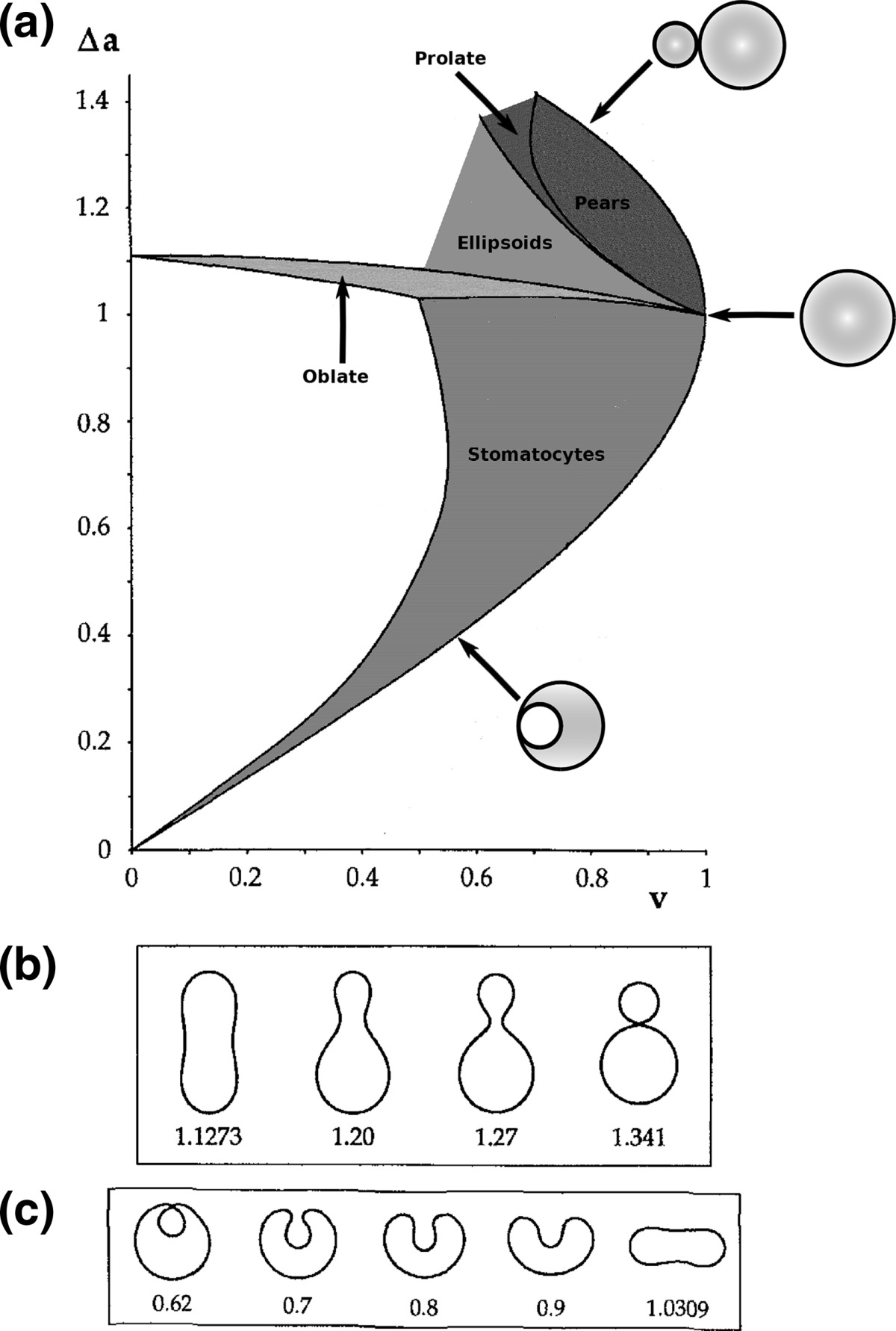
|
a Phase diagram of the strict-bilayer-coupling model showing different classes of shapes. Areas in white were not explored. Note that a sphere is the point 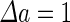 and and 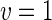 . b Different stable shapes along the shape-trajectory from a prolate to two spheres connected by a narrow neck through the pears class for the line . b Different stable shapes along the shape-trajectory from a prolate to two spheres connected by a narrow neck through the pears class for the line 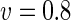 . The value of . The value of 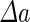 for each shape is stated below it. c Different stable shapes along the shape-trajectory from a prolate to a sphere inside a sphere shape through the stomatocyte class for the line for each shape is stated below it. c Different stable shapes along the shape-trajectory from a prolate to a sphere inside a sphere shape through the stomatocyte class for the line 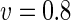 . The value of . The value of 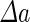 for each shape is stated below it. All panels were modified with permissions from reference Seifert et al. (1991) for each shape is stated below it. All panels were modified with permissions from reference Seifert et al. (1991)  (1991) American Physical Society. Panel a was compiled by the aid of reference Svetina (2009). (1991) American Physical Society. Panel a was compiled by the aid of reference Svetina (2009).Deformation of a 2-D surface embedded in a 3-D space can involve either stretching or bending of the surface. These reversible deformation modes cost elastic energy. Since the exposure of the hydrophobic moieties of the molecules in an amphiphatic membrane to the aqueous environments costs a relatively large amount of energy, the main deformation mode of membranes is bending, that is, changes in the curvature of the membrane. Differential-geometry considerations show that, for a general surface, one can define two principles curvatures, 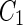 and and 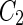 that are associated with the two perpendicular tangents to the surface. Two measures for the curvature of a shape can be constructed from that are associated with the two perpendicular tangents to the surface. Two measures for the curvature of a shape can be constructed from 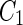 and and 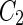 , the mean curvature , the mean curvature 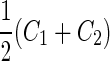 and the Gaussian curvature and the Gaussian curvature 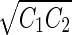 . Form these two measures, one can calculate the bending energy of a piece of surface ( . Form these two measures, one can calculate the bending energy of a piece of surface (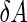 ) as ) as 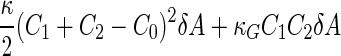 , where , where 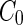 is the local spontaneous curvature and is the local spontaneous curvature and 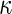 and and 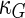 are the mean and Gaussian bending moduli, respectively. The total elastic energy in this case is the integral of these two terms over the whole surface of the vesicle. It was shown, however, that these two terms alone cannot account for the behavior of lipid membranes. The reason is that a lipid bilayer membrane is composed of two leaflets, so that when it bends, one layer is stretched and the other one is compressed. Hence, an additional term is added to the elastic energy that describes the coupling between the two layers. One thus arrives at the so-called, area-difference-elasticity model, where the elastic energy ( are the mean and Gaussian bending moduli, respectively. The total elastic energy in this case is the integral of these two terms over the whole surface of the vesicle. It was shown, however, that these two terms alone cannot account for the behavior of lipid membranes. The reason is that a lipid bilayer membrane is composed of two leaflets, so that when it bends, one layer is stretched and the other one is compressed. Hence, an additional term is added to the elastic energy that describes the coupling between the two layers. One thus arrives at the so-called, area-difference-elasticity model, where the elastic energy (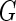 ) equals ) equals |
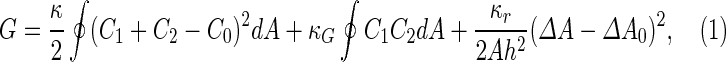
where 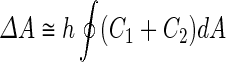 is the area difference between the two leaflets for a given shape, is the area difference between the two leaflets for a given shape, 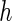 is the width of the membrane, is the width of the membrane, 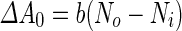 the area difference that is associated with the number of molecules in the outer ( the area difference that is associated with the number of molecules in the outer (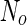 ) and inner ( ) and inner (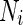 ) leaflets and the preferred area per molecule due to the hydrophobic packing ( ) leaflets and the preferred area per molecule due to the hydrophobic packing (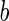 ), and ), and 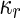 the non-local bending modulus. Since the integral of the Gaussian curvature is topology invariant and changes only when fusion or fission events add or removes holes in a vesicle, it is usually omitted from the calculation. By defining the non-local bending modulus. Since the integral of the Gaussian curvature is topology invariant and changes only when fusion or fission events add or removes holes in a vesicle, it is usually omitted from the calculation. By defining 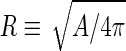 , it is customary to work in reduced dimensionless variables , it is customary to work in reduced dimensionless variables 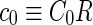 , , 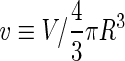 , , 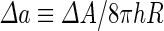 and and 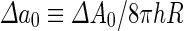 . The ratio . The ratio 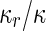 determines the relative contribution of the local and non-local bending terms. For the case determines the relative contribution of the local and non-local bending terms. For the case 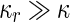 , one can omit the last term of the elastic energy, and impose instead another hard constrain , one can omit the last term of the elastic energy, and impose instead another hard constrain 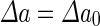 . This limit is called the strict-bilayer-coupling model. The other limit where . This limit is called the strict-bilayer-coupling model. The other limit where 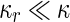 is called the spontaneous-curvature model, and in that case the elastic energy is simply taken to be is called the spontaneous-curvature model, and in that case the elastic energy is simply taken to be 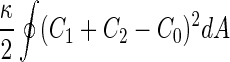 . Interestingly, this limit is identical to the original model that was introduced by Canham Helfrich and Evans for the understanding of the shape of red blood cells and which led to the development of the full area-difference-elasticity model (Canham 1970; Helfrich 1973; Evans 1974). The result of these models is a phase diagram in the . Interestingly, this limit is identical to the original model that was introduced by Canham Helfrich and Evans for the understanding of the shape of red blood cells and which led to the development of the full area-difference-elasticity model (Canham 1970; Helfrich 1973; Evans 1974). The result of these models is a phase diagram in the 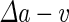 (or (or  ) space with a gallery of shapes like spheres, prolates, oblates, pears, cigars and stomatocytes, as can be seen in the figure. For a continuous change in the values of ) space with a gallery of shapes like spheres, prolates, oblates, pears, cigars and stomatocytes, as can be seen in the figure. For a continuous change in the values of 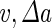 (or (or 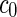 ), the shape of the vesicle changes along a shape-trajectory in the phase diagram ), the shape of the vesicle changes along a shape-trajectory in the phase diagram | |
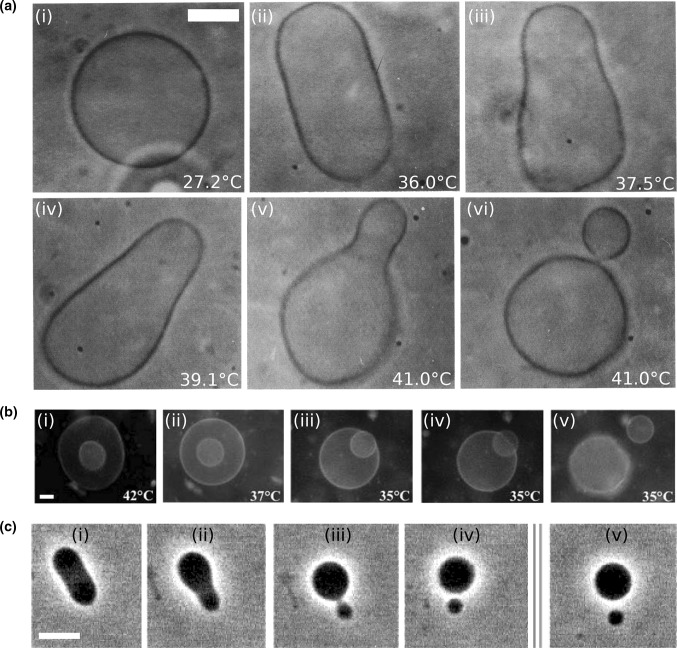
An interesting experimental demonstration of such a shape trajectory is shown in Fig. 1a (Käs and Sackmann 1991) where a spherical vesicle is transformed into a pear and subsequently into two vesicles that are still connected by a narrow neck. In this experiment, shape transformation was realized by an increase in temperature, which caused a larger thermal expansion of the vesicle surface relative to its volume. It should be noted that the shape trajectory of a vesicle depends on its pre-transformed state, especially if the vesicle is flaccid or turgid.
Fig. 1.
Shape trajectories for vesicles that are physically manipulated. a A DPMC vesicle is heated from 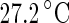 (i) to
(i) to 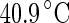 (v). A further increase of the temperature by
(v). A further increase of the temperature by 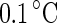 transferred the shape of the vesicle from a pear to the limiting shape of two spheres that stay connected by a narrow neck (without abscission of the neck). Area increased from 2,570 to 2,820
transferred the shape of the vesicle from a pear to the limiting shape of two spheres that stay connected by a narrow neck (without abscission of the neck). Area increased from 2,570 to 2,820 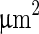 , while volume changed only slightly from 12,200 to 12,000
, while volume changed only slightly from 12,200 to 12,000 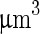 . Note that before the heating cycle the vesicle was stiff and did not have any excess surface area. Scale bar 10
. Note that before the heating cycle the vesicle was stiff and did not have any excess surface area. Scale bar 10 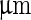 . b Time series of the birthing of an inner vesicle through the membrane of a mother vesicle. A DLPE:DPPC 3:7 vesicle was cooled down from
. b Time series of the birthing of an inner vesicle through the membrane of a mother vesicle. A DLPE:DPPC 3:7 vesicle was cooled down from 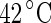 (i) to
(i) to 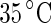 (iii–v). The cooling process created an inner pressure that caused an inner bud (that had formed previously due to a heating process) to be ejaculated from the lumen of the mother vesicle. Scale bar 5
(iii–v). The cooling process created an inner pressure that caused an inner bud (that had formed previously due to a heating process) to be ejaculated from the lumen of the mother vesicle. Scale bar 5 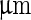 . c Fission of a DPPC:cholesterol vesicle due to the incorporation of lyso-PC. Budding and fission were induced by the local injection of a solution of 1
. c Fission of a DPPC:cholesterol vesicle due to the incorporation of lyso-PC. Budding and fission were induced by the local injection of a solution of 1 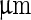 palmitoyl-lyso-PC. Time after injection are 0, 17, 18, 40, and 240 s for (i)–(v), respectively. Scale bar 10
palmitoyl-lyso-PC. Time after injection are 0, 17, 18, 40, and 240 s for (i)–(v), respectively. Scale bar 10 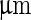 . a Modified with permissions from reference Käs and Sackmann (1991)
. a Modified with permissions from reference Käs and Sackmann (1991)  (1991) Elsevier Limited. b Modified with permissions from reference Sakuma and Imai (2011)
(1991) Elsevier Limited. b Modified with permissions from reference Sakuma and Imai (2011)  (2011) American Physical Society. c Modified with permission from reference Tanaka et al. (2004)
(2011) American Physical Society. c Modified with permission from reference Tanaka et al. (2004)  (2004) American Chemical Society
(2004) American Chemical Society
Using shape trajectories for inducing the division of synthetic cells is an appealing concept, but it is not sufficient. The reason, as seen in Fig. 1a, is that the two vesicles stayed connected and some other mechanism should drive the destabilization of the resulting neck and thus the final separation into daughter cells. One possible mechanism for this abscission involves the phase of the lipids in the membrane. Bilayer-forming lipids can be obtained in several distinct phases that are classified according to the hydrocarbon chain order and the lateral mobility of the lipids. It is common to define the main phase transition temperature between the liquid-disordered phase (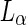 ), in which the hydrocarbon chains are randomly oriented and the lipids are highly mobile, to the ordered-gel phase (
), in which the hydrocarbon chains are randomly oriented and the lipids are highly mobile, to the ordered-gel phase (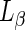 ), in which the order parameter of the hydrocarbon chains is high and their mobility is low (Meer et al. 2008). For example, heating of bovine brain sphingomyelins (SM) vesicles across the transition temperature resulted in the formation of large excess area and the pinching off of small vesicles from the mother one (Döbereiner et al. 1993). Similarly, when 1,2-dipalmitoyl-sn-glycero-3-phosphocholine:1,2-dimyristoyl-sn-glycero-3-phosphocholine (DPPC:DPMC) vesicles were heated above the transition temperature, inwards or outwards buds were formed, without control on the bud direction. These buds were pinched off of the mother vesicles by subsequent cooling below the transition temperature (Leirer et al. 2009). By contrast, for DPPC vesicles with a low fraction of 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine (DLPE), a heating-cooling cycle across the transition temperature induced only outer-bud formation and abscission (Sakuma and Imai 2011). For high DLPE fractions, a similar temperature cycle resulted in the formation of inward buds that were ejaculated by ’birthing’ out of the mother vesicle due to inner pressure buildup that resulted in the formation of transient pores in the mother vesicle membrane (see Fig. 1b). The temperature cycle could be repeated up to five times to create more and more daughter vesicles with the same lipid composition as that of the mother one.
), in which the order parameter of the hydrocarbon chains is high and their mobility is low (Meer et al. 2008). For example, heating of bovine brain sphingomyelins (SM) vesicles across the transition temperature resulted in the formation of large excess area and the pinching off of small vesicles from the mother one (Döbereiner et al. 1993). Similarly, when 1,2-dipalmitoyl-sn-glycero-3-phosphocholine:1,2-dimyristoyl-sn-glycero-3-phosphocholine (DPPC:DPMC) vesicles were heated above the transition temperature, inwards or outwards buds were formed, without control on the bud direction. These buds were pinched off of the mother vesicles by subsequent cooling below the transition temperature (Leirer et al. 2009). By contrast, for DPPC vesicles with a low fraction of 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine (DLPE), a heating-cooling cycle across the transition temperature induced only outer-bud formation and abscission (Sakuma and Imai 2011). For high DLPE fractions, a similar temperature cycle resulted in the formation of inward buds that were ejaculated by ’birthing’ out of the mother vesicle due to inner pressure buildup that resulted in the formation of transient pores in the mother vesicle membrane (see Fig. 1b). The temperature cycle could be repeated up to five times to create more and more daughter vesicles with the same lipid composition as that of the mother one.
Another route for neck abscission is by the use of lipids in the liquid-ordered (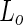 ) phase, where the mobility of the lipids is still high although the hydrocarbon chains are tightly packed. If the membrane is composed out of
) phase, where the mobility of the lipids is still high although the hydrocarbon chains are tightly packed. If the membrane is composed out of 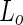 as well as
as well as 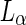 lipids, regions of high lipid order can coexist with regions of low lipid order. The boundary between these domain regions is accompanied by a line tension, and its minimization can drive shape transformations and neck abscission. For example, changing the
lipids, regions of high lipid order can coexist with regions of low lipid order. The boundary between these domain regions is accompanied by a line tension, and its minimization can drive shape transformations and neck abscission. For example, changing the 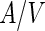 ratio by heating (or by increasing the osmotic pressure) of egg-SM:1,2-dioleoyl-sn-glycero-3-phosphocholine:cholesterol (egg-SM:DOPC:cholesterol) vesicles resulted in the fission of the vesicles at the domain boundaries (see Fig. 2a) (Baumgart et al. 2003). Indeed, the calculated lateral tension was large enough to guillotine the resulted neck. A somewhat different behavior was observed when vesicles of DOPC:DPPC:cholesterol encapsulated a polyethyleneglycol:dextran aqueous two-phase system (ATPS) (Andes-Koback and Keating 2011). At low temperature, dextran and polyethyleneglycol (PEG) phase separate within the vesicle lumen. In parallel, the lipids phase separate to
ratio by heating (or by increasing the osmotic pressure) of egg-SM:1,2-dioleoyl-sn-glycero-3-phosphocholine:cholesterol (egg-SM:DOPC:cholesterol) vesicles resulted in the fission of the vesicles at the domain boundaries (see Fig. 2a) (Baumgart et al. 2003). Indeed, the calculated lateral tension was large enough to guillotine the resulted neck. A somewhat different behavior was observed when vesicles of DOPC:DPPC:cholesterol encapsulated a polyethyleneglycol:dextran aqueous two-phase system (ATPS) (Andes-Koback and Keating 2011). At low temperature, dextran and polyethyleneglycol (PEG) phase separate within the vesicle lumen. In parallel, the lipids phase separate to 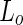 and
and 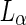 phases within the membrane. Due to the incorporation of head-group PEGylated lipids into the
phases within the membrane. Due to the incorporation of head-group PEGylated lipids into the 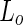 phase, the
phase, the 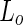 lipid phase preferentially wet the PEG-rich aqueous phase. Thus, the dextran was found to be mainly encapsulated by
lipid phase preferentially wet the PEG-rich aqueous phase. Thus, the dextran was found to be mainly encapsulated by 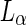 membrane while the PEG was encapsulated by the
membrane while the PEG was encapsulated by the 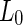 phase membrane. Addition of sucrose to induce an osmotic pressure resulted in a transformation to a pear shape and subsequently fission at the
phase membrane. Addition of sucrose to induce an osmotic pressure resulted in a transformation to a pear shape and subsequently fission at the 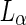 domain boundary. Interestingly, when a mismatch existed between the domain areas and the amount of membrane needed to encapsulate each polymeric aqueous phase, fission occurred at a location that maximized the separation between the two ATPS phases, leaving excess
domain boundary. Interestingly, when a mismatch existed between the domain areas and the amount of membrane needed to encapsulate each polymeric aqueous phase, fission occurred at a location that maximized the separation between the two ATPS phases, leaving excess 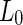 or
or 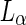 membrane in one of the daughter vesicles. Thus, in this approach, the driving force for abscission resulted from the interplay between the encapsulated material and the membrane rather than from forces that acted within the membrane planes alone (see Fig. 2b).
membrane in one of the daughter vesicles. Thus, in this approach, the driving force for abscission resulted from the interplay between the encapsulated material and the membrane rather than from forces that acted within the membrane planes alone (see Fig. 2b).
Fig. 2.
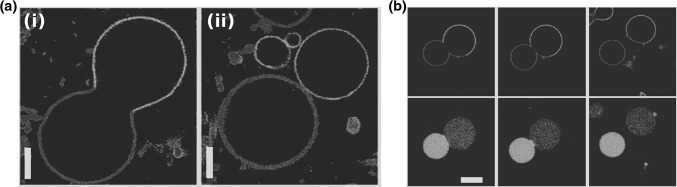
Fission of buds in liposomes with liquid-disordered phase membranes. a Fission of a bud at the liquid-ordered/liquid-disordered separation line upon heating from 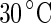 (i) to
(i) to 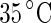 (ii). Vesicle composition 0.615:0.135:0.25 SM:DOPC:cholesterol.
(ii). Vesicle composition 0.615:0.135:0.25 SM:DOPC:cholesterol. 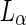 phase was imaged in blue by incorporating small amount of a Dil dye,
phase was imaged in blue by incorporating small amount of a Dil dye, 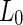 phase was imaged in red by incorporating a small amount of N-lissamine rhodamine dipalmitoylphosphoethanolamine. Scale bar 5
phase was imaged in red by incorporating a small amount of N-lissamine rhodamine dipalmitoylphosphoethanolamine. Scale bar 5 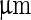 . b Fission of vesicle due to an encapsulation of an ATPS system. 0.35:0.35:0.3 POPC:DPPC:cholesterol vesicle encapsulating a PEG:dextran ATPS. Osmotic pressure increases form left to right. Blue indicates lectin SBA Alexa 647 (dextran phase); red indicates
. b Fission of vesicle due to an encapsulation of an ATPS system. 0.35:0.35:0.3 POPC:DPPC:cholesterol vesicle encapsulating a PEG:dextran ATPS. Osmotic pressure increases form left to right. Blue indicates lectin SBA Alexa 647 (dextran phase); red indicates 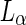 domain lipid (DOPE-rhodamine); and green indicates
domain lipid (DOPE-rhodamine); and green indicates 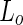 domain through streptavidin-Alexa488 (bound to the lipid DSPE-PEG-2K-biotin). Scale bar 10 microns. a Modified with permissions from reference Baumgart et al. (2003)
domain through streptavidin-Alexa488 (bound to the lipid DSPE-PEG-2K-biotin). Scale bar 10 microns. a Modified with permissions from reference Baumgart et al. (2003)  (2003) Nature Publishing Group. b Modified with permission from reference Andes-Koback and Keating (2011)
(2003) Nature Publishing Group. b Modified with permission from reference Andes-Koback and Keating (2011)  (2011) American Chemical Society. (Color figure online)
(2011) American Chemical Society. (Color figure online)
As noted before, varying the area difference 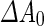 between the two leaflets can trigger shape transformations. Indeed, tubulation, budding and pearling have been observed by inducing flipping of lipids from the inner to the outer leaflet (Mui et al. 1995), by adding dextran coupled to hydrophobic anchors to the outside of a multilamellar membranes (Tsafrir et al. 2003), or by adsorption of fatty acids from an oleic acid suspension (Peterlin et al. 2009; Budin et al. 2012). Coupling of this shape-transformation mechanism with destabilization of the shape neck(s) at the
between the two leaflets can trigger shape transformations. Indeed, tubulation, budding and pearling have been observed by inducing flipping of lipids from the inner to the outer leaflet (Mui et al. 1995), by adding dextran coupled to hydrophobic anchors to the outside of a multilamellar membranes (Tsafrir et al. 2003), or by adsorption of fatty acids from an oleic acid suspension (Peterlin et al. 2009; Budin et al. 2012). Coupling of this shape-transformation mechanism with destabilization of the shape neck(s) at the  phase can trigger vesicle separation. For example, upon locally adding phospholipase
phase can trigger vesicle separation. For example, upon locally adding phospholipase 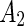 , (an enzyme that catalyzes the ester bond hydrolysis of glycerophospholipids to lyso-phospholipids and a fatty acids) to SM:cholesterol:phosphocholine (PC) vesicles induced budding followed by abscission at the domain boundary (Staneva et al. 2004). Likewise, local addition of lyso-PC, as well as of other single long chain detergents, below the minimal micelle concentration to DPPC:cholesterol or SM:cholesterol vesicles resulted in budding and subsequent separation to daughter vesicles (see Fig. 1c) (Tanaka et al. 2004; Staneva et al. 2005; Inaoka and Yamazaki 2007). In both cases, these phenomena were explained based on budding induced by material incorporation to the outer leaflet (Lyso-PC or phospholipase
, (an enzyme that catalyzes the ester bond hydrolysis of glycerophospholipids to lyso-phospholipids and a fatty acids) to SM:cholesterol:phosphocholine (PC) vesicles induced budding followed by abscission at the domain boundary (Staneva et al. 2004). Likewise, local addition of lyso-PC, as well as of other single long chain detergents, below the minimal micelle concentration to DPPC:cholesterol or SM:cholesterol vesicles resulted in budding and subsequent separation to daughter vesicles (see Fig. 1c) (Tanaka et al. 2004; Staneva et al. 2005; Inaoka and Yamazaki 2007). In both cases, these phenomena were explained based on budding induced by material incorporation to the outer leaflet (Lyso-PC or phospholipase 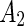 ), followed by the creation of membrane defects (due to the high order of the
), followed by the creation of membrane defects (due to the high order of the 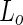 lipids), and finally, defect-induced attraction of the two opposite bilayers that resulting in abscission. Thus, local targeting of
lipids), and finally, defect-induced attraction of the two opposite bilayers that resulting in abscission. Thus, local targeting of 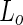 lipids to the division site of synthetic cells may promote their separation.
lipids to the division site of synthetic cells may promote their separation.
A variation on the  fission theme was observed when long (about 10
fission theme was observed when long (about 10 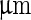 ) and narrow (about 100 nm) tubes were pulled from DOPC:SM:Cholesterol vesicles using kinesin biomolecular motors (Roux et al. 2005). In this case, the tubes were highly enriched in the
) and narrow (about 100 nm) tubes were pulled from DOPC:SM:Cholesterol vesicles using kinesin biomolecular motors (Roux et al. 2005). In this case, the tubes were highly enriched in the 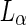 DOPC lipids due to a lipid sorting mechanism between the highly curved tubes and the less curved vesicles. Such lipid sorting is caused since the
DOPC lipids due to a lipid sorting mechanism between the highly curved tubes and the less curved vesicles. Such lipid sorting is caused since the 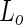 phase is more rigid than the
phase is more rigid than the 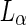 one. Thus, under the constant tension that is created by the pulling molecular motors, the elastic energy can by minimized by enriching the highly curved tubes with the less rigid lipid phase. However, also in this case, tubes abscission only occurred at
one. Thus, under the constant tension that is created by the pulling molecular motors, the elastic energy can by minimized by enriching the highly curved tubes with the less rigid lipid phase. However, also in this case, tubes abscission only occurred at  domain boundaries that were created after light-induced oxidation of the cholesterol resulted in the domain segregation. Note that theoretical considerations suggested another mechanism for membrane splitting that involves sorting of lipids with a different Gaussian bending modulus within a lipid mixture if the minor-component lipid prefer to stay at regions with a large positive Gaussian curvature (Chen et al. 1997).
domain boundaries that were created after light-induced oxidation of the cholesterol resulted in the domain segregation. Note that theoretical considerations suggested another mechanism for membrane splitting that involves sorting of lipids with a different Gaussian bending modulus within a lipid mixture if the minor-component lipid prefer to stay at regions with a large positive Gaussian curvature (Chen et al. 1997).
Finally, yet another interesting mechanism to induce shape transformation and splitting of vesicles is by inducing tension in the membrane. In that case, the total energy of the vesicles consists of its elastic energy plus the term 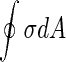 , where
, where 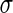 is the surface tension. Indeed, when optical tweezers induced tension in long tube-like vesicles, a pearling instability was observed (Bar-Ziv and Moses 1994). Such pearling instability is a type of Rayleigh instability where long tubes are transferred to a string of spheres (that remain connected by narrow necks), in order to reduce the surface area under a constant volume. Similarly, when optical tweezers induced tension in giant unilamellar vesicles, it suppressed their thermal fluctuations and over-pressurized them. Upon turning the optical tweezers off, the release of the pressure resulted in ’birthing’ of inner vesicles (Bar-Ziv et al. 1995). A similar pearling phenomenon was observed with long tubes of oleate vesicles after light-induced oxidation of thiols (Zhu et al. 2012). It was suggested that the oxidation increased the surface tension of the membrane thus triggering the separation of the small pearls. However, the exact physical mechanism remained somewhat unclear in this case.
is the surface tension. Indeed, when optical tweezers induced tension in long tube-like vesicles, a pearling instability was observed (Bar-Ziv and Moses 1994). Such pearling instability is a type of Rayleigh instability where long tubes are transferred to a string of spheres (that remain connected by narrow necks), in order to reduce the surface area under a constant volume. Similarly, when optical tweezers induced tension in giant unilamellar vesicles, it suppressed their thermal fluctuations and over-pressurized them. Upon turning the optical tweezers off, the release of the pressure resulted in ’birthing’ of inner vesicles (Bar-Ziv et al. 1995). A similar pearling phenomenon was observed with long tubes of oleate vesicles after light-induced oxidation of thiols (Zhu et al. 2012). It was suggested that the oxidation increased the surface tension of the membrane thus triggering the separation of the small pearls. However, the exact physical mechanism remained somewhat unclear in this case.
Vesicle growth and fission by incorporation of external mater
Obviously, proliferation of synthetic cells requires both growth and division. It is therefore interesting to ask what are the possible routes to couple these two processes. Since an excellent review about the subject was recently published (Stano and Luisi 2010), we will dwell upon this only briefly. In this section, we review growth-division processes that are induced by external addition of amphiphatic molecules.
In principle, lipid vesicles or synthetic cells can grow by incorporating lipids from the environment. However, under most relevant experimental conditions, the use of such process is prevented due to the large hydrophobic energy that is associated with (1) creating membrane defects that are large enough to accommodate the incorporation of new lipids, and (2) the relatively low critical aggregation concentration of lipids in aqueous solution. This situation is radically changed with fatty acids. Unlike lipids, fatty acids have only one hydrophobic tail and a relatively small hydrophilic head group. In comparison to lipids, fatty acids therefore have a much higher critical aggregation concentration and a much higher probability of exchange between any amphiphatic aggregate and the aqueous solution. In particular, the average residence time of the typical fatty acids in the membranes is of the order of milliseconds to seconds, depending on the fatty acid chain length. As a result of this fast dynamics, the probability for the incorporation of fatty acids into a vesicle membrane from the environment is relatively high, which can be used for growth of the vesicle shell.
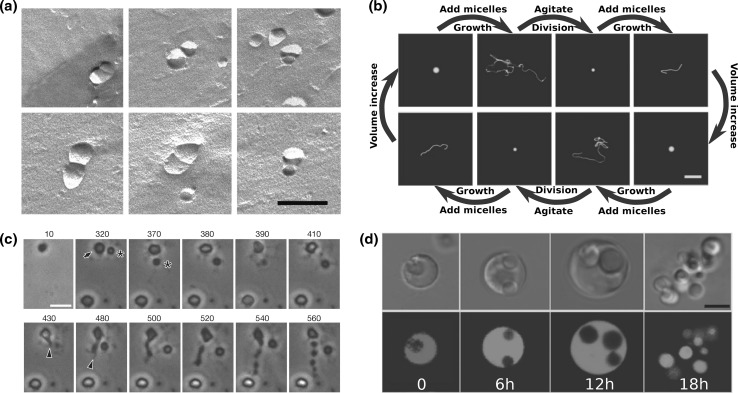
The growth of oleic acid/oleate vesicles as well as of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) vesicles by fatty acids incorporation was demonstrated when oleate micelles were added to the solution with pre-formed vesicles (Blöchliger et al. 1998; Lonchin et al. 1999). Addition of oleate micelles from a solution with a high pH to a solution with a lower pH resulted in growth of oleic acid/oleate vesicles with a broad size distribution. By contrast, the existence of pre-formed vesicles caused a bias of the new vesicle-size distribution towards the size of the pre-existing ones, and also induced vesicles division as was inferred from electron microscopy and light-scattering measurements (see Fig. 3a) (Berclaz et al. 2001; Stano et al. 2006). This ability of pre-formed vesicle to influence the growth of oleate micelle was termed the ’matrix effect’. It can be envisioned that similar mechanisms might be harnessed for inducing growth and division of synthetic cells. However, since for bottom-up synthetic cells field, lipids are highly preferred over fatty acids as they create a much better barrier for the translocation of small molecules across the membrane, such a mechanism probably would have to be supported by a way to catalyze the conversion of the absorbed fatty acids to lipids.
Fig. 3.
Division due to membrane growth of liposomes and cells. a Freeze-fracture electron microscopy images of twin-vesicles that appear to have a septum between them, formed from the addition of oleate micelles to pre-formed oleic acid/oleate vesicles. Scale bar 200 nm. b Coupled growth and division cycle of a multilamellar oleate vesicle, induced by the incorporation of oleate micelles followed by division of the resulted tubes into small spherical vesicles by gentle hydrodynamic forces. Scale bar 20  . c Division of L-form
. c Division of L-form  B. subtilis cells by an extrusion–resolution mechanism. The cell marked with an arrow at
B. subtilis cells by an extrusion–resolution mechanism. The cell marked with an arrow at  minutes start to form protrusions after 430 min which resolved into a string of six cells. The cell marked with an asterisk did not divide. Scale bar 500 nm. d Phase-contrast (upper row) and fluorescence microscopy (lower row) of different phases of the L-form L. monocytogenes growth and multiplication. Intra-cellular bodies are formed within the mother cell. After they grow, the mother cell ruptures. Upon release, most of the intra-cellular bodies gain metabolic activity. Metabolic function was followed by synthesis and/or subsequent maturation of intracellular GFP (green). Time is in hours after incubation on soft agar. a Modified with permission of IOP Publishing, from reference Stano et al. (2006)
minutes start to form protrusions after 430 min which resolved into a string of six cells. The cell marked with an asterisk did not divide. Scale bar 500 nm. d Phase-contrast (upper row) and fluorescence microscopy (lower row) of different phases of the L-form L. monocytogenes growth and multiplication. Intra-cellular bodies are formed within the mother cell. After they grow, the mother cell ruptures. Upon release, most of the intra-cellular bodies gain metabolic activity. Metabolic function was followed by synthesis and/or subsequent maturation of intracellular GFP (green). Time is in hours after incubation on soft agar. a Modified with permission of IOP Publishing, from reference Stano et al. (2006)  (2006) IOP Publishing. doi:10.1088/0953-8984/18/33/S37. All rights reserved. b Modified with permission from Zhu and Szostak (2009) Copyright (2009) American Chemical Society. c Modified with permission from reference Leaver et al. (2009)
(2006) IOP Publishing. doi:10.1088/0953-8984/18/33/S37. All rights reserved. b Modified with permission from Zhu and Szostak (2009) Copyright (2009) American Chemical Society. c Modified with permission from reference Leaver et al. (2009)  (2009) Nature Publishing Group. d Modified with permission from reference Dell’Era et al. (2009)
(2009) Nature Publishing Group. d Modified with permission from reference Dell’Era et al. (2009)  (2009) John Wiley and Sons, Inc. (Color figure online)
(2009) John Wiley and Sons, Inc. (Color figure online)
Interestingly, the addition of oleic acid micelles to pre-formed multilamellar spherical oleate vesicles resulted in vesicle tubulation into long thread-like shapes due to a relatively fast growth of the vesicle surface area combined with an osmotically restricted growth of its volume (see Fig. 3b) (Zhu and Szostak 2009). Subsequent gentle shearing caused separation of these thread-like tubules to many small spherical vesicles with little content lost. The resulting small vesicles could further grow by the incorporation of additional oleate micelles, and subsequently tubulate again, thus showing cycles of growth and division.
As noted above, growth of liposomes by incorporation of small lipid aggregates or free lipids is highly unfavorable. Other mechanisms can, however, be applied in order to achieve liposome growth by lipid incorporation. One example is vesicle fusion that can be triggered by various factors such as multivalent cations (Tanaka and Yamazaki 2004) or small peptides (Nomura et al. 2004). In particular, electroporation-induced fusion was used in order to study growth-division process of liposomes (Terasawa et al. 2012). Alignment of POPC:POPG:cholesterol vesicles [POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)] in an alternating electric field following by electroporation using a high DC electric pulse resulted in fusion of multiple vesicles into larger multilamellar spherical vesicle. When PEGs was encapsulated inside the vesicles prior to the electroporation, the fused vesicles had various shapes including horseshoes, tori and elongated tubes. After a period of relaxation, these elongated vesicles developed buds, and finally, the buds separated from the mother vesicles. The driving force for this shape transformation was a decrease in the translational entropy of the PEG polymers due to encapsulation. Since the center of mass of a polymer cannot be located closer to a wall than its hydrodynamic radius, a depletion zone is created next to membranes that limits the polymers translational entropy. The volume of this depletion zone is proportional to the size of the polymer as well as to the 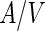 ratio of the vesicle. Minimization of the free energy of the combined polymers and vesicle system requires minimization of the volume of the depletion zone. Since the size of the depletion zone can be minimized by changes in the
ratio of the vesicle. Minimization of the free energy of the combined polymers and vesicle system requires minimization of the volume of the depletion zone. Since the size of the depletion zone can be minimized by changes in the 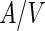 ratio, the vesicles tend to elongate. A further decrease in the system free energy can be achieved by forming buds that are connected through narrow necks, which provide the thermodynamic force for the observed shape transformation. Apparently, the inner osmotic pressure of the PEGs was in this case large enough to cause neck rupture as well.
ratio, the vesicles tend to elongate. A further decrease in the system free energy can be achieved by forming buds that are connected through narrow necks, which provide the thermodynamic force for the observed shape transformation. Apparently, the inner osmotic pressure of the PEGs was in this case large enough to cause neck rupture as well.
The coupling between vesicle growth and division can also be studied from a theoretical point of view using a combination of (i) an equation that describes the relation between the rate of change of the vesicle volume and the flow of solutes and water molecules into the growing vesicles, (ii) an exponential growth law for the membrane surface with doubling time 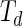 , and (iii) shape trajectories of the vesicles in their shape phase diagram. Assuming a zero solute concentration and only water molecules penetrating through the membrane,
, and (iii) shape trajectories of the vesicles in their shape phase diagram. Assuming a zero solute concentration and only water molecules penetrating through the membrane, 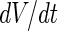 was taken to be
was taken to be 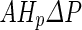 , where
, where 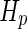 is the hydraulic permeability of the water molecules and
is the hydraulic permeability of the water molecules and 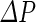 the pressure difference between the inside and the outside of the vesicle (Boźić and Svetina 2004). Using the spontaneous-curvature model for the vesicle bending energy, it was, thus, found that the condition
the pressure difference between the inside and the outside of the vesicle (Boźić and Svetina 2004). Using the spontaneous-curvature model for the vesicle bending energy, it was, thus, found that the condition 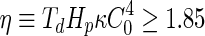 must be fulfilled in order to achieve the condition that the shape of a spherical vesicle will move through the phase diagram in such a way that it ends up as two spheres connected by a narrow neck. Incorporation of solute flow similarly showed that vesicle self-reproduction occurs only for certain combinations of the hydraulic and solute membrane permeability values and external solute concentration (Božič and Svetina 2007). Thus, this approach shows that only a limited set of conditions can support growth-induced shape transformation that end up as two vesicles connected by a narrow neck, which can subsequently divide.
must be fulfilled in order to achieve the condition that the shape of a spherical vesicle will move through the phase diagram in such a way that it ends up as two spheres connected by a narrow neck. Incorporation of solute flow similarly showed that vesicle self-reproduction occurs only for certain combinations of the hydraulic and solute membrane permeability values and external solute concentration (Božič and Svetina 2007). Thus, this approach shows that only a limited set of conditions can support growth-induced shape transformation that end up as two vesicles connected by a narrow neck, which can subsequently divide.
An extension of this approach was introduced by incorporating into the equation for 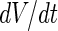 two terms that are derived from the theory of linear non-equilibrium thermodynamics (Morris et al. 2010; Morris and McKane 2011): One term describing the lateral tension in the membrane and a second term describing the contribution of the bending energy to the change in the free energy of the system. Using this approach, the stability of spherical vesicles to perturbations during their growth was studied. It was found that spherical vesicles are unstable for prolate shape perturbations if their radius is smaller than a critical radius,
two terms that are derived from the theory of linear non-equilibrium thermodynamics (Morris et al. 2010; Morris and McKane 2011): One term describing the lateral tension in the membrane and a second term describing the contribution of the bending energy to the change in the free energy of the system. Using this approach, the stability of spherical vesicles to perturbations during their growth was studied. It was found that spherical vesicles are unstable for prolate shape perturbations if their radius is smaller than a critical radius, 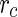 , and are unstable for oblate shape perturbations if their radius is larger than
, and are unstable for oblate shape perturbations if their radius is larger than 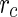 . Here the critical radius
. Here the critical radius 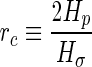 with
with 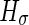 a constant that describes the contribution of the lateral tension to the flow of water molecules through the membrane.
a constant that describes the contribution of the lateral tension to the flow of water molecules through the membrane.
Division driven by active chemical synthesis of materials
One of the hallmarks of biological systems is metabolic activity, that is, the catalytic synthesis of the constituting molecules of the cell. Growth of natural cells importantly includes the synthesis of cell-envelope material. Since active membrane synthesis can change physical parameters such as 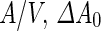 or
or 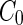 , it can drive shape transformation of synthetic cells and also destabilize narrow membrane necks. The ability to control this process can potentially couple growth and division cycles and provide a reproduction mechanism for synthetic cells that is more robust than merely passive adsorption of shell material.
, it can drive shape transformation of synthetic cells and also destabilize narrow membrane necks. The ability to control this process can potentially couple growth and division cycles and provide a reproduction mechanism for synthetic cells that is more robust than merely passive adsorption of shell material.
In one of the first attempts to follow this approach, oleic anhydride droplets were placed next to oleic acid vesicles (Wick et al. 1995). Under these conditions, the oleic acid vesicles catalyzed the hydrolysis of oleic anhydride to oleic acid and oleate molecules. Subsequent absorption of these molecules into the vesicle shells caused their growth. Note that this example does not yet feature a machinery to synthesize membrane molecules from within the vesicle, but importantly, it involves active catalysis, which goes beyond mere passive vesicle growth. Interestingly, for multilamellar oleic acid vesicles, the self-catalyzed growth caused the formation of inclusion vesicles, probably due to separation of the outer and inner lamellae. These inclusions were subsequently expelled outwards through the membrane of the mother vesicle, leading to a ‘birthing process’ to form daughter vesicles. A similar birthing process was observed when an artificial amphiphile (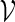 ) bearing an imine group assembled into vesicles with a small amount of cholesterol as a stabilizer (Takakura et al. 2003). Encapsulation of molecules of the two
) bearing an imine group assembled into vesicles with a small amount of cholesterol as a stabilizer (Takakura et al. 2003). Encapsulation of molecules of the two 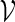 -precursors with catalyst molecules inside
-precursors with catalyst molecules inside 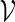 -made vesicles resulted in the inner production of membrane molecules that self-assembled to form daughter vesicles. When these inner vesicles were large enough, they were expelled out of the mother vesicles.
-made vesicles resulted in the inner production of membrane molecules that self-assembled to form daughter vesicles. When these inner vesicles were large enough, they were expelled out of the mother vesicles.
Budding and separation due to auto-catalyzed membrane synthesis was also documented. In a variation of the previous approach, vesicles made of a different synthetic cationic amphiphile together with amphiphilic acidic catalyst and POPC:POPG led to vesicles that could incorporate a bolaamphiphilic precursor of the cationic amphiphile from solution (Kurihara et al. 2011). In principle, this precursor can convert into the amphiphile by the activity of the catalyst on the membrane, with the caveat of a high probability for the precursor to dissolve back into the aqueous solution. When short DNA oligos were encapsulated inside the vesicles and were amplified by PCR, budding and pinching off of the buds was observed after the addition of the amphiphilic precursor. The authors suggested that interactions between the negatively charged DNA and the cationic amphiphiles disrupted the lamellarity of the membrane, causing entrapment of the bolaamphiphile precursor in the membrane. This, in turn, caused budding and abscission of the neck after the conversion of the bolaamphiphile precursor to the cationic amphiphile.
Of course, a straightforward route for accomplishing a growth-division mechanism of vesicles or synthetic cells is to use reconstituted systems of proteins that can catalyze lipid formation from within. Initial attempts in this direction have indeed been made, but with a limited success so far (Schmidli et al. 1991; Kuruma et al. 2009; Murtas 2010).
Stepping out of the abiotic-vesicles world into the biological world, two examples were recently documented for division mechanisms that operate through the internal synthesis of excess membrane material. Both reports involved L-form bacteria, bacterial cells that have lost their cell wall. In the first example, a stable Bacillus subtilis L-form was produced by transcription inhibition of an operon that encodes several genes for cell-wall precursor synthesis and selection with a cell-wall targeted antibiotic (Leaver et al. 2009). The resulting L-form strain was viable and was able to proliferate independent of the regular cell-division machinery. Time-lapse microscopy observations showed the formation of blunt protrusions that dynamically emerged out of the membrane and retracted back. After some time, a stable protrusion emerged, elongated, and resolved into one or several small membrane-encapsulated cell bodies (see Fig. 3c). In other cases, many small buds emerged out of the membrane surface, some of which also erupted from the surface. Recently, a possible genetic mechanism behind this behavior was identified (Mercier et al. 2013). Upregulation of the fatty-acid-synthase-type-II enzyme system sufficed in order to produce a similar behavior in a transient L-form B. subtilis strain, where the cell-wall synthesis was under an inducible promoter. It was suggested that the overproduction of membrane material caused an imbalanced 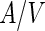 ratio that resulted in the release of the excess surface area in the form of protrusions or buds. Furthermore, it was shown that in branched-chain-fatty-acid-synthesis-deficient L-form cells, formation of membrane protrusion can still occur, but fission cannot (Mercier et al. 2012). Since branched-chain lipids have a much lower transition temperature and are usually found in the
ratio that resulted in the release of the excess surface area in the form of protrusions or buds. Furthermore, it was shown that in branched-chain-fatty-acid-synthesis-deficient L-form cells, formation of membrane protrusion can still occur, but fission cannot (Mercier et al. 2012). Since branched-chain lipids have a much lower transition temperature and are usually found in the 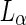 phase, this observation stresses the importance of membrane fluidity for membrane-synthesis-dependent reproduction. It should be noted that, unlike the previous examples of membrane budding and tubulation, the cells were very flaccid in this case.
phase, this observation stresses the importance of membrane fluidity for membrane-synthesis-dependent reproduction. It should be noted that, unlike the previous examples of membrane budding and tubulation, the cells were very flaccid in this case.
In the second example, a stable L-form Listeria monocytogenes strain was obtained that produced cell-wall precursors but nevertheless lacked a mature cell wall, likely due to the some mutation in the cell-wall synthesis pathway (Dell’Era et al. 2009). Surprisingly, large intracellular vesicles emerged, which grew next to sites of disrupted unilamellarity and phospholipid accumulation on the mother cell membrane (Briers et al. 2012a). These intracellular vesicles contained part of the mother cell cytoplasm as well as DNA. Since large L. monocytogenes L-form cells contain multiple chromosomes, some of them could partition into the intracellular vesicles. Following the growth of several intracellular vesicles, the mother cell ruptured and the daughter vesicles were released (see Fig. 3d). Importantly, though many intracellular vesicles were metabolically silent and lacked the membrane potential necessary for ATP generation, they did over time, upon release from the mother cell, acquire the absent membrane potential and became metabolically active. Thus, it was corroborated that these vesicles are a viable reproductive units of this strain.
Interestingly, other L-form members of the genuses Listeria and Entrococcus show similar phenotypes and multiplication modes. In light of the fact that many L-form bacteria reproduce by “budding-like” mechanisms (Gilpin and Nagy 1976; Martin 2010), membrane synthesis may be a good reproduction strategy for simple synthetic cells. Indeed, multiple authors have suggested that this mechanism represents an ancient reproduction mode that might have been active during the prebiotic times, dating back to the origin of life (Leaver et al. 2009; Chen 2009; Briers et al. 2012b; Koonin and Mulkidjanian 2013).
Division processes based on common biological mechanisms
The vast majority of natural cells use intricate protein machineries for their division. Hence, division of synthetic cells can probably be developed by use of minimal forms of these machineries, especially those from a prokaryotic origin. In this section, we survey some of these.
FtsZ-ring-based division
The bacterial division machinery (divisome) of E. coli is composed of about twenty proteins that are recruited to the division site, synthesize the cell wall material of the new poles and exert force to invaginate the membrane (Aarsman et al. 2005; Kirkpatrick and Viollier 2011; Egan and Vollmer 2013). A key component in this process is a protein called FtsZ (filamentous temperature sensitive protein Z). FtsZ is a highly conserved GTPase that in most bacteria and euryarchaea forms the core of the divisome, recruiting downstream division proteins to the division site (de Boer et al. 1992). From conventional fluorescence and transmission electron microscopy data, it has been found that, prior to cytokinesis, FtsZ forms a ring at the future division site that is called the Z-ring. In Gram-negative bacteria, the size of this ring shrinks in concert with the septum growth and invagination (see Fig. 4a) (Bi and Lutkenhaus 1991; Sun and Margolin 1998; Anderson et al. 2004). The fact that in some cell-wall-less bacteria, such as members of the mycoplasma genus, most of the downstream proteins are missing led to the hypothesis that FtsZ by itself can be both the scaffold and force-generating unit of the divisome (Erickson 1997). If this indeed is the case, FtsZ is an ideal machine to promote binary fission of cell-wall-less synthetic cells.
Fig. 4.
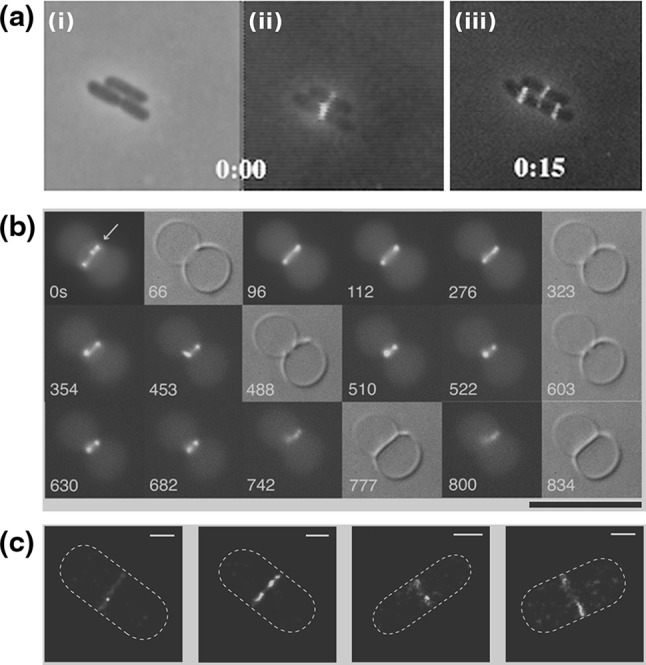
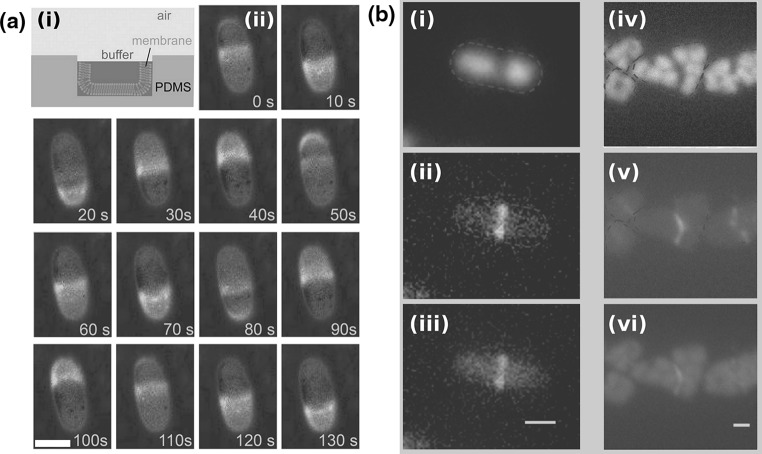
FtsZ in the cytokinesis of cells and liposomes. a Localization of the Z-ring during the cell cycle of E. coli. i Phase-contrast image of three cells. ii Overlay of a phase-contrast image and a conventional fluorescence microscopy image of FtsZ–GFP. iii Overlay of a phase-contrast image and a conventional fluorescence microscopy image of FtsZ–GFP after 15 min. Note that initially one FtsZ ring appears in the lower cell, while two new rings appears in the two daughter cells after the mother cell has divided. b Time lapse recording of the formation of a septum between two liposomes containing equimolar FtsZ–YFP and a mutant 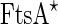 proteins, together with GTP and ATP. Time represents seconds after the start of the recording. Black background images are epi-fluorescence images while gray background images are DIC ones. c Four high-resolution images of the Z-ring in E. coli cells (PALM images of FtsZ–Emos2). Note the inhomogeneity of the fluorescence intensity, indicating that the Z-ring is not a perfect continuous filament. Scale bar 500 nm. Initially, an FtsZ arc is seen localized at a constriction site. After several spatial reorganization steps, the system seems to end up in an intra-vesicular septum. Scale bar 10
proteins, together with GTP and ATP. Time represents seconds after the start of the recording. Black background images are epi-fluorescence images while gray background images are DIC ones. c Four high-resolution images of the Z-ring in E. coli cells (PALM images of FtsZ–Emos2). Note the inhomogeneity of the fluorescence intensity, indicating that the Z-ring is not a perfect continuous filament. Scale bar 500 nm. Initially, an FtsZ arc is seen localized at a constriction site. After several spatial reorganization steps, the system seems to end up in an intra-vesicular septum. Scale bar 10  . a Modified with permission from reference Sun and Margolin (1998)
. a Modified with permission from reference Sun and Margolin (1998)  (1998) American Society of Microbiology. b Modified with permission from reference Osawa and Erickson (2013)
(1998) American Society of Microbiology. b Modified with permission from reference Osawa and Erickson (2013)  (2013) National Academy of Sciences, USA. c Modified from reference Fu et al. (2010) under the Creative Commons Attribution (CC BY) license http://creativecommons.org/licenses/by/3.0/. (Color figure online)
(2013) National Academy of Sciences, USA. c Modified from reference Fu et al. (2010) under the Creative Commons Attribution (CC BY) license http://creativecommons.org/licenses/by/3.0/. (Color figure online)
Several lines of evidence support this hypothesis. First, FtsZ is a structural homolog of tubulin with which it forms a distinct family of GTPases (Nogales et al. 1998). Like tubulin, FtsZ can polymerize to protofilaments in a GTP-dependent manner (Mukherjee and Lutkenhaus 1994), and like tubulin it forms different types of polymeric condensates under different polymerization conditions (Popp et al. 2009; González et al. 2003). However, in contrast to tubulin, FtsZ filaments show no sign of cooperative depolymerization, i.e. a dynamical instability (Mateos-Gil et al. 2012). In particular, the curvature of the protofilaments seems to be associated with the nucleotide state (Erickson 2009). In the absence of GTP hydrolysis, the protofilaments are slightly curved with an average angle of about 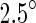 between subunits, an angle that corresponds to a ring with a diameter of approximately 200 nm. However, when all monomers are bound to GDP and in the presence of a cationic lipid bilayer or polycationic-dextran, the protofilaments form highly curved rings with an average angle of
between subunits, an angle that corresponds to a ring with a diameter of approximately 200 nm. However, when all monomers are bound to GDP and in the presence of a cationic lipid bilayer or polycationic-dextran, the protofilaments form highly curved rings with an average angle of 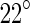 between the subunits and typical diameter of about 24 nm (Lu et al. 2000). These facts may support the Z-ring-centric hypothesis that the constriction force is generated by a GTP-hydrolysis-induced conformational change. Structurally, FtsZ is composed of two domains that are connected by an
between the subunits and typical diameter of about 24 nm (Lu et al. 2000). These facts may support the Z-ring-centric hypothesis that the constriction force is generated by a GTP-hydrolysis-induced conformational change. Structurally, FtsZ is composed of two domains that are connected by an 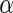 -helix (Löwe and Amos 1998). It is natural to assume that the conformational change occurs by a rotation of the two domains around this
-helix (Löwe and Amos 1998). It is natural to assume that the conformational change occurs by a rotation of the two domains around this 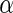 -helix. However, no indication for a conformational change, either around this
-helix. However, no indication for a conformational change, either around this 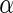 -helix, or elsewhere in the monomer, was detected when examining different crystal structures of FtsZ monomers that binds either GTP, GDP or non-hydrolyzable analogs of GTP (Oliva et al. 2007). This poses a puzzle for the Z-ring force-generating hypothesis. One possible solution is that the conformational change is a characteristic of the polymer rather than of separate monomers, i.e., that it occurs by changing the polymerization angle between neighboring monomers. Indeed, recently molecular dynamics simulation suggested a hinge-like mechanism with an angle difference of
-helix, or elsewhere in the monomer, was detected when examining different crystal structures of FtsZ monomers that binds either GTP, GDP or non-hydrolyzable analogs of GTP (Oliva et al. 2007). This poses a puzzle for the Z-ring force-generating hypothesis. One possible solution is that the conformational change is a characteristic of the polymer rather than of separate monomers, i.e., that it occurs by changing the polymerization angle between neighboring monomers. Indeed, recently molecular dynamics simulation suggested a hinge-like mechanism with an angle difference of 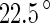 between the states of two monomers in the GTP- and GDP-bound forms (Hsin et al. 2012).
between the states of two monomers in the GTP- and GDP-bound forms (Hsin et al. 2012).
By itself, FtsZ is a cytoplasmic protein, and it must be anchored to the membrane by an auxiliary protein. In E. coli, two such proteins were identified, viz., FtsA and ZipA, which both interact with the C-terminal end of FtsZ. FtsA is an abundant protein in many bacteria that is related to actin and can bind ATP, while ZipA is found mainly in the 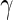 -protobacteria class (Adams and Errington 2009; de Boer 2010). A second and prominent support for the FtsZ force-generating hypothesis comes from research where the C-terminal of FtsZ was replaced with a YFP–MTS moiety (YFP—yellow fluorescence protein; MTS—a membrane targeting sequence from the MinD protein) (Osawa et al. 2008). When the FtsZ–YFP–MTS chimera was incubated, together with GTP, on the outside of tubular multilamellar liposomes, it penetrated into the vesicles lumen and formed dynamic rings on the inner surface of the liposomes. Coalescence of the dynamic rings resulted in broad rings that had a tendency to get localized on visible inward indentations of the membranes, indicating possible forces that were applied by the FtsZ. When the FtsZ–YFP–MTS was incubated onto spherical multilamelar liposomes, it tubulated the liposomes by inducing the growth of protrusion with a diameter of approximately 200 nm (Osawa et al. 2009). Similarly, the FtsZ–YFP–MTS chimera formed concave depressions on unilamellar liposomes. However, when the MTS was transferred to the N-terminal of FtsZ rather than its C-terminal, the chimeric protein MTS–FtsZ–YFP created convex protrusions, thus suggesting that the membrane deformation depends on the direction of the filaments anchoring relative to the membrane. Moreover, without GTP, MTS–FtsZ–YFP created fixed-size rings that wrapped around the outside of tubular multilamellar liposomes, and tended to avoid the more bulgy parts of the liposomes (Osawa and Erickson 2011). These results were interpreted as suggesting a preferential curvature of the FtsZ protofilaments that enables it to exert forces on membranes.
-protobacteria class (Adams and Errington 2009; de Boer 2010). A second and prominent support for the FtsZ force-generating hypothesis comes from research where the C-terminal of FtsZ was replaced with a YFP–MTS moiety (YFP—yellow fluorescence protein; MTS—a membrane targeting sequence from the MinD protein) (Osawa et al. 2008). When the FtsZ–YFP–MTS chimera was incubated, together with GTP, on the outside of tubular multilamellar liposomes, it penetrated into the vesicles lumen and formed dynamic rings on the inner surface of the liposomes. Coalescence of the dynamic rings resulted in broad rings that had a tendency to get localized on visible inward indentations of the membranes, indicating possible forces that were applied by the FtsZ. When the FtsZ–YFP–MTS was incubated onto spherical multilamelar liposomes, it tubulated the liposomes by inducing the growth of protrusion with a diameter of approximately 200 nm (Osawa et al. 2009). Similarly, the FtsZ–YFP–MTS chimera formed concave depressions on unilamellar liposomes. However, when the MTS was transferred to the N-terminal of FtsZ rather than its C-terminal, the chimeric protein MTS–FtsZ–YFP created convex protrusions, thus suggesting that the membrane deformation depends on the direction of the filaments anchoring relative to the membrane. Moreover, without GTP, MTS–FtsZ–YFP created fixed-size rings that wrapped around the outside of tubular multilamellar liposomes, and tended to avoid the more bulgy parts of the liposomes (Osawa and Erickson 2011). These results were interpreted as suggesting a preferential curvature of the FtsZ protofilaments that enables it to exert forces on membranes.
Additional support for the FtsZ force-generating picture was demonstrated by reconstituting the Z-ring together with its native auxiliary proteins inside vesicles. When FtsZ–YFP was incubated inside synthetic liposomes together with 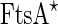 , GTP and ATP, a progressive formation of an inter-luman septum was observed in a small fraction of the vesicles (1.3 %) (see Fig. 5b) (Osawa and Erickson 2013). It was suggested that in these group of vesicles, the activity of the anchored FtsZ protofilaments was the driving force for the septum formation. However, it should be noted that the septa that were formed did not constrict, and that the vesicles did not separate into two daughter vesicles. Here,
, GTP and ATP, a progressive formation of an inter-luman septum was observed in a small fraction of the vesicles (1.3 %) (see Fig. 5b) (Osawa and Erickson 2013). It was suggested that in these group of vesicles, the activity of the anchored FtsZ protofilaments was the driving force for the septum formation. However, it should be noted that the septa that were formed did not constrict, and that the vesicles did not separate into two daughter vesicles. Here, 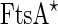 is a hypermorphic mutant of FtsA that is known to depolymerize FtsZ protofilamets in vitro in an ATP-dependent manner and which can support E. coli growth in vivo in the absence of ZipA (Geissler et al. 2003; Beuria et al. 2009). Similarly, when FtsZ was incubated with His-tagged truncated ZipA that could bind Ni-modified lipids in vesicles, shrinkage and sometimes rupturing of the vesicles was observed (Cabré et al. 2013). These two results strongly suggest that FtsZ can exert force on membranes, cause their deformation and probably may function as a binary-fission machine. Interestingly, when FtsZ was incubated with GTP and FtsA inside vesicles that were formed from E. coli inner-membrane and thus also contained some amount of ZipA, FtsA was dislodged from the membrane by FtsZ, suggesting that the type of anchoring has an important influence on the ability of the protofilaments to deform the membrane (Jiménez et al. 2011).
is a hypermorphic mutant of FtsA that is known to depolymerize FtsZ protofilamets in vitro in an ATP-dependent manner and which can support E. coli growth in vivo in the absence of ZipA (Geissler et al. 2003; Beuria et al. 2009). Similarly, when FtsZ was incubated with His-tagged truncated ZipA that could bind Ni-modified lipids in vesicles, shrinkage and sometimes rupturing of the vesicles was observed (Cabré et al. 2013). These two results strongly suggest that FtsZ can exert force on membranes, cause their deformation and probably may function as a binary-fission machine. Interestingly, when FtsZ was incubated with GTP and FtsA inside vesicles that were formed from E. coli inner-membrane and thus also contained some amount of ZipA, FtsA was dislodged from the membrane by FtsZ, suggesting that the type of anchoring has an important influence on the ability of the protofilaments to deform the membrane (Jiménez et al. 2011).
Fig. 5.
Binary fission of cells driven by ESCRT/Cdv or mechanical forces. a Three-dimensional reconstruction of ESCRT-III treated giant unilaminar vesicles (GUV). i Membrane staining of the intra-vesicular bodies that were formed after reconstitution of the eukaryote ESCRT-III complex on the outside of a GUV. ii Z-stack confocal image of the same GUV showing that the intra-vesicular bodies were filled with the extra-vesicular content as a result of ESCRT-III-induced inward budding. Scale bar 5  . b In situ immunofluorescence localization of CdvA (red middle column) and CdvB (green right column) during the constriction of four S. acidocaldarius cells. Note that both CdvA and CdvB are localized between the two segregated chromosomes (blue stained with DAPI). Left column phase-contrast illumination. c Stages in the division of
. b In situ immunofluorescence localization of CdvA (red middle column) and CdvB (green right column) during the constriction of four S. acidocaldarius cells. Note that both CdvA and CdvB are localized between the two segregated chromosomes (blue stained with DAPI). Left column phase-contrast illumination. c Stages in the division of  M. genitalium. Newborn cells possess a single terminal organelle. Division starts when the terminal organelle duplicates, leading to second organelle at the other cell pole. Cytoskeleton filaments then elongate the cell, and create a constricted tube at the cell middle. After the formation of a chain of filaments, some cells are torn off the chain and can start a new reproduction cycle. Scale bar 500 nm. a Modified with permission from reference Wollert et al. (2009)
M. genitalium. Newborn cells possess a single terminal organelle. Division starts when the terminal organelle duplicates, leading to second organelle at the other cell pole. Cytoskeleton filaments then elongate the cell, and create a constricted tube at the cell middle. After the formation of a chain of filaments, some cells are torn off the chain and can start a new reproduction cycle. Scale bar 500 nm. a Modified with permission from reference Wollert et al. (2009)  (2009) Nature Publishing Group. b Modified with permission from reference Samson et al. (2008)
(2009) Nature Publishing Group. b Modified with permission from reference Samson et al. (2008)  (2008) National Academy of Sciences, USA. c Modified with permission from reference Lluch-Senar et al. (2010)
(2008) National Academy of Sciences, USA. c Modified with permission from reference Lluch-Senar et al. (2010)  (2010) John Wiley and Sons, Inc. (Color figure online)
(2010) John Wiley and Sons, Inc. (Color figure online)
A different view regarding the ability of FtsZ to exert forces on membranes emerged from in vitro study of the polymerization of FtsZ-YFP-MTS on curved surfaces. In that case it was shown that the FtsZ protofilamets do not only bend but also twist, and that if a twist is taken into account, the calculated force that a protofilament can apply on a membrane due to its preferred curvature is quite low (about 0.06 pN) (Arumugam et al. 2011). At this point in time, the question whether the curvature of the protofilaments by themselves can drive the deformation of membranes thus still remains under debate.
Advance microscopy imaging studies showed that the Z-ring is not formed by one single continuous FtsZ filament that encircles the cell periphery, as well as that it has a somewhat different structures in different bacteria, thus questioning the curvature-induced-force-generation picture. For example, cryoelectron tomography of FtsZ in Caulobacter crescentus detected only sparsely unconnected short (100 nm) protofilaments near the division site (Li et al. 2007). 3D structured-illumination fluorescence microscopy of B. subtilis and Staphylococous aureus showed that the Z-ring is not homogeneous but has a structure of several high density regions that are connected by low density regions (Strauss et al. 2012). Photo-activated-localization fluorescence microscopy (PALM) in E. coli similarly indicated that the Z-ring is not a single homogenous structure, as well as that the protofilaments assemble into a wide bundle-like structure (see Fig. 5c) (Fu et al. 2010). Finally, polarized fluorescence microscopy showed that, in E. coli but not in C. crescentus, a substantial number of the FtsZ protofilaments are oriented on the membrane parallel to the long axis of the cell rather than, as expected, perpendicular to it (Si et al. 2013). All these observations stress the importance of lateral interactions between the FtsZ protofilaments for the Z-ring organization. In vivo, these lateral interactions are mediated by multiple positive regulators (Huang et al. 2013). Furthermore, in vitro the protofilaments are typically only about 100 nm long, while in vivo the Z-ring is a highly dynamic structure that constantly exchanges material with the cytoplasmic pool (Anderson et al. 2004). Taking all these facts into account, the emerging picture of the Z-ring is that of many short protofilaments, which interact with the membrane and with each other, rather than of a complete continuous ring that simply shrinks during the division process. In order to cope with the question of how such a set of short protofilaments that only weakly interact laterally can drive cell division, a new class of models was suggested. In these models, the driving force for constriction results from protofilaments condensation (Hörger et al. 2008; Lan et al. 2009; Sun and Jiang 2011), though the applicability of these models is still under debate (Erickson 2009). Yet, even if binary fission of bacterial cells by the divisome is much more complicated and is not yet fully understood, the fact that FtsZ (assisted by FtsA, ZipA, or some other membrane-attachment method) can deform membranes makes it a highly attractive machine for binary fission of synthetic cells.
One important concern regarding the use of FtsZ to drive binary fission of synthetic cells is the question of the abscission. Even if FtsZ can exert a force that is large enough to constrict membranes, it is at this point unclear if it can promote the abscission that is needed to separate the mother cell into two daughter cells. Assuming that the highest curvature structure that these FtsZ protofilaments can form is the 24 nm ring that was observed under certain in vitro conditions, and recalling that the Z-ring has to be attached to the membrane by axillary proteins one can estimate that the total diameter of the membrane that is coupled to the Z-ring will be about 55 nm in the most condensed state (Erickson et al. 2010). This means that, even in its most condensed state, the separation between the two membranes on both sides of the Z-ring is too large to promote a spontaneous destabilization of the neck. Hence, as was discussed above, another mechanism should be used to promote the abscission of the neck. Though in vivo this process may be related to cell-wall synthesis, it is not yet clear what simple mechanism can induce the neck destabilization in a Z-ring-based division of synthetic cells.
ESCRT/Cdv-based division
In contrast to the Z-ring, the archaeal Cdv system, which is homologous to the eukaryotic Endosomal Sorting Complex Required for Transport (ESCRT), may be a good candidate for both forcing membrane deformation and fission. Organisms belonging to the archaeal crenarcheal phylum usually lack an FtsZ homolog but instead, they possess a homologous system to the ESCRT one. In eukaryote, the ESCRT system plays a key role in producing inward-forming buds from the endosome membrane that mature to form the multivesicular bodies [see Samson and Bell (Samson and Bell 2009) and references therein]. The core budding machine consists of the ESCRT-III complex that is responsible for the membrane deformation and which forms a contractile ring on the bud neck. The complex is supported by Vps4 that is needed for the ESCRT-III complex disassembly and recycling. In addition to its role in vesicle trafficking, the ESCRT system also participates in the membrane abscission during cytokinesis (Morita et al. 2007). In vitro, a reconstitution assay of the ESRCT-III complex together with giant unilamellar vesicles (GUVs) has shown that the complex can form buds from the GUVs membrane as well as that it can pinch them off (see Fig. 5a) (Wollert et al. 2009).
Archaea feature an ESCRT-like system which appears to be simpler. The archaeal species Sulfolobus acidocaldarius possess an operon-like locus with three genes CdvABC. CdvB and CdvC are homologs of the ESCRT-III complex and Vsp4, respectively. Intriguingly, the transcription and protein levels of CdvB are regulated in a cell-cycle-dependent manner and they peak at the cell-division phase (Samson et al. 2008). Moreover, CdvB forms a ring-like structure at midcell and CdvC is recruited to the same location after chromosome segregation (see Fig. 5b) (Lindås et al. 2008). CdvB interacts with CdvC, and overexpression of a catalytically inactive CdvC mutant inhibits cell division. CdvA does not have a close eukaryotic homolog. CdvA mRNA level shows a peak prior to the CdvB mRNA peak and, like CdvC, CdvA proteins interacts with CdvB to co-form a ring-like structure at mid-cell that decreases in length as the cell division progresses (Samson et al. 2011). In vitro assays showed that C-terminus-deleted CdvB forms filamentous polymers, CdvC assembles as a homopolymer structure and CdvA forms polymers in association with DNA (Moriscot et al. 2011). CdvA can also wrap around liposomes and, upon recruiting CdvB, deform the liposomes to a network of small tubes (Samson et al. 2011; Dobro et al. 2013).
These evidences suggest that the Crenarcheal Cdv system is a good candidate to be used as a protein-based machinery for division of synthetic cells, likely even better than FtsZ. This conclusion is further supported by the observation that cell division seems to be executed by the Cdv system rather than by the FtsZ one in Thaumarchaeon nitrosopumilus, a member of the Thaumarcheota phylum of archaea that possesses both FtsZ and Cdv (Pelve et al. 2011). One can ask whether the ESCRT system might not be preferred over Cdv. The Cdv system appears to have two advantages over the ECRST one: First, it appears to be composed of less proteins, and second, it is directly involved is binary cell fission rather than in budding. However, at this point, there is a direct evidence only for the ESCRT system that it can by itself cause separation of one vesicle from another.
Alternative division mechanisms based on cytoskeleton proteins
Another interesting, though very different, possibility to divide synthetic cells is by the use of force-generating proteins that tear the cell into two by pushing on the membrane to the opposite sides of the cell. Actin or tubulin systems lend themselves naturally for this purpose since in vivo they participate in structures like the filopodia, the lamellipodia and the mitotic spindle. Indeed, in vitro reconstitution systems that are based on actin or tubulin structures are well established (Bausch and Kroy 2006; Liu and Fletcher 2009). For example, assembly of either actin or tubulin inside vesicles resulted in their deformation and the growth of protrusions (Fygenson et al. 1997; Miyata et al. 1999). More recently, protein cortices made up of actin and its associated proteins were reconstituted inside vesicles (Liu and Fletcher 2006; Merkle et al. 2008; Pontani et al. 2009; Tsai et al. 2011; López-Montero et al. 2012). These cortices increase the mechanical stability of vesicles. However, the ability to implement these types of in vitro reconstituted systems in order to achieve cytokinesis seems to be still far away. Recalling, in addition, the large number of actin and tubulin associated protein that are tailored in vivo for different tasks, we anticipate that it may be hard to engineer a division system based on actin or tubulin associated protein networks, while striving to base synthetic cells on a simple minimal system.
In contrast to the common eukaryotic cytoskeleton, the cytoskeleton system of reduced-genome mycoplasma bacteria may provide a good substitute. Recently, it was shown that a  Mycoplasma genitalium can still divide through a binary fission mechanism (Lluch-Senar et al. 2010). When FtsZ was deleted, M. genitalium cells propagated by utilizing their gliding apparatus and cytoskeleton polymers (see Fig. 5c). Under native conditions, Mycoplasma attaches to surfaces through a terminal organelle that is located at one of its ends, and they use polymerization of their cytoskeleton in order to subsequently move. When FtsZ was deleted, the M. genitalium cells produced two terminal organelles at opposite ends of the cell. After attachment of these to the surface, the action of the gliding machinery resulted in a force that tore cells into two sibling cells. In fact, this reproduction mode is very similar to that of Mycoplasma mobile, a close relative of M. genitalium, which naturally lacks FtsZ (Rosengarten and Kirchhoff 1989). Though the molecular mechanism of
Mycoplasma genitalium can still divide through a binary fission mechanism (Lluch-Senar et al. 2010). When FtsZ was deleted, M. genitalium cells propagated by utilizing their gliding apparatus and cytoskeleton polymers (see Fig. 5c). Under native conditions, Mycoplasma attaches to surfaces through a terminal organelle that is located at one of its ends, and they use polymerization of their cytoskeleton in order to subsequently move. When FtsZ was deleted, the M. genitalium cells produced two terminal organelles at opposite ends of the cell. After attachment of these to the surface, the action of the gliding machinery resulted in a force that tore cells into two sibling cells. In fact, this reproduction mode is very similar to that of Mycoplasma mobile, a close relative of M. genitalium, which naturally lacks FtsZ (Rosengarten and Kirchhoff 1989). Though the molecular mechanism of 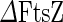 M. genitalium division is not clear, the fact that M. genitalium is already used in top-down approaches as a starting point for minimal cells, suggests that this form of reproduction will receive more attention in future research on synthetic cell division.
M. genitalium division is not clear, the fact that M. genitalium is already used in top-down approaches as a starting point for minimal cells, suggests that this form of reproduction will receive more attention in future research on synthetic cell division.
Controlling the position of the division site
Above we reviewed some basic mechanisms that may serve to proliferate synthetic cells by division. It is important to realize that, in order to achieve a viable and robust methodology for synthetic cell division, much more is needed beyond merely demonstrating fission of vesicles. When a cell reproduces, its content is divided between its daughter cells. Obviously, if this process is not tightly regulated, some of the progenies can lack one or more of the essential components and die. This process, which is termed as death by dilution, is a serious threat to the vitality of any synthetic cell line. This calls for the need to include well-controlled synthesis of components as well as to incorporate spatial and temporal mechanisms for controlling the site for cell division in synthetic cells. Here we briefly discuss the three most characterized division-site-regulation mechanisms in bacteria with the hope that they can inspire the engineering principles of synthetic cells. Though these examples all work by antagonizing the assembly of the FtsZ ring, they suggest some general engineering principles for division-site selection.
The best studied division regulation mechanism in bacteria is the Min system. Its name is derived from the mini-cells, a phenotype where division leads to small cells that miss the basic chromatin, which is observed in the absence of Min proteins. In E. coli, the Min system is composed of three proteins, MinCDE. MinC is an antagonist of FtsZ ring-assembly (Hu et al. 1999). It has two domains (Cordell et al. 2001), a C-terminus domain that prevents the lateral interaction between FtsZ protofilaments and thus the maturation of the FtsZ ring, and an N-terminus domain that weakens the binding between FtsZ monomers (Dajkovic et al. 2008). By itself, MinC is a cytoplasmic protein, but it is targeted to the membrane by MinD, an ATPase from the ParA/MinD protein family with a membrane-targeting domain at its C-terminus. MinD binds to the membrane as a dimer only in its ATP-bound form (Hu and Lutkenhaus 1999; Lackner et al. 2003). In the absence of MinE, the MinCD complex is distributed homogeneously over the membrane and the FtsZ ring formation is blocked all along the cell. MinE, which is the topological regulator of the Min system, binds MinD on a binding site overlapping to that for MinC (Wu et al. 2011) and thus can displace MinC from the MinD-covered surface back to the cytoplasm. Moreover, MinE stimulates the rather marginal ATPase activity of MinD and releases it from the membrane (Hu and Lutkenhaus 2001).
The outcome of this reaction-diffusion network of interactions is a remarkable dynamical pattern for the spatiotemporal organization of the Min protein, where the Min proteins display pole-to-pole oscillations (Raskin and de Boer 1999). One oscillation cycle starts when MinCD forms a cup at one of the cell poles, and a MinE ring forms at the end of the cup close to the mid-cell (Hale et al. 2001). The MinE ring causes the release of the MinD proteins and the associated shrinking of the cup. In the cytoplasm, MinD proteins exchange their ADP with ATP, diffuse to the other pole and start forming a new cup there that again extends to midcell. Assembly of a MinE ring at the rim of the new cup causes the process to repeat itself, resulting in an oscillatory motion with a period of about 1 minute. Interestingly, it was suggested that transient interactions of MinE with the membrane following the release of MinD are essential for formation of this pole-to-pole oscillations (Hsieh et al. 2010; Park et al. 2011). Due to this dynamical behavior, the time-averaged MinD concentration on the membrane (and hence also that of MinC) is the highest next to the poles and the lowest around midcell (Kanthang et al. 2011). Due to this concentration profile of MinC, the FtsZ ring will be prevented to form near the poles (i.e. avoiding the formation of useless mini-cells) and tend to form somewhere near the middle of the cell. Notably, in B. subtilis, MinE is absent. Instead, the MinCD complex is targeted permanently (i.e. without temporal oscillations) to the curved poles, as well as to the highly curved area next to the newly formed septum, by the auxiliary protein MinJ (Patrick and Kearns 2008; Bramkamp et al. 2008). MinJ in turn interacts with DivIVA, a protein that was reported to sense negatively curved membranes (Lenarcic et al. 2009; Eswaramoorthy et al. 2011). Thus, also in B. subtilis a gradient of MinC is created with a minimum at midcell and the FtsZ formation is blocked near the poles.
Importantly, the E. coli Min system was reconstituted in an in vitro assay both on a planar supported lipid membrane (SLB) (Loose et al. 2008, 2011; Schweizer et al. 2012) and recently in micro-cavities (see Fig. 6a) (Zieske and Schwille 2013; Caspi et al. 2014, to be published). On planar SLB, the reconstituted system forms running waves with a very long wavelength of several tens of microns [as well as other dynamical patterns (Ivanov and Mizuuchi 2010)], while in open micro-cavities it shows an oscillatory behavior similar to the in vivo one, albeit with the long in vitro wavelength (which is much longer than the in vivo length scale). This suggests that the Min system may function in a straightforward manner in synthetic cells. Moreover, it may be possible to use the dynamical Min system in synthetic cells with shapes different from the rods, since, as was recently show in our lab, it possesses a preferable length scale and is active in E. coli cells with non-natural shapes (Wu et al. 2014 to be published).
Fig. 6.
Mechanisms for division apparatus localization. a
i Assay for the reconstitution of MinDE in microchambers (side view). Microchambers are fabricated in PDMS, coated with a supported lipid bilayer, and buffer with MinD, MinE and ATP is placed on top. ii
Top-view images of MinDE dynamic localization in one of the microchambers of a. Time is in seconds; 10 % of the MinE proteins are labeled with Alexa-488. Scale bar 10 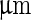 . b Localization of the nucleoid and FtsZ-GFP in rod shaped (i–iii
left column) and anomalously shaped E. coli cells (iv–vi
right column). Nucleoid is labeled with the nucleoid-associated protein HupA conjugated to RFP. Red dashed line represents the outline of the cells. i, iv Chromosomes profile. ii, v FtsZ-GFP. iii, vi Overlay of the HupA-RPF (orange) and the FtsZ-GFP (green) images. Scale bar in both cases is 1
. b Localization of the nucleoid and FtsZ-GFP in rod shaped (i–iii
left column) and anomalously shaped E. coli cells (iv–vi
right column). Nucleoid is labeled with the nucleoid-associated protein HupA conjugated to RFP. Red dashed line represents the outline of the cells. i, iv Chromosomes profile. ii, v FtsZ-GFP. iii, vi Overlay of the HupA-RPF (orange) and the FtsZ-GFP (green) images. Scale bar in both cases is 1 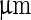 . a Modified with permission from reference Zieske and Schwille (2013)
. a Modified with permission from reference Zieske and Schwille (2013)  (2013) John Wiley and Sons, Inc. b Modified with permission from reference Männik et al. (2012)
(2013) John Wiley and Sons, Inc. b Modified with permission from reference Männik et al. (2012)  (2012) National Academy of Sciences, USA. (Color figure online)
(2012) National Academy of Sciences, USA. (Color figure online)
Although the Min system seems to be a good candidate for a division-site selection in synthetic cells, and although transient interactions between MinD and the chromosome were reported (Akerlund et al. 2002; Di Ventura et al. 2013), it lacks an important feature—coordination of division-site selection with chromosome segregation. In many bacteria, this is achieved by the Nucleoid Occlusion (NO) mechanism, which assures that the FtsZ ring will not form over unsegregated chromosomes (Mulder and Woldringh 1989). In E. coli, NO is mediated by SlmA, a member of the TetR DNA-binding protein family (Bernhardt and De Boer 2005). SlmA binds the chromosome in a sequence-specific manner and its binding sites are distributed all along the chromosome, excluding the terminus region (Ter) (Cho et al. 2011; Tonthat et al. 2011). These binding sites serve as nucleation site for the polymerization of SlmA into filaments distributed among the chromosome. Importantly, essential for its function, SlmA can bind simultaneously to DNA and to FtsZ, and, upon activation by the chromosome binding, disassemble FtsZ polymers and higher-order FtsZ polymeric structures (Tonthat et al. 2013). In B. subtilis, a similar mechanism is mediated by an unrelated protein Noc, that possesses a ParB-like DNA binding site (Wu and Errington 2004). Like SlmA, Noc binds DNA and its binding sites are distributed mainly away from the Ter site on the chromosome (Wu et al. 2009). However, since no direct interaction has been shown between Noc and FtsZ, the exact mode of Z-ring inhibition by Noc is still unclear. Yet, the fact that for both SlmA and Noc, the DNA binding sites are distributed away from the Ter region suggests a coupling between the division site selection and the timing of the start of the division execution, as the Ter region is the last to segregate. Thus, NO appears to dictate both the location and the timing of the FtsZ ring formation, where it ensures that the Z-ring forms only in between the chromosomes and only after the replication propagated towards its end.
It was suggested that the two mechanisms together, i.e. both Min and NO, are essential for a tight regulation of the division site selection, though a study in E. coli with distorted shapes suggested that the NO occlusion is the more determinant one (see Fig. 6b) (Männik et al. 2012). The combination of the Min and NO mechanisms show the three basic requirements of a division-site selection mechanism in synthetic cells; (1) it should inhibit the action of the division machinery away from undesirable locations, (2) it should direct it to desirable locations, and (3) it should control the timing of division during the cell cycle. In fission yeast, a similar triple-purpose mechanism exist where negative cues from the cell poles inhibit the contractile ring assembly, while positive cues from the nucleus influence the timing and position of the contractile ring assembly at midcell (Pollard 2010). Interestingly, in C. crescentus, a two-protein (MipZ and ParB) mechanism combines the features of a dynamical Min system with the principle of NO inhibition, and accomplishes these three targets simultaneously (Thanbichler and Shapiro 2006). MipZ is an ATPase and a member of the ParA/MinD family. In its dimeric form, it can interact with FtsZ and DNA to inhibit FtsZ ring assembly (Kiekebusch et al. 2012). After ATP hydrolysis, the dimer disassembles and is released from the DNA. The ADP-to-ATP exchange rate of MipZ is rather low, but it is much accelerated when MipZ monomers interact with ParB, a DNA-binding protein that binds the parS DNA site next to the origin of replication (Ori). Since the Ori is attached to one of the cell poles in C. crescentus, the net result is the formation of a MipZ dimer gradient from one of the cell poles towards the other, and this gradient pushes the FtsZ towards that end. When the chromosome of C. crescentus replicates, one of the parS sites migrates towards the other pole together with the Ori. Thus, after chromosome replication, a bidirectional gradient of MipZ dimers forms, which has a valley between the two poles. As result, the FtsZ ring will tend to assemble at that point, but only after replication is almost completed. This again stresses the necessity of both spatial and temporal cues for division-site selection, and show that their most prominent function is to ensure that the genetic material is distributed evenly between the next generation of siblings. These principles can be used to guide the engineering of division site selection in synthetic cells.
Outlook
In this review we discussed some possibilities for the engineering of the division of synthetic cells that use lipid membranes as their shell. Due to the necessity of reproduction for cellular life, these principles are highly important for any successful future (even if still far away) attempts to construct synthetic cells via a bottom-up approach. However, they can also be applicable for top-down approaches that try to push natural cells towards their minimal state.
We emphasized the fact that the physical properties of amphiphiles may promote a variety of robust deformation mechanisms that are based on factors like shape transformations, absorption of shell molecules, synthesis of shell molecules, and growth. However, these mechanisms must be supplemented by mechanisms for the abscission of small necks in membranes that finally leads to the separation of the newly born cells. We discussed factors like the lipid composition or the forces that are impinged on the membrane from simple protein machines that can provide this necessary link. As a second step, processes that control the division site placement and its temporal regulation should also be incorporated in order to create viable and robust synthetic cell lines.
As was shown, the range of possible mechanisms that can promote the division of synthetic cells can be inspired from both physical, chemical and biological principles. It is interesting to note that in spite of the extensive work that was done on shape transformation of vesicles and its possible relation to cell division, there are still factors that were less considered such as the influence of the spontaneous curvature  on membrane neck abscission. In particular, studies on the function in cell division and vesicle splitting of cardiolipin, a bacterial lipid with high
on membrane neck abscission. In particular, studies on the function in cell division and vesicle splitting of cardiolipin, a bacterial lipid with high  (Mileykovskaya and Dowhan 2009) and an ability to form non-lamellar lipid phases (Matsumoto et al. 2006), may be beneficial for the engineering of fission of synthetic cells. Likewise, proteins that curve the membrane (Zimmerberg and Kozlov 2006) or assisting in the cutting off of buds (Pucadyil and Schmid 2008; Daumke et al. 2014), even if they are from a eukaryotic origin, may also be valuable for engineering non-natural pathways for division of synthetic cells. For example, it was recently demonstrated that expression of caveolae in E. coli can promote the emergence of buds from the inner membrane of the bacterium as well as their pinching off from the membrane (Walser et al. 2012). We furthermore expect that advances in the understanding of new bacteria and archaea cells that do not reproduce using the FtsZ ring and the ESCRT mechanisms (Angert 2005; Sonobe et al. 2010; Bernander and Ettema 2010), will expand our ability to engineer the division of synthetic cells under different environmental conditions.
(Mileykovskaya and Dowhan 2009) and an ability to form non-lamellar lipid phases (Matsumoto et al. 2006), may be beneficial for the engineering of fission of synthetic cells. Likewise, proteins that curve the membrane (Zimmerberg and Kozlov 2006) or assisting in the cutting off of buds (Pucadyil and Schmid 2008; Daumke et al. 2014), even if they are from a eukaryotic origin, may also be valuable for engineering non-natural pathways for division of synthetic cells. For example, it was recently demonstrated that expression of caveolae in E. coli can promote the emergence of buds from the inner membrane of the bacterium as well as their pinching off from the membrane (Walser et al. 2012). We furthermore expect that advances in the understanding of new bacteria and archaea cells that do not reproduce using the FtsZ ring and the ESCRT mechanisms (Angert 2005; Sonobe et al. 2010; Bernander and Ettema 2010), will expand our ability to engineer the division of synthetic cells under different environmental conditions.
In addition, we would like to mention the possible importance of cell-wall-like structures for the robustness of synthetic cells. Recently, it was suggested that the invention of the cell wall caused the massive radiation of prokaryotes during the origin of life (Errington 2013). The cell wall confers many advantages to living organisms and may enable them to live in many different environmental niches without the need for protective external conditions. Likewise, if the field of synthetic cells also wants to confer synthetic cells with biotechnological value, their encapsulation in cell-wall-like structures will be important to provide them with any reasonable level of robustness. Synthetic cell walls have so far hardly been explored. Evidently, the encapsulation of synthetic cells in cell-wall-like structures will also deepen our understanding regarding the relations between cells and environmental processes. Thus, we estimate that engineering mechanisms to grow and divide these cell-wall-like structures will become one of the important future challenges in the field of synthetic cells.
From prehistoric times until today, mankind has been busy in creating and improving forms of life that it can utilize through cross-breeding and directed evolution. Today, many scientists and philosophers predict that we stand on a doorsill of history where attempts to create man-made synthetic life, and in particular synthetic cells, may soon succeed and maturate. Whether this will represent a revolutionary step forward or merely an evolutionary development, such research will be of great interest to the basic understanding of the fundamental concepts of life. Indeed, such an understanding is bound to come from the engineering of cellular elements. As Richard Feynman famously said: ’What I cannot create, I do not understand.’ Possible solutions for synthetic-cell division, which we have discussed here, will stand at the forefront of the endeavors to construct synthetic cells. Indeed, we predict that, the exciting coming years will display many foundational and highly interesting results for ways to divide synthetic cells, as well as ways to combine their division with other essential synthetic modules, thus constituting the essential steps in building a synthetic cell.
Acknowledgments
This work was supported by European Research Council research grant NanoforBio (No. 247072) and the EuroSYNBIO grant SynDiv. We acknowledge Fabai Wu and Siddharth Deshpande for a critical reading of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Traditionally, this quote is attributed to Rudolph Virchow (1821–1902), one of the founders of the cell theory that made his seminal contribution mainly during the fourth decade of 19th centenary. However, in fact Virshow adopted this epigram from Raspail (1825) and only popularized it (Wright and Poulsom 2012).
References
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Akerlund T, Gullbrand B, Nordström K. Effects of the Min system on nucleoid segregation in Escherichia coli. Microbiology. 2002;148:3213–3222. doi: 10.1099/00221287-148-10-3213. [DOI] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes-Koback M, Keating CD. Complete budding and asymmetric division of primitive model cells to produce daughter vesicles with different interior and membrane compositions. J Am Chem Soc. 2011;133:9545–9555. doi: 10.1021/ja202406v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angert ER. Alternatives to binary fission in bacteria. Nat Rev Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- Arumugam S, Chwastek G, Schwille P. Protein-membrane interactions: the virtue of minimal systems in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2011;3:269–280. doi: 10.1002/wsbm.119. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Srinivasan R, Huang Y, Ng KH. Comparing contractile apparatus-driven cytokinesis mechanisms across kingdoms. Cytoskeleton. 2012;69:942–956. doi: 10.1002/cm.21082. [DOI] [PubMed] [Google Scholar]
- Bar-Ziv R, Moses E. Instability and pearling states produced in tubular membranes by competition of curvature and tension. Phys Rev Lett. 1994;73:1392. doi: 10.1103/PhysRevLett.73.1392. [DOI] [PubMed] [Google Scholar]
- Bar-Ziv R, Frisch T, Moses E, et al. Entropic expulsion in vesicles. Phys Rev Lett. 1995;75:3481–3484. doi: 10.1103/PhysRevLett.75.3481. [DOI] [PubMed] [Google Scholar]
- Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- Bausch AR, Kroy K. A bottom-up approach to cell mechanics. Nat Phys. 2006;2:231–238. [Google Scholar]
- Berclaz N, Müler M, Walde P, Luisi PL. Growth and transformation of vesicles studied by Ferritin labeling and cryotransmission electron microscopy. J Phys Chem B. 2001;105:1056–1064. [Google Scholar]
- Bernander R, Ettema TJ. FtsZ-less cell division in archaea and bacteria. Curr Opin Microbiol. 2010;13:747–752. doi: 10.1016/j.mib.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, De Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuria TK, Mullapudi S, Mileykovskaya E, Sadasivam M, Dowhan W, Margolin W (2009) Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem 284:14079–14086 [DOI] [PMC free article] [PubMed]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Blöchliger E, Blocher M, Walde P, Luisi PL. Matrix effect in the size distribution of fatty acid vesicles. J Phys Chem B. 1998;102:10383–10390. [Google Scholar]
- Boźić B, Svetina S. A relationship between membrane properties forms the basis of a selectivity mechanism for vesicle self-reproduction. Eur Biophys J. 2004;33:565–571. doi: 10.1007/s00249-004-0404-5. [DOI] [PubMed] [Google Scholar]
- Božič B, Svetina S. Vesicle self-reproduction: the involvement of membrane hydraulic and solute permeabilities. Eur Phys J E. 2007;24:79–90. doi: 10.1140/epje/i2007-10217-1. [DOI] [PubMed] [Google Scholar]
- Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor minCD. Mol Microbiol. 2008;70:1556–1569. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- Briers Y, Staubli T, Schmid MC, Wagner M, Schuppler M, Loessner MJ. Intracellular vesicles as reproduction elements in cell wall-deficient L-form bacteria. PLoS One. 2012;7:e38514. doi: 10.1371/journal.pone.0038514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briers Y, Walde P, Schuppler M, Loessner MJ. How did bacterial ancestors reproduce? lessons from L-form cells and giant lipid vesicles: multiplication similarities between lipid vesicles and L-form bacteria. Bioessays. 2012;34:1078–1084. doi: 10.1002/bies.201200080. [DOI] [PubMed] [Google Scholar]
- Budin I, Debnath A, Szostak JW. Concentration-driven growth of model protocell membranes. J Am Chem Soc. 2012;134:20812–20819. doi: 10.1021/ja310382d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabré EJ, Sánchez-Gorostiaga A, Carrara P, Ropero N, Casanova M, Palacios P, Stano P, Jiménez M, Rivas G, Vicente M. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J Biol Chem. 2013;288:26625–26634. doi: 10.1074/jbc.M113.491688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham PB. The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J Theor Biol. 1970;26:61–81. doi: 10.1016/s0022-5193(70)80032-7. [DOI] [PubMed] [Google Scholar]
- Chen CM, Higgs P, MacKintosh F. Theory of fission for two-component lipid vesicles. Phys Rev Lett. 1997;79:1579. [Google Scholar]
- Chen IA. Cell division: breaking up is easy to do. Curr Biol. 2009;19:R327–R328. doi: 10.1016/j.cub.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci U S A. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell SC, Anderson RE, Löwe J. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 2001;20:2454–2461. doi: 10.1093/emboj/20.10.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- Daumke O, Roux A, Haucke V. Bar domain scaffolds in dynamin-mediated membrane fission. Cell. 2014;156:882–892. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Era S, Buchrieser C, Couvé E, Schnell B, Briers Y, Schuppler M, Loessner MJ. Listeria monocytogenes L-forms respond to cell wall deficiency by modifying gene expression and the mode of division. Mol Microbiol. 2009;73:306–322. doi: 10.1111/j.1365-2958.2009.06774.x. [DOI] [PubMed] [Google Scholar]
- Division-Proteins; E. coli divisome proteins–(i) FtsZ, (ii) FtsA, (iii) ZipA, (iv) FtsN, (v) FtsW, (vi) FtsX, (vii) FtsE, (viii) FtsK, (ix) FtsQ, (x) FtsL, (xi) FtsB, (xii) FtsP, (xiii) PBP1B, (xiv) PBP3, (xv) Tol-Pal, (xvi) MltA, (xvii) MipA, (xviii) LpoB, (xix) NlpD, (xx) AmiA/B, (xi) AmiC, (xii) EnvC; E. coli FtsZ regulators- (i) MinC, (ii) MinD, (iii) MinE, (iv) SlmA, (v) SulA, (vi) ClipX, (viii) ZapA, (viii) ZapB, (ix) ZapC, (x) ZapD, (xi) OpgH
- Di Ventura B, Knecht B, Andreas H, Godinez WJ, Fritsche M, Rohr K, Nickel W, Heermann DW, Sourjik V. Chromosome segregation by the Escherichia coli Min system. Mol Syst Biol. 2013;9:686. doi: 10.1038/msb.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döbereiner HG, Käs J, Noppl D, Sprenger I, Sackmann E. Budding and fission of vesicles. Biophys J. 1993;65:1396–1403. doi: 10.1016/S0006-3495(93)81203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PLG, Bell SD, Jensen GJ. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–2327. doi: 10.1091/mbc.E12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DA, Way JC, Silver PA. Designing biological systems. Genes Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- Dzieciol AJ, Mann S. Designs for life: protocell models in the laboratory. Chem Soc Rev. 2012;41:79–85. doi: 10.1039/c1cs15211d. [DOI] [PubMed] [Google Scholar]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Modeling the physics of FtsZ assembly and force generation. Proc Natl Acad Sci U S A. 2009;106:9238–9243. doi: 10.1073/pnas.0902258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. L-form bacteria, cell walls and the origins of life. Open Biol. 2013;3(120):143. doi: 10.1098/rsob.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. MBio. 2011;2:e0025711. doi: 10.1128/mBio.00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA. Bending resistance and chemically induced moments in membrane bilayers. Biophys J. 1974;14:923–931. doi: 10.1016/S0006-3495(74)85959-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisunov GY, Alexeev DG, Bazaleev NA, Ladygina VG, Galyamina MA, Kondratov IG, Zhukova NA, Serebryakova MV, Demina IA, Govorun VM. Core proteome of the minimal cell: comparative proteomics of three mollicute species. PLoS One. 2011;6:e21964. doi: 10.1371/journal.pone.0021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fygenson DK, Marko JF, Libchaber A. Mechanics of microtubule-based membrane extension. Phys Rev Lett. 1997;79:4497–4500. [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in FtsA bypasses the requirement for the essential cell division gene ZipA in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin RW, Nagy SS. Time-lapse photography of Bacillus subtilis L-forms replicating in liquid medium. J Bacteriol. 1976;127:1018. doi: 10.1128/jb.127.2.1018-1021.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, III, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Jiménez M, Vélez M, Mingorance J, Andreu JM, Vicente M, Rivas G. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J Biol Chem. 2003;278:37664–37671. doi: 10.1074/jbc.M305230200. [DOI] [PubMed] [Google Scholar]
- Hale CA, Meinhardt H, de Boer PA. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Henry C, Overbeek R, Stevens RL. Building the blueprint of life. Biotechnol J. 2010;5:695–704. doi: 10.1002/biot.201000076. [DOI] [PubMed] [Google Scholar]
- Hörger I, Velasco E, Mingorance J, Rivas G, Tarazona P, Vélez M. Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;77:011902. doi: 10.1103/PhysRevE.77.011902. [DOI] [PubMed] [Google Scholar]
- Hsieh CW, Lin TY, Lai HM, Lin CC, Hsieh TS, Shih YL. Direct MinE–membrane interaction contributes to the proper localization of MinDE in E. coli. Mol Microbiol. 2010;75:499–512. doi: 10.1111/j.1365-2958.2009.07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J, Gopinathan A, Huang KC. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proc Natl Acad Sci U S A. 2012;109:9432–9437. doi: 10.1073/pnas.1120761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KH, Durand-Heredia J, Janakiraman A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Inaoka Y, Yamazaki M. Vesicle fission of giant unilamellar vesicles of liquid-ordered-phase membranes induced by amphiphiles with a single long hydrocarbon chain. Langmuir. 2007;23:720–728. doi: 10.1021/la062078k. [DOI] [PubMed] [Google Scholar]
- Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci U S A. 2010;107:8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MC, Forster AC. Update on designing and building minimal cells. Curr Opin Biotechnol. 2010;21:697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez M, Martos A, Vicente M, Rivas G. Reconstitution and organization of Escherichia coli proto-ring elements (FtsZ and FtsA) inside giant unilamellar vesicles obtained from bacterial inner membranes. J Biol Chem. 2011;286:11236–11241. doi: 10.1074/jbc.M110.194365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthang P, Ngamsaad W, Nuttavut N, Triampo W, Triampo D, Krittanai C. Biophysical approach for studying the MinD protein dynamics and energy landscape: a novel use of the spot tracking technique. The Eur Phys J Appl Phys. 2011;55:11201. [Google Scholar]
- Käs J, Sackmann E. Shape transitions and shape stability of giant phospholipid vesicles in pure water induced by area-to-volume changes. Biophys J. 1991;60:825–844. doi: 10.1016/S0006-3495(91)82117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekebusch D, Michie KA, Essen LO, Löwe J, Thanbichler M. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell. 2012;46:245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Curr Opin Microbiol. 2011;14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Koonin EV. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Mulkidjanian AY. Evolution of cell division: from shear mechanics to complex molecular machineries. Cell. 2013;152:942–944. doi: 10.1016/j.cell.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Kurihara K, Tamura M, Shohda KI, Toyota T, Suzuki K, Sugawara T. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat Chem. 2011;3:775–781. doi: 10.1038/nchem.1127. [DOI] [PubMed] [Google Scholar]
- Kuruma Y, Stano P, Ueda T, Luisi PL. A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim Biophys Acta Biomembr. 2009;1788:567–574. doi: 10.1016/j.bbamem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Raskin DM, de Boer PAJ. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol. 2003;185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci U S A. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- Leirer C, Wunderlich B, Myles VM, Schneider MF. Phase transition induced fission in lipid vesicles. Biophys Chem. 2009;143:106–109. doi: 10.1016/j.bpc.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R (2008), A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA 105:18942–18946. [DOI] [PMC free article] [PubMed]
- Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AP, Fletcher DA. Biology under construction: in vitro reconstitution of cellular function. Nat Rev Mol Cell Biol. 2009;10:644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M, Querol E, Piñol J. Cell division in a minimal bacterium in the absence of FtsZ. Mol Microbiol. 2010;78:278–289. doi: 10.1111/j.1365-2958.2010.07306.x. [DOI] [PubMed] [Google Scholar]
- Lonchin S, Luisi PL, Walde P, Robinson BH. A matrix effect in mixed phospholipid/fatty acid vesicle formation. J Phys Chem B. 1999;103:10910–10916. [Google Scholar]
- Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- Loose M, Fischer-Friedrich E, Herold C, Kruse K, Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat Struct Mol Biol. 2011;18:577–583. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- López-Montero I, Rodríguez-García R, Monroy F. Artificial spectrin shells reconstituted on giant vesicles. J Phys Chem Lett. 2012;3:1583–1588. doi: 10.1021/jz300377q. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi PL, Ferri F, Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften. 2006;93:1–13. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- Männik J, Wu F, Hol FJ, Bisicchia P, Sherratt DJ, Keymer JE, Dekker C. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc Natl Acad Sci U S A. 2012;109:6957–6962. doi: 10.1073/pnas.1120854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HH. L-forms and the unusual formation of progeny in cell wall-less bacteria: recognizing the old roots of new science. Arch Microbiol. 2010;192:235–236. doi: 10.1007/s00203-009-0540-5. [DOI] [PubMed] [Google Scholar]
- Mateos-Gil P, Paez A, Hörger I, Rivas G, Vicente M, Tarazona P, Mlez Vélez M. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc Natl Acad Sci U S A. 2012;109:8133–8138. doi: 10.1073/pnas.1204844109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- Mazzarello P. A unifying concept: the history of cell theory. Nat Cell Biol. 1999;1:E13–E15. doi: 10.1038/8964. [DOI] [PubMed] [Google Scholar]
- Mercier R, Dominguez-Cuevas P, Errington J. Crucial role for membrane fluidity in proliferation of primitive cells. Cell Rep. 2012;1:417–423. doi: 10.1016/j.celrep.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Mercier R, Kawai Y, Errington J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell. 2013;152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- Merkle D, Kahya N, Schwille P. Reconstitution and anchoring of cytoskeleton inside giant unilamellar vesicles. Chembiochem. 2008;9:2673–2681. doi: 10.1002/cbic.200800340. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H, Nishiyama S, Akashi K, Kinosita K., Jr Protrusive growth from giant liposomes driven by actin polymerization. Proc Natl Acad Sci U S A. 1999;96:2048–2053. doi: 10.1073/pnas.96.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, Schoehn G, Forterre P, Weissenhorn W, Renesto P. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS One. 2011;6(e21):921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RG, McKane AJ. Stability of growing vesicles. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:061151. doi: 10.1103/PhysRevE.83.061151. [DOI] [PubMed] [Google Scholar]
- Morris RG, Fanelli D, McKane AJ. Dynamical description of vesicle growth and shape change. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82:031125. doi: 10.1103/PhysRevE.82.031125. [DOI] [PubMed] [Google Scholar]
- Moya A, Gil R, Latorre A, Peretó J, Pilar Garcillán-Barcia M . Toward minimal bacterial cells: evolution vs. design. FEMS Microbiol Rev. 2009;33:225–235. doi: 10.1111/j.1574-6976.2008.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui B, Döbereiner H, Madden T, Cullis P. Influence of transbilayer area asymmetry on the morphology of large unilamellar vesicles. Biophys J. 1995;69:930–941. doi: 10.1016/S0006-3495(95)79967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E, Woldringh CL. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G. Internal lipid synthesis and vesicle growth as a step toward self-reproduction of the minimal cell. Syst Synth Biol. 2010;4:85–93. doi: 10.1007/s11693-009-9048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G. Early self-reproduction, the emergence of division mechanisms in protocells. Mol BioSyst. 2013;9:195–204. doi: 10.1039/c2mb25375e. [DOI] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci U S A. 2011;108:3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura F, Inaba T, Ishikawa S, Nagata M, Takahashi S, Hotani H, Takiguchi K. Microscopic observations reveal that fusogenic peptides induce liposome shrinkage prior to membrane fusion. Proc Natl Acad Sci U S A. 2004;101:3420–3425. doi: 10.1073/pnas.0304660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva MA, Trambaiolo D, Löwe J. Structural insights into the conformational variability of FtsZ. J Mol Biol. 2007;373:1229–1242. doi: 10.1016/j.jmb.2007.08.056. [DOI] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Inside-out Z rings-constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81:571–579. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP (2013), Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA 110:11000–11004. [DOI] [PMC free article] [PubMed]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KT, Wu W, Battaile KP, Lovell S, Holyoak T, Lutkenhaus J. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146:396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol. 2008;70:1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- Pelve EA, Lindås AC, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol. 2011;82:555–566. doi: 10.1111/j.1365-2958.2011.07834.x. [DOI] [PubMed] [Google Scholar]
- Peterlin P, Arrigler V, Kogej K, Svetina S, Walde P. Growth and shape transformations of giant phospholipid vesicles upon interaction with an aqueous oleic acid suspension. Chem Phys Lipids. 2009;159:67–76. doi: 10.1016/j.chemphyslip.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontani LL, van der Gucht J, Salbreux G, Heuvingh J, Joanny JF, Sykes C. Reconstitution of an actin cortex inside a liposome. Biophys J. 2009;96:192–198. doi: 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Iwasa M, Narita A, Erickson HP, Maéda Y. FtsZ condensates: an in vitro electron microscopy study. Biopolymers. 2009;91:340–350. doi: 10.1002/bip.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcar M, Danchin A, de Lorenzo V, dos Santos VA, Krasnogor N, Rasmussen S, Moya A. The ten grand challenges of synthetic life. Syst Synth Biol. 2011;5:1–9. doi: 10.1007/s11693-011-9084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Chen L, Deamer D, Krakauer DC, Packard NH, Stadler PF, Bedau MA. Transitions from nonliving to living matter. Science. 2004;303:963–965. doi: 10.1126/science.1093669. [DOI] [PubMed] [Google Scholar]
- Raspail FV (1825) Développement de la fécule dans les orgabes de la fructification des céréales et analyse microscopique de la fécule suivie d’exprériences propres à en expliquer la conversion en gomme. Annals des Sciences Naturelles 224–39:384–427. Cited from: Weiner DB (1968) Raspail: scientist and reformer. Columbia University Press, Columbia
- Rosengarten R, Kirchhoff H. Growth morphology of Mycoplasma mobile 163K on solid surfaces: reproduction, aggregation, and microcolony formation. Curr Microbiol. 1989;18:15–22. [Google Scholar]
- Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24:1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Imai M. Model system of self-reproducing vesicles. Phys Rev Lett. 2011;107(198):101. doi: 10.1103/PhysRevLett.107.198101. [DOI] [PubMed] [Google Scholar]
- Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Hodgson B, Shaw MK, Chong PLG, Williams RL, Bell SD. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidli PK, Schurtenberger P, Luisi PL. Liposome-mediated enzymatic synthesis of phosphatidylcholine as an approach to self-replicating liposomes. J Am Chem Soc. 1991;113:8127–8130. [Google Scholar]
- Schrum JP, Zhu TF, Szostak JW. The origins of cellular life. Cold Spring Harb Perspect Biol. 2010;2:a002212. doi: 10.1101/cshperspect.a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J, Loose M, Bonny M, Kruse K, Mönch I, Schwille P. Geometry sensing by self-organized protein patterns. Proc Natl Acad Sci U S A. 2012;109:15283–15288. doi: 10.1073/pnas.1206953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P. Bottom-up synthetic biology: engineering in a tinkerer’s world. Science. 2011;333:1252–1254. doi: 10.1126/science.1211701. [DOI] [PubMed] [Google Scholar]
- Seifert U, Berndl K, Lipowsky R. Shape transformations of vesicles: Phase diagram for spontaneous-curvature and bilayer-coupling models. Phys Rev A. 1991;44:1182–1202. doi: 10.1103/physreva.44.1182. [DOI] [PubMed] [Google Scholar]
- Si F, Busiek K, Margolin W, Sun SX. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys J. 2013;105:1976–1986. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonobe S, Aoyama K, Suzuki C, Saito Kh, Nagata K, Shimmen T, Nagata Y. Proliferation of the hyperthermophilic archaeon Pyrobaculum islandicum by cell fission. Extremophiles. 2010;14:403–407. doi: 10.1007/s00792-010-0321-8. [DOI] [PubMed] [Google Scholar]
- Staneva G, Angelova MI, Koumanov K. Phospholipase A2 promotes raft budding and fission from giant liposomes. Chem Phys Lipids. 2004;129:53–62. doi: 10.1016/j.chemphyslip.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Staneva G, Seigneuret M, Koumanov K, Trugnan G, Angelova MI. Detergents induce raft-like domains budding and fission from giant unilamellar heterogeneous vesicles: a direct microscopy observation. Chem Phys Lipids. 2005;136:55–66. doi: 10.1016/j.chemphyslip.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Stano P, Luisi PL. Achievements and open questions in the self-reproduction of vesicles and synthetic minimal cells. Chem Commun. 2010;46:3639–3653. doi: 10.1039/b913997d. [DOI] [PubMed] [Google Scholar]
- Stano P, Luisi PL. Semi-synthetic minimal cells: origin and recent developments. Curr Opin Biotechnol. 2013;24:633–638. doi: 10.1016/j.copbio.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Stano P, Wehrli E, Luisi P. Insights into the self-reproduction of oleate vesicles. J Phys Condens Matter. 2006;18:S2231. [Google Scholar]
- Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SX, Jiang H. Physics of bacterial morphogenesis. Microbiol Mol Biol Rev. 2011;75:543–565. doi: 10.1128/MMBR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetina S. Vesicle budding and the origin of cellular life. ChemPhysChem. 2009;10:2769–2776. doi: 10.1002/cphc.200900577. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Takakura K, Toyota T, Sugawara T. A novel system of self-reproducing giant vesicles. J Am Chem Soc. 2003;125:8134–8140. doi: 10.1021/ja029379a. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yamazaki M. Membrane fusion of giant unilamellar vesicles of neutral phospholipid membranes induced by La3+ Langmuir. 2004;20:5160–5164. doi: 10.1021/la049681s. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Sano R, Yamashita Y, Yamazaki M. Shape changes and vesicle fission of giant unilamellar vesicles of liquid-ordered phase membrane induced by lysophosphatidylcholine. Langmuir. 2004;20:9526–9534. doi: 10.1021/la049481g. [DOI] [PubMed] [Google Scholar]
- Terasawa H, Nishimura K, Suzuki H, Matsuura T, Yomo T. Coupling of the fusion and budding of giant phospholipid vesicles containing macromolecules. Proc Natl Acad Sci U S A. 2012;109:5942–5947. doi: 10.1073/pnas.1120327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci U S A. 2013;110:10586–10591. doi: 10.1073/pnas.1221036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir I, Caspi Y, Guedeau-Boudeville MA, Arzi T, Stavans J. Budding and tubulation in highly oblate vesicles by anchored amphiphilic molecules. Phys Rev Lett. 2003;91(138):102. doi: 10.1103/PhysRevLett.91.138102. [DOI] [PubMed] [Google Scholar]
- Tsai FC, Stuhrmann B, Koenderink GH. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir. 2011;27:10061–10071. doi: 10.1021/la201604z. [DOI] [PubMed] [Google Scholar]
- Turner W. The cell theory, past and present. J Anat Physiol. 1890;24:253–287. [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser PJ, Ariotti N, Howes M, Ferguson C, Webb R, Schwudke D, Leneva N, Cho KJ, Cooper L, Rae J, Floetenmeyer M, Oorschot VMJ, Skoglund U, Simons K, Hancock JF, Parton RG. Constitutive formation of Caveolae in a bacterium. Cell. 2012;150:752–763. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Wick R, Walde P, Luisi PL. Light microscopic investigations of the autocatalytic self-reproduction of giant vesicles. J Am Chem Soc. 1995;117:1435–1436. [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright NA, Poulsom R. Omnis cellula e cellula revisited: cell biology as the foundation of pathology. J Pathol. 2012;226:145–147. doi: 10.1002/path.3030. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Park KT, Holyoak T, Lutkenhaus J. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Mol Microbiol. 2011;79:1515–1528. doi: 10.1111/j.1365-2958.2010.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TF, Szostak JW. Coupled growth and division of model protocell membranes. J Am Chem Soc. 2009;131:5705–5713. doi: 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TF, Adamala K, Zhang N, Szostak JW. Photochemically driven redox chemistry induces protocell membrane pearling and division. Proc Natl Acad Sci U S A. 2012;109:9828–9832. doi: 10.1073/pnas.1203212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske K, Schwille P. Reconstitution of pole-to-pole oscillations of min proteins in microengineered polydimethylsiloxane compartments. Angew Chem Int Ed Engl. 2013;52:459–462. doi: 10.1002/anie.201207078. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]